SERS-TLC Device for Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Milk
Abstract
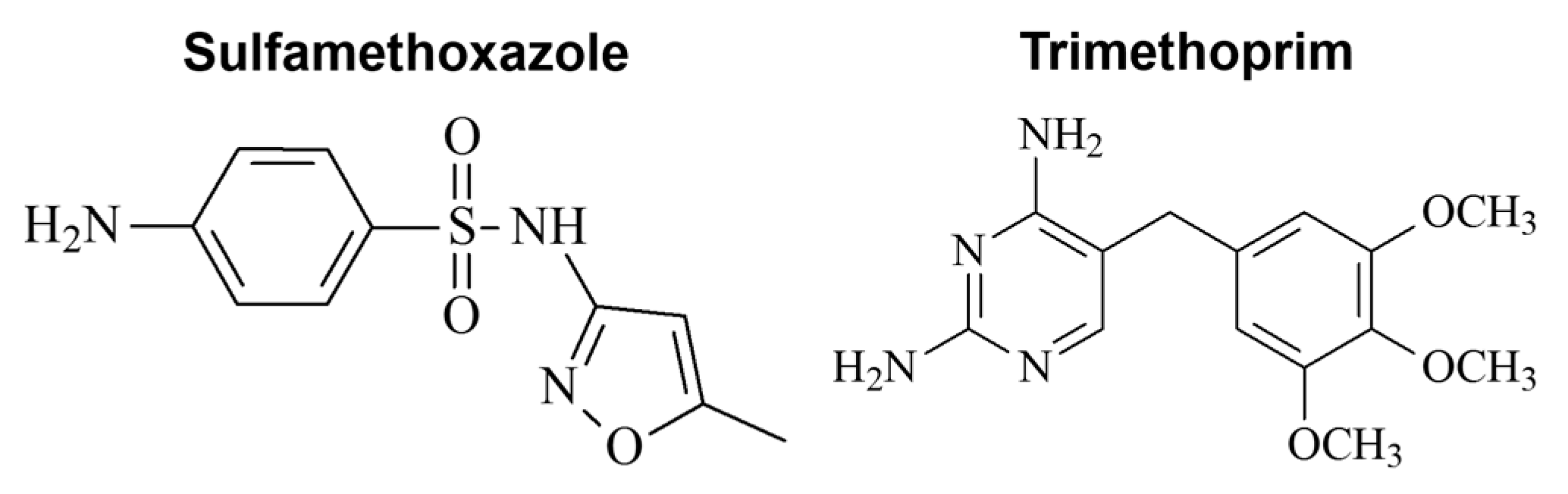
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Nanoparticle Synthesis
2.3. Preparation of Solutions and Samples
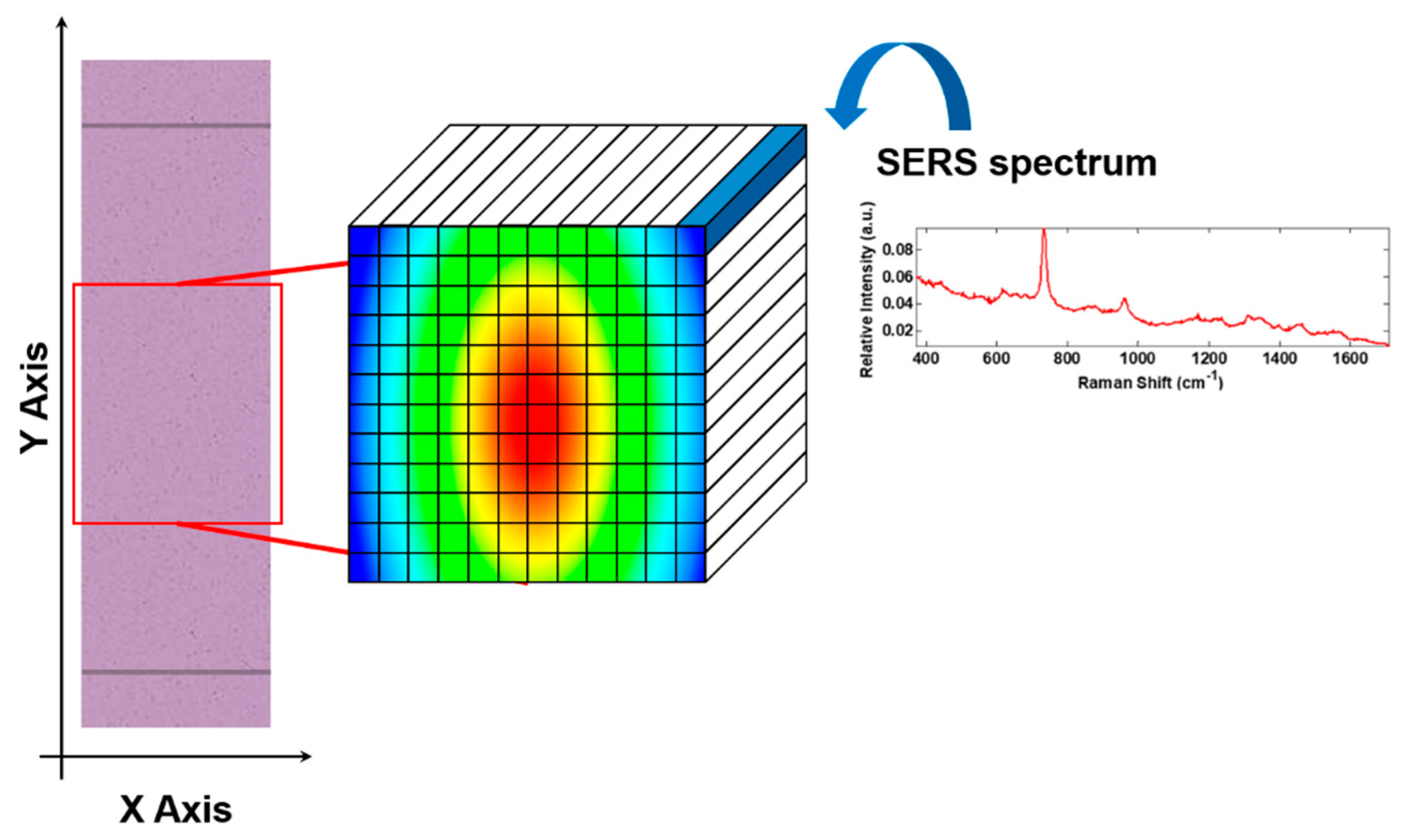
2.4. TLC-SERS Device Procedure
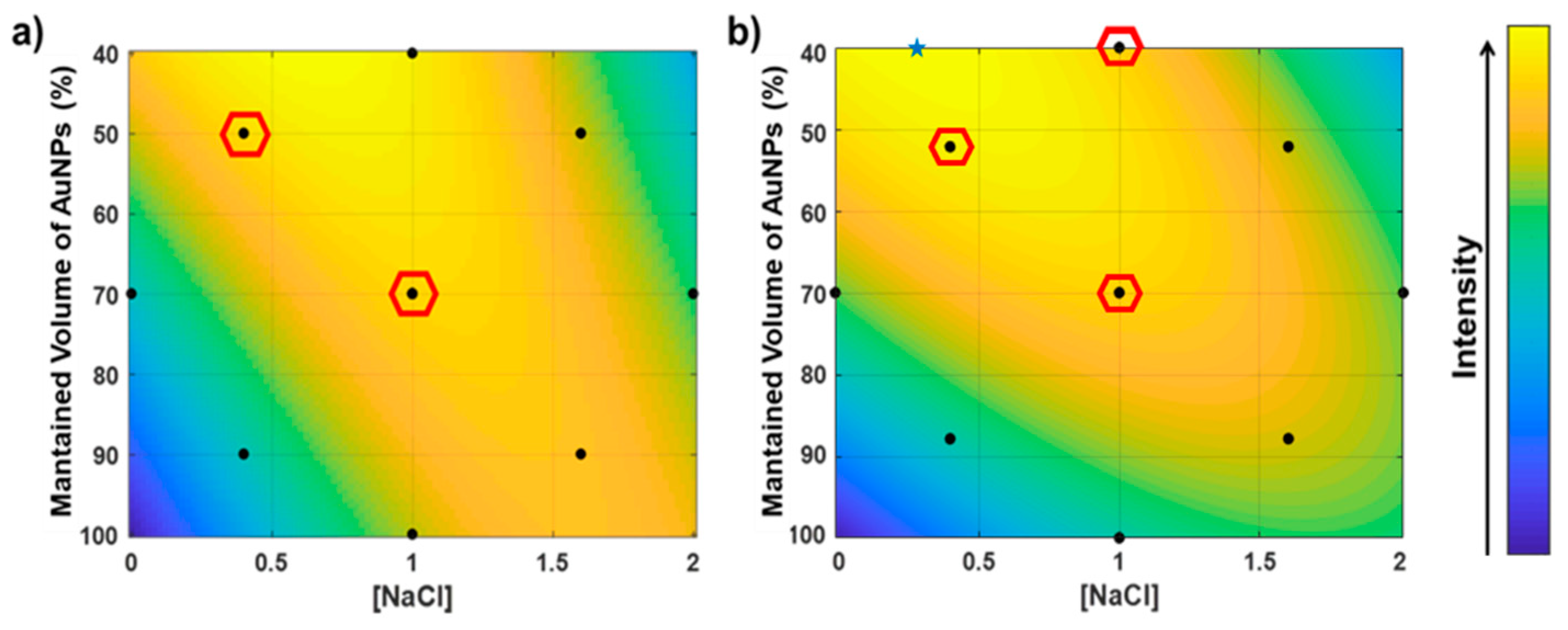
2.5. Analysis Optimization
2.6. Chemometric Methods
3. Results and Discussion
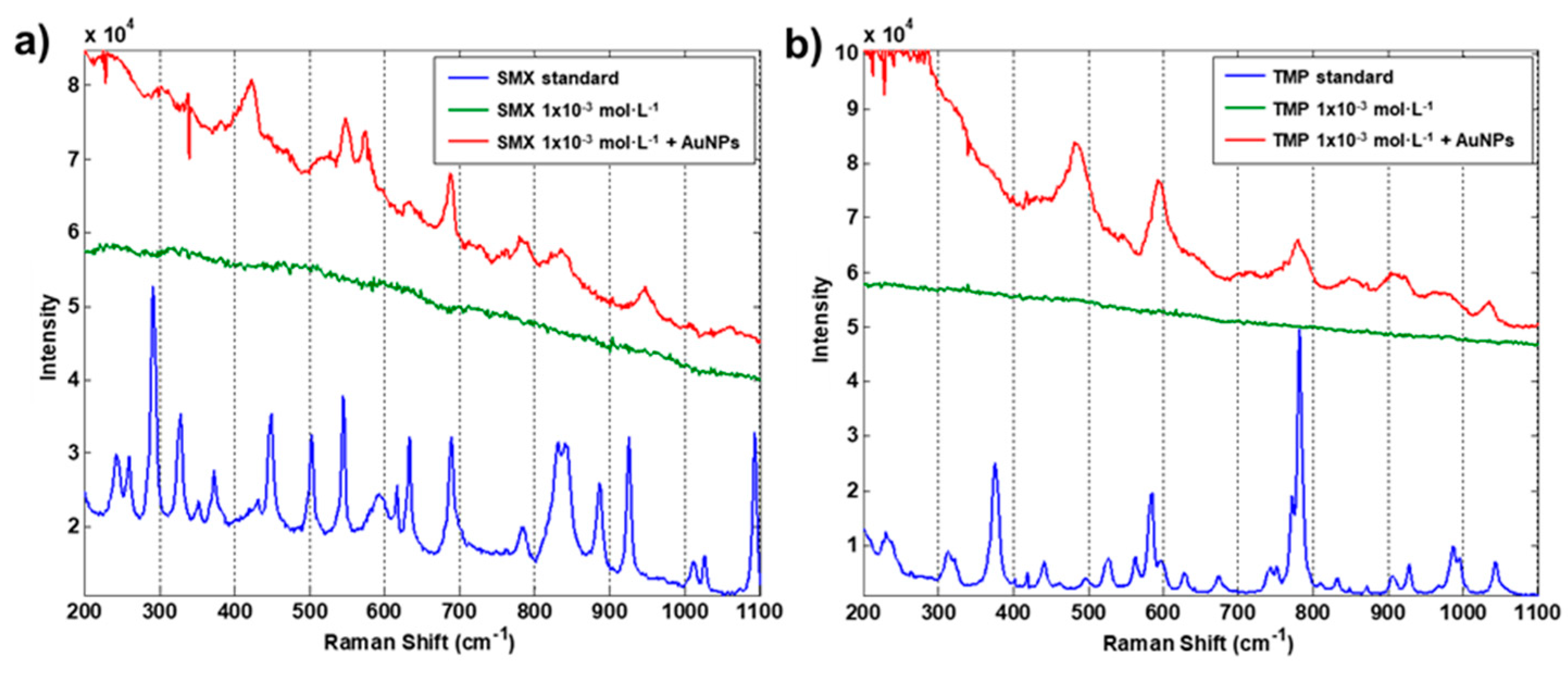
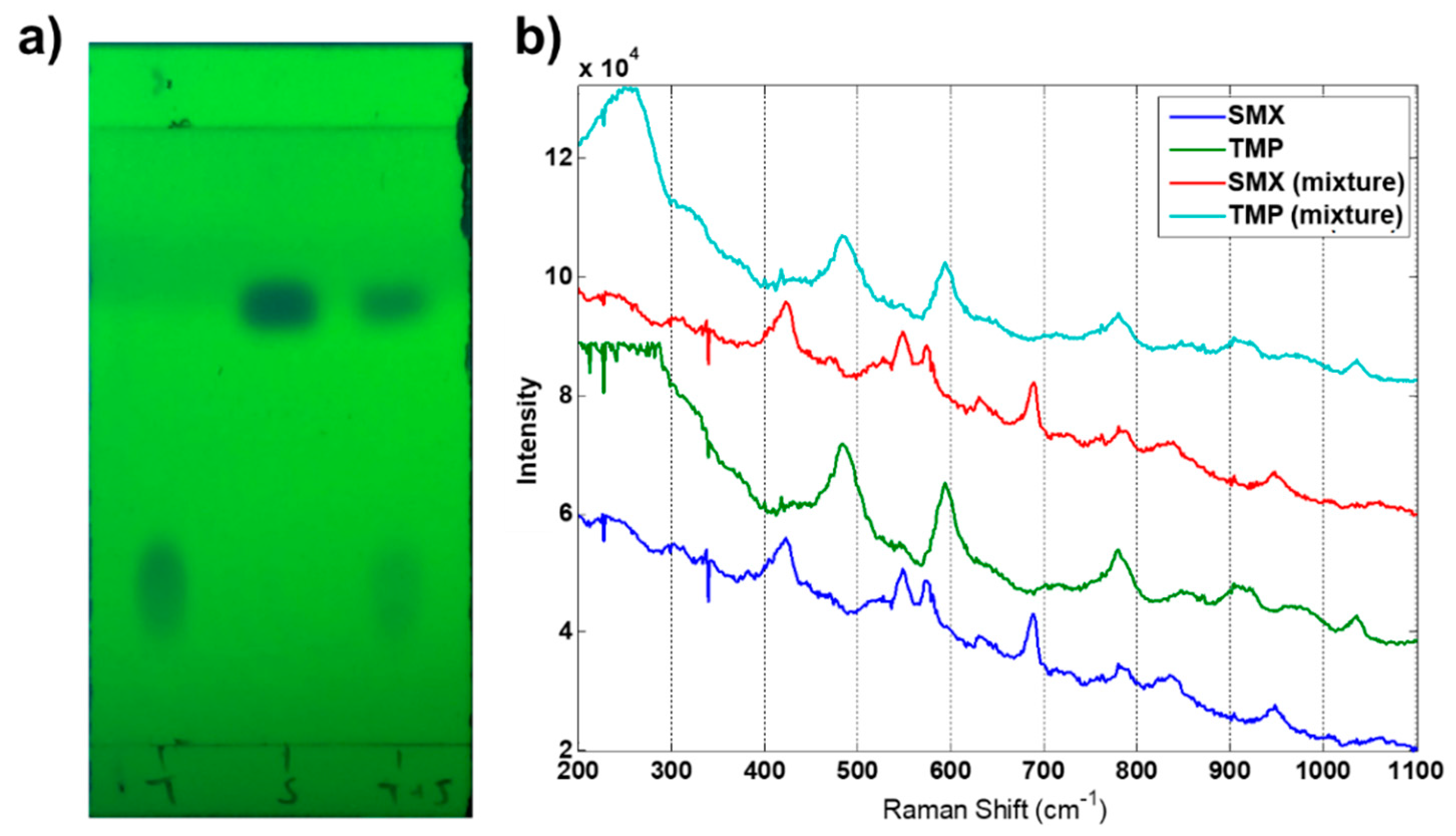
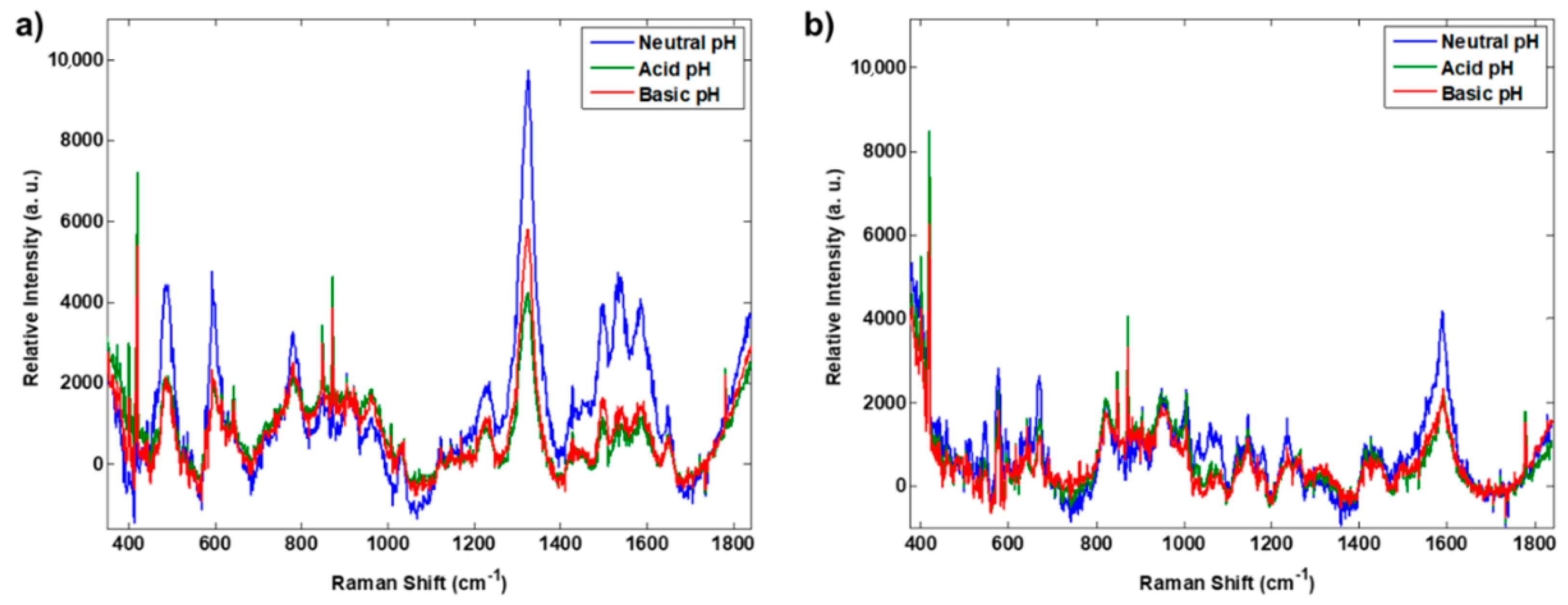
3.1. Preliminary Tests
3.2. Device Optimization
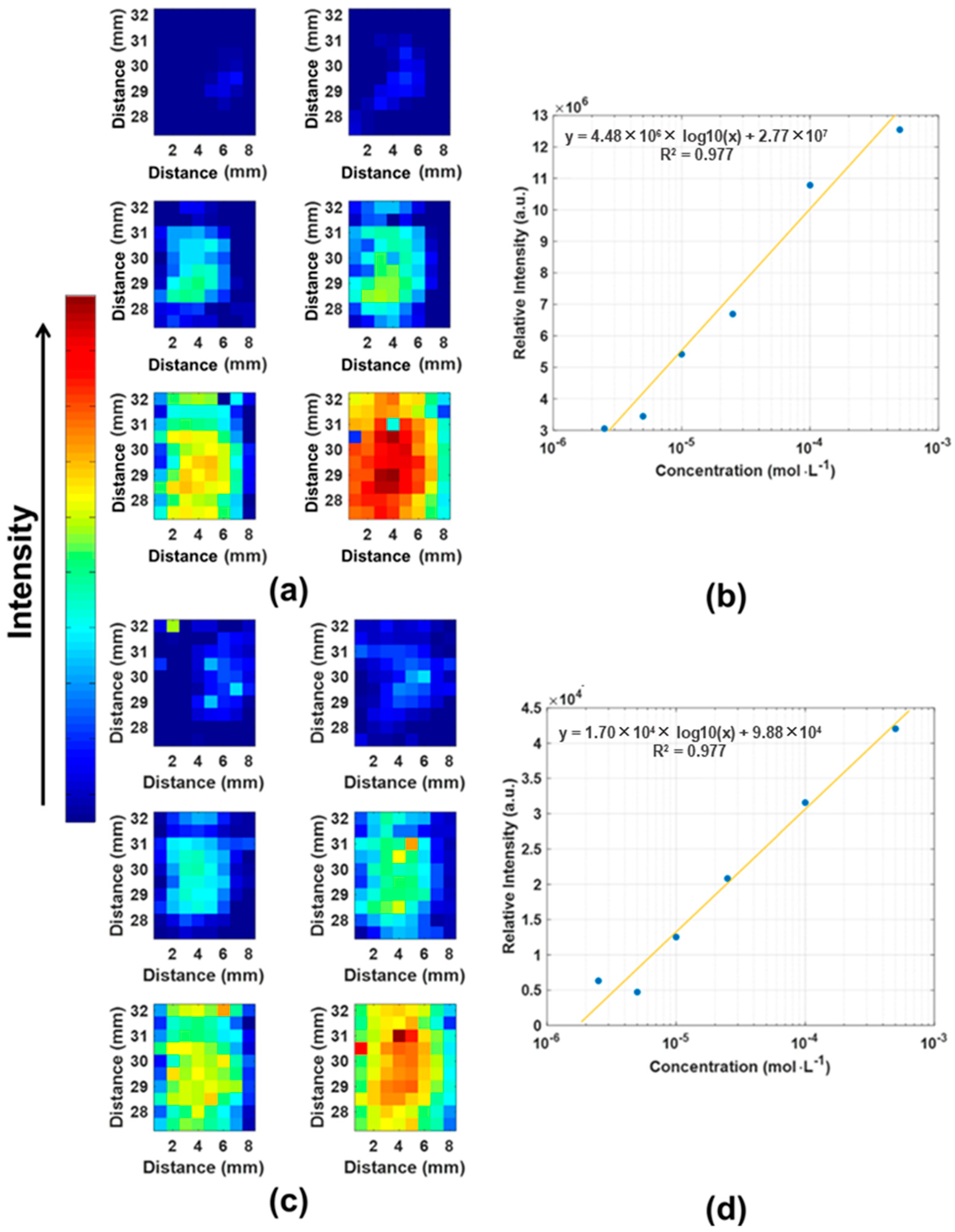
3.3. SMX Quantification
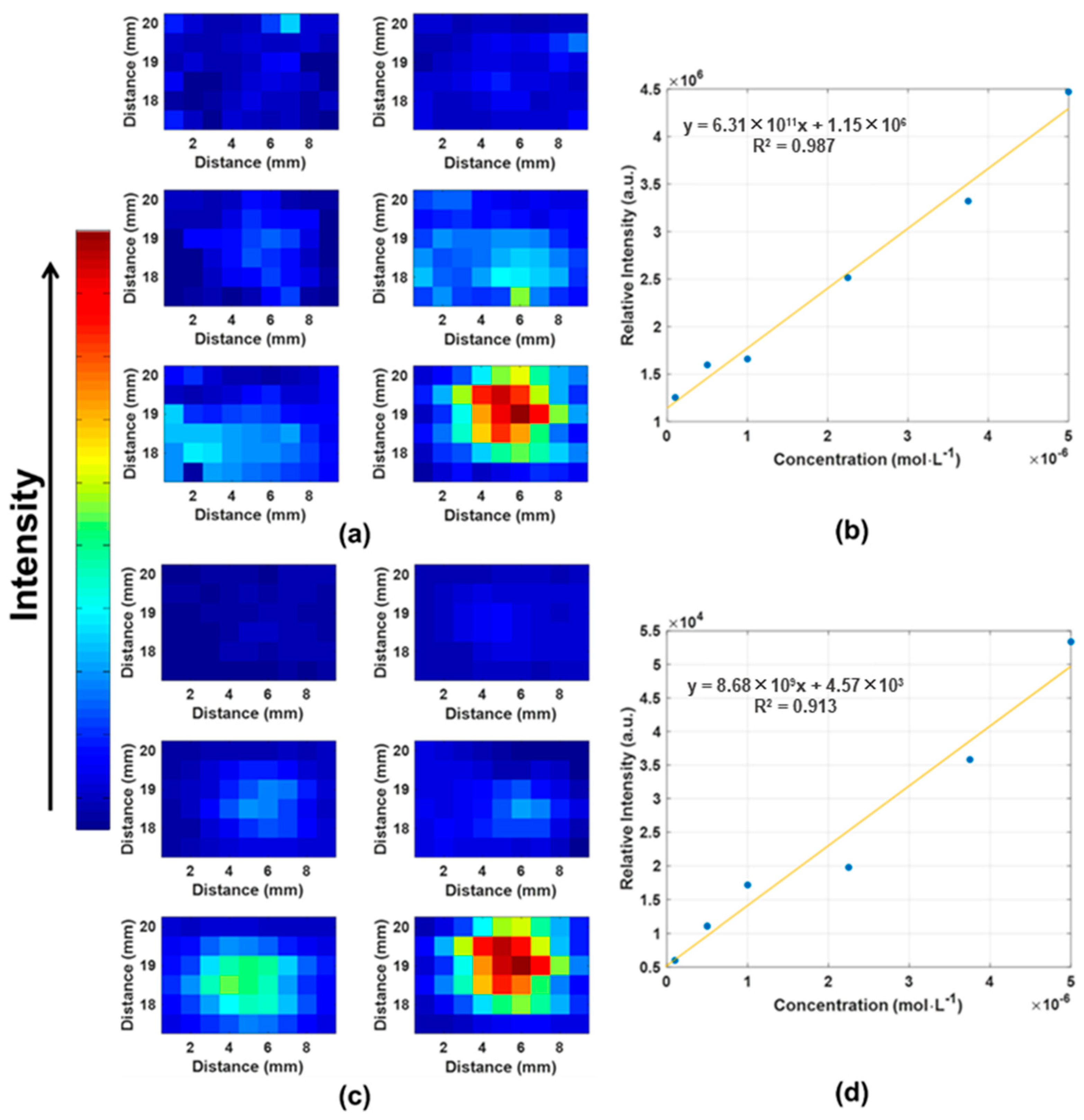
3.4. TMP Quantification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Andrade, L.S.; de Moraes, M.C.; Rocha-Filho, R.C.; Fatibello-Filho, O.; Cass, Q.B. A Multidimensional High Performance Liquid Chromatography Method Coupled with Amperometric Detection Using a Boron-Doped Diamond Electrode for the Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Bovine Milk. Anal. Chim. Acta 2009, 654, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Campbell, W.C. History of the Discovery of Sulfaquinoxaline as a Coccidiostat. J. Parasitol. 2008, 94, 934–945. [Google Scholar] [CrossRef] [PubMed]
- Bousova, K.; Senyuva, H.; Mittendorf, K. Quantitative Multi-Residue Method for Determination Antibiotics in Chicken Meat Using Turbulent Flow Chromatography Coupled to Liquid Chromatography–Tandem Mass Spectrometry. J. Chromatogr. A 2013, 1274, 19–27. [Google Scholar] [CrossRef] [PubMed]
