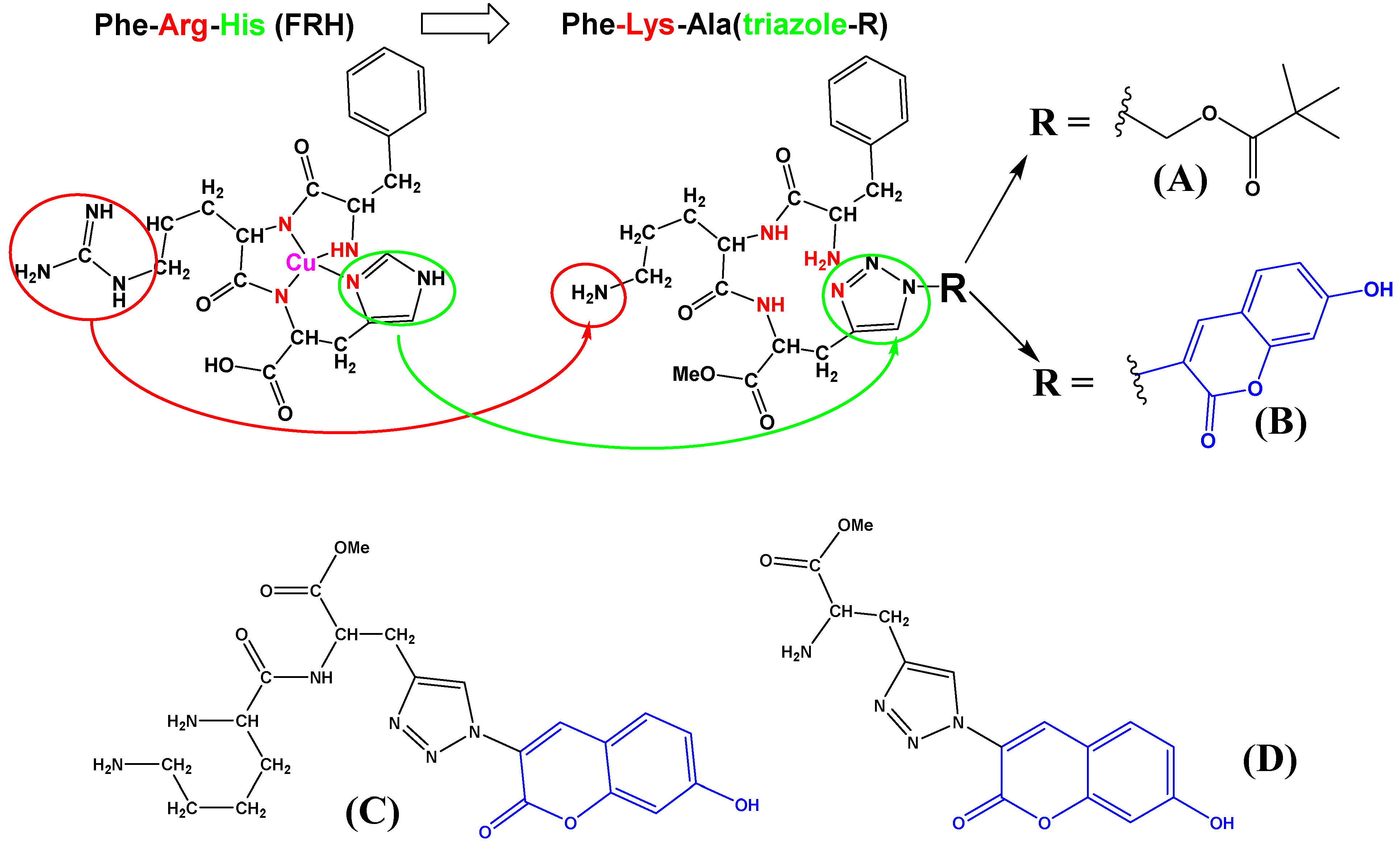
Fluorescent Analogues of FRH Peptide: Cu(II) Binding and Interactions with ds-DNA/RNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. General Information
2.2. Spectrophotometric Measurements
2.3. Study of DNA/RNA Interactions
3. Results
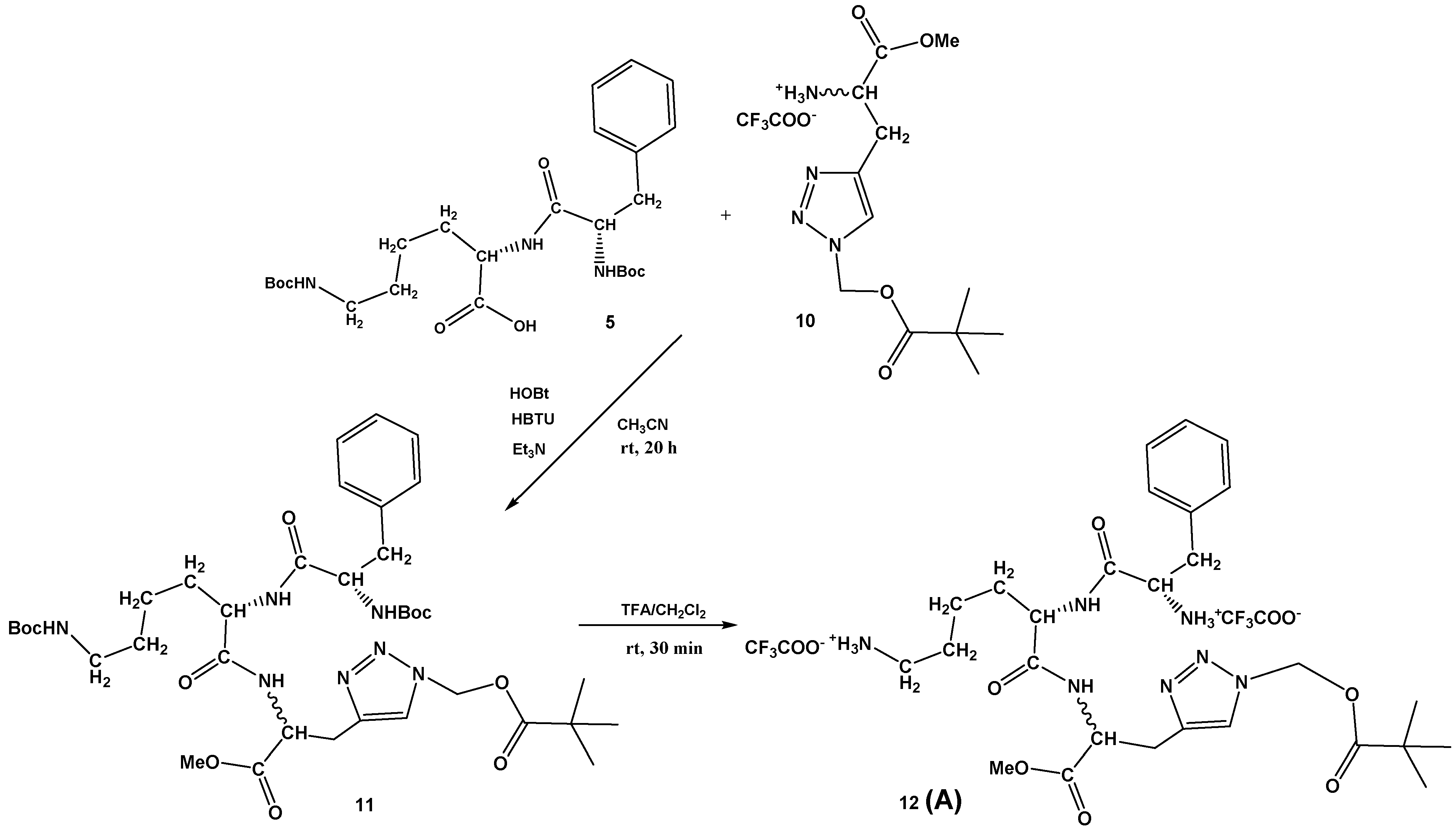
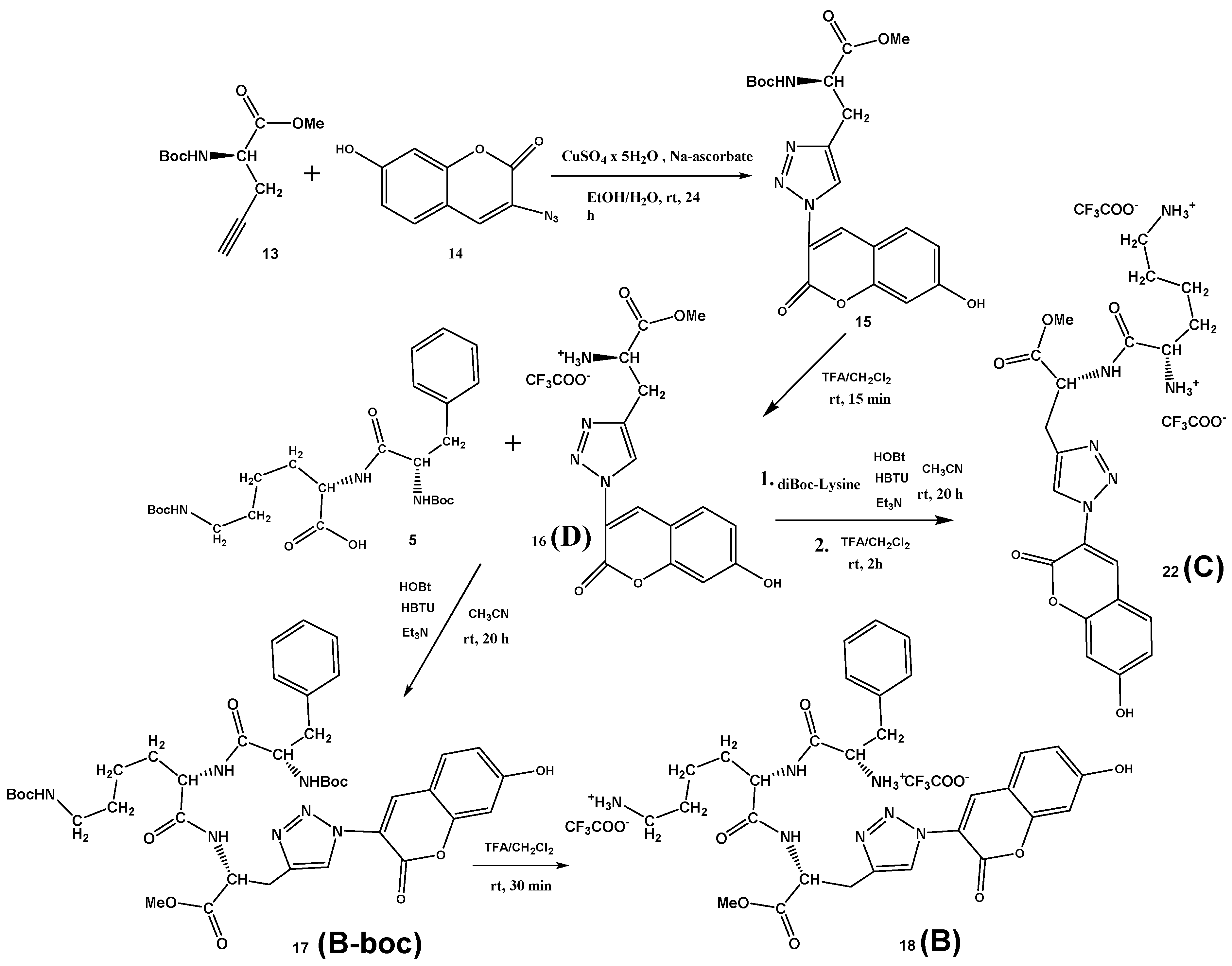
3.1. Synthesis
3.2. Photophysical Characterization
3.3. Binding of Cu2+ Cation
3.4. Study of Interactions of A, B, B-boc, C and D with ds-DNA and ds-RNA
3.4.1. Thermal Denaturation Experiments
3.4.2. Spectrophotometric Titrations
3.4.3. Circular Dichroism (CD) Experiments
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rodzik, A.; Pomastowski, P.; Sagandykova, G.N.; Buszewski, B. Interactions of Whey Proteins