- de Paula, F.C.C.R.; de Pietro, A.C.; Cass, Q.B. Simultaneous Quantification of Sulfamethoxazole and Trimethoprim in Whole Egg Samples by Column-Switching High-Performance Liquid Chromatography Using Restricted Access Media Column for on-Line Sample Clean-Up. J. Chromatogr. A 2008, 1189, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Hajian, R.; Mousavi, E.; Shams, N. Net Analyte Signal Standard Addition Method for Simultaneous Determination of Sulphadiazine and Trimethoprim in Bovine Milk and Veterinary Medicines. Food Chem. 2013, 138, 745–749. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.V.; Cass, Q.B. High-Performance Liquid Chromatography Method for the Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Bovine Milk Using an on-Line Clean-up Column. J. Chromatogr. B 2005, 826, 139–146. [Google Scholar] [CrossRef]
- Ministério da Agricultura, Pecuária e Abastecimento. Instrução Normativa No 09. Available online: http://alimentusconsultoria.com.br/instrucao-normativa-9-fevereiro-2017/ (accessed on 25 September 2022).
- Almeida, M.R.; Correa, D.N.; Zacca, J.J.; Logrado, L.P.L.; Poppi, R.J. Detection of Explosives on the Surface of Banknotes by Raman Hyperspectral Imaging and Independent Component Analysis. Anal. Chim. Acta 2015, 860, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.; Shadi, I.T.; Xu, Y.; Goodacre, R. Quantitative Analysis of the Banned Food Dye Sudan-1 Using Surface Enhanced Raman Scattering with Multivariate Chemometrics. J. Phys. Chem. C 2010, 114, 7285–7290. [Google Scholar] [CrossRef]
- Jarvis, R.M.; Goodacre, R. Discrimination of Bacteria Using Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2004, 76, 40–47. [Google Scholar] [CrossRef]
- Mamián-López, M.B.; Poppi, R.J. Standard Addition Method Applied to the Urinary Quantification of Nicotine in the Presence of Cotinine and Anabasine Using Surface Enhanced Raman Spectroscopy and Multivariate Curve Resolution. Anal. Chim. Acta 2013, 760, 53–59. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-Enhanced Raman Spectroscopy: Concepts and Chemical Applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Sivashanmugan, K.; Zhao, Y.; Zhang, B.; Wang, A.X. Multiplex Sensing of Complex Mixtures by Machine Vision Analysis of TLC-SERS Images. Sens. Actuators B Chem. 2022, 357, 131355. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Fan, Q.; Yu, Q.; Wang, R.; Wang, H.; Kong, X. Facile Detection of Carbendazim in Food Using TLC-SERS on Diatomite Thin Layer Chromatography. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 247, 119037. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, S.; Zheng, J.; He, L. Surface-Enhanced Raman Spectroscopy (SERS) Combined Techniques for High-Performance Detection and Characterization. TrAC Trends Anal. Chem. 2017, 90, 1–13. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, H.; Yu, Q.; Li, Q.; Lu, X.; Kong, X. On-Site Separation and Identification of Polycyclic Aromatic Hydrocarbons from Edible Oil by TLC-SERS on Diatomite Photonic Biosilica Plate. Microchem. J. 2021, 160, 105672. [Google Scholar] [CrossRef]
- Li, H.; Zhu, Q.X.; Chwee, T.S.; Wu, L.; Chai, Y.F.; Lu, F.; Yuan, Y.F. Detection of Structurally Similar Adulterants in Botanical Dietary Supplements by Thin-Layer Chromatography and Surface Enhanced Raman Spectroscopy Combined with Two-Dimensional Correlation Spectroscopy. Anal. Chim. Acta 2015, 883, 22–31. [Google Scholar] [CrossRef]
- Zhang, Z.-M.; Liu, J.-F.; Liu, R.; Sun, J.-F.; Wei, G.-H. Thin Layer Chromatography Coupled with Surface-Enhanced Raman Scattering as a Facile Method for On-Site Quantitative Monitoring of Chemical Reactions. Anal. Chem. 2014, 86, 7286–7292. [Google Scholar] [CrossRef]
- Xie, Z.; Wang, Y.; Chen, Y.; Xu, X.; Jin, Z.; Ding, Y.; Yang, N.; Wu, F. Tuneable Surface Enhanced Raman Spectroscopy Hyphenated to Chemically Derivatized Thin-Layer Chromatography Plates for Screening Histamine in Fish. Food Chem. 2017, 230, 547–552. [Google Scholar] [CrossRef]
- Tan, A.; Zhao, Y.; Sivashanmugan, K.; Squire, K.; Wang, A.X. Quantitative TLC-SERS Detection of Histamine in Seafood with Support Vector Machine Analysis. Food Control 2019, 103, 111–118. [Google Scholar] [CrossRef]
- Sha, X.; Han, S.; Fang, G.; Li, N.; Lin, D.; Hasi, W. A Novel Suitable TLC-SERS Assembly Strategy for Detection of Rhodamine B and Sudan I in Chili Oil. Food Control 2022, 138, 109040. [Google Scholar] [CrossRef]
- Carneiro, R.L.; Poppi, R.J. Infrared Imaging Spectroscopy and Chemometric Tools for in Situ Analysis of an Imiquimod Pharmaceutical Preparation Presented as Cream. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 118, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, R.L.; Poppi, R.J. A Quantitative Method Using near Infrared Imaging Spectroscopy for Determination of Surface Composition of Tablet Dosage Forms: An Example of Spirolactone Tablets. J. Braz. Chem. Soc. 2012, 23, 1570–1576. [Google Scholar] [CrossRef][Green Version]
- Carneiro, R.L.; Poppi, R.J. Homogeneity Study of Ointment Dosage Forms by Infrared Imaging Spectroscopy. J. Pharm. Biomed. Anal. 2012, 58, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Araya, J.A.; Carneiro, R.L.; Arévalo, C.; Freer, J.; Castillo, R. del P. Single Pixel Quantification Strategies Using Middle Infrared Hyperspectral Imaging of Lignocellulosic Fibers and MCR-ALS Analysis. Microchem. J. 2017, 134, 164–172. [Google Scholar] [CrossRef]
- Saha, D.; Manickavasagan, A. Machine Learning Techniques for Analysis of Hyperspectral Images to Determine Quality of Food Products: A Review. Curr. Res. Food Sci. 2021, 4, 28–44. [Google Scholar] [CrossRef]
- Mamián-López, M.B.; Poppi, R.J. SERS Hyperspectral Imaging Assisted by MCR-ALS for Studying Polymeric Microfilms Loaded with Paracetamol. Microchem. J. 2015, 123, 243–251. [Google Scholar] [CrossRef]
- Alexandrino, G.L.; Khorasani, M.R.; Amigo, J.M.; Rantanen, J.; Poppi, R.J. Monitoring of Multiple Solid-State Transformations at Tablet Surfaces Using Multi-Series near-Infrared Hyperspectral Imaging and Multivariate Curve Resolution. Eur. J. Pharm. Biopharm. 2015, 93, 224–230. [Google Scholar] [CrossRef]
- Piqueras, S.; Duponchel, L.; Tauler, R.; de Juan, A. Monitoring Polymorphic Transformations by Using in Situ Raman Hyperspectral Imaging and Image Multiset Analysis. Anal. Chim. Acta 2014, 819, 15–25. [Google Scholar] [CrossRef]
- Laborde, A.; Puig-Castellví, F.; Jouan-Rimbaud Bouveresse, D.; Eveleigh, L.; Cordella, C.; Jaillais, B. Detection of Chocolate Powder Adulteration with Peanut Using Near-Infrared Hyperspectral Imaging and Multivariate Curve Resolution. Food Control 2021, 119, 107454. [Google Scholar] [CrossRef]
- Fornasaro, S.; Vicario, A.; de Leo, L.; Bonifacio, A.; Not, T.; Sergo, V. Potential Use of MCR-ALS for the Identification of Coeliac-Related Biochemical Changes in Hyperspectral Raman Maps from Pediatric Intestinal Biopsies. Integr. Biol. 2018, 10, 356–363. [Google Scholar] [CrossRef]
- Rutledge, D.N.; Jouan-Rimbaud Bouveresse, D. Independent Components Analysis with the JADE Algorithm. TrAC Trends Anal. Chem. 2013, 50, 22–32. [Google Scholar] [CrossRef]
- Hashemi-Nasab, F.S.; Parastar, H. Vis-NIR Hyperspectral Imaging Coupled with Independent Component Analysis for Saffron Authentication. Food Chem. 2022, 393, 133450. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Meisel, D. Adsorption and Surface-Enhanced Raman of Dyes on Silver and Gold Sols. J. Phys. Chem. 1982, 86, 3391–3395. [Google Scholar] [CrossRef]
- Aguilera-Luiz, M.M.; Vidal, J.L.M.; Romero-González, R.; Frenich, A.G. Multi-Residue Determination of Veterinary Drugs in Milk by Ultra-High-Pressure Liquid Chromatography–Tandem Mass Spectrometry. J. Chromatogr. A 2008, 1205, 10–16. [Google Scholar] [CrossRef]
- Chen, Y.; Schwack, W. Planar Chromatography Mediated Screening of Tetracycline and Fluoroquinolone Antibiotics in Milk by Fluorescence and Mass Selective Detection. J. Chromatogr. A 2013, 1312, 143–151. [Google Scholar] [CrossRef]
- Jähnke, R.W.O. A Concise Quality Control Guide on Essential Drugs and Other Medicines—Volume II Thin Layer Chromatographic Tests; Global Pharma Health Fund: Frankfurt, Germany, 2008. [Google Scholar]
- Soares, F.L.F.; Ardila, J.A.; Carneiro, R.L. Thin-Layer Chromatography–Surface-Enhanced Raman Spectroscopy and Chemometric Tools Applied to Pilsner Beer Fingerprint Analysis. J. Raman Spectrosc. 2017, 48, 943–950. [Google Scholar] [CrossRef]
- Junior, B.R.A.; Soares, F.L.F.; Ardila, J.A.; Durango, L.G.C.; Forim, M.R.; Carneiro, R.L. Determination of B-Complex Vitamins in Pharmaceutical Formulations by Surface-Enhanced Raman Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 188, 589–595. [Google Scholar] [CrossRef]
- Jaumot, J.; de Juan, A.; Tauler, R. MCR-ALS GUI 2.0: New Features and Applications. Chemom. Intell. Lab. Syst. 2015, 140, 1–12. [Google Scholar] [CrossRef]
- Kassouf, A.; Jouan-Rimbaud Bouveresse, D.; Rutledge, D.N. Determination of the Optimal Number of Components in Independent Components Analysis. Talanta 2018, 179, 538–545. [Google Scholar] [CrossRef]
- Vijaya Chamundeeswari, S.P.; James Jebaseelan Samuel, E.; Sundaraganesan, N. Molecular Structure, Vibrational Spectra, NMR and UV Spectral Analysis of Sulfamethoxazole. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 118, 1–10. [Google Scholar] [CrossRef]
- Ungurean, A.; Leopold, N.; David, L.; Chiş, V. Vibrational Spectroscopic and DFT Study of Trimethoprim. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2013, 102, 52–58. [Google Scholar] [CrossRef] [PubMed]
- van Rhijn, J.A.; Lasaroms, J.J.P.; Berendsen, B.J.A.; Brinkman, U.A.T. Liquid Chromatographic–Tandem Mass Spectrometric Determination of Selected Sulphonamides in Milk. J. Chromatogr. A 2002, 960, 121–133. [Google Scholar] [CrossRef] [PubMed]



| Codified Values | Real Values | Responses | ||||
|---|---|---|---|---|---|---|
| Experiment | NaCl | AuNPs M.V. ¹ (%) | NaCl (mol·L−1) | AuNPs M.V. ¹ (%) | SMX | TMP |
| 1 | −1 | −1 | 0.4 | 87.15 | 257 | 1930 |
| 2 | 1 | −1 | 1.6 | 87.15 | 1193 | 4975 |
| 3 | −1 | 1 | 0.4 | 52.14 | 1337 | 9842 |
| 4 | 1 | 1 | 1.6 | 52.14 | 942 | 6686 |
| 5 | 0 | 0 | 1 | 70 | 1818 | 8701 |
| 6 | 0 | 0 | 1 | 70 | 1457 | 6668 |
| 7 | 0 | 0 | 1 | 70 | 1236 | 5813 |
| 8 | −1.68 | 0 | 0 | 70 | 1369 | 5841 |
| 9 | 1.68 | 0 | 2 | 70 | 360 | 6201 |
| 10 | 0 | −1.68 | 1 | 100 | 354 | 4899 |
| 11 | 0 | 1.68 | 1 | 40 | 1661 | 2263 |
| Recovering (%) | ||
|---|---|---|
| MCR-ALS | ICA | |
| Milk A—S0 | N.D. | N.D. |
| Milk B—S0 | N.D. | N.D. |
| Milk C—S0 | N.D. | N.D. |
| Milk A—S1 | 98 ± 6% | 106 ± 15% |
| Milk B—S1 | 119 ± 24% | 100 ± 15% |
| Milk C—S1 | 110 ± 2% | 82 ± 17% |
| Milk A—S2 | 102 ± 22% | 83 ± 1% |
| Milk B—S2 | 111 ± 19% | 96 ± 8% |
| Milk C—S2 | 107 ± 15% | 96 ± 13% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, F.L.F.; Junior, B.R.d.A.; Carneiro, R.L. SERS-TLC Device for Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Milk. Chemosensors 2022, 10, 528. https://doi.org/10.3390/chemosensors10120528
Soares FLF, Junior BRdA, Carneiro RL. SERS-TLC Device for Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Milk. Chemosensors. 2022; 10(12):528. https://doi.org/10.3390/chemosensors10120528
Chicago/Turabian StyleSoares, Frederico Luis Felipe, Benedito Roberto de Alvarenga Junior, and Renato Lajarim Carneiro. 2022. "SERS-TLC Device for Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Milk" Chemosensors 10, no. 12: 528. https://doi.org/10.3390/chemosensors10120528
APA StyleSoares, F. L. F., Junior, B. R. d. A., & Carneiro, R. L. (2022). SERS-TLC Device for Simultaneous Determination of Sulfamethoxazole and Trimethoprim in Milk. Chemosensors, 10(12), 528. https://doi.org/10.3390/chemosensors10120528