with Metal Ions. Int. J. Mol. Sci. 2020, 21, 2156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Copper in diseases and treatments and copper-based anticancer strategies. Med. Res. Rev. 2010, 30, 708–749. [Google Scholar] [CrossRef]
- Sathyadevi, P.; Krishnamoorthy, P.; Jayanthi, E.; Butorac, R.R.; Cowley, A.H.; Dharmaraj, N. Studies on the effect of metal ions of hydrazone complexes on interaction with nucleic acids, bovine serum albumin and antioxidant properties. Inorg. Chim. Acta 2012, 384, 83–96. [Google Scholar] [CrossRef]
- Harding, M.M.; Nowicki, M.W.; Walkinshaw, M.D. Metals in protein structures: A review of their principal features. Crystallogr. Rev. 2010, 16, 247–302. [Google Scholar] [CrossRef]
- Zou, J.; Sugimoto, N. Complexation of peptide with Cu2+ responsible to inducing and enhancing the formation of α-helix conformation. BioMetals 2000, 13, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M. Metal Ions in Proteins: Structural and Functional Roles; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1987; Volume 52, pp. 579–585. [Google Scholar]
- Horn, D.; Barrientos, A. Mitochondrial copper metabolism and delivery to cytochromec oxidase. IUBMB Life 2008, 60, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Katoh, S. Early research on the role of plastocyanin in photosynthesis. Photosynth. Res. 2003, 76, 255–261. [Google Scholar] [CrossRef]
- Robinett, N.G.; Peterson, R.L.; Culotta, V.C. Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains. J. Biol. Chem. 2017, 293, 4636–4643. [Google Scholar] [CrossRef] [Green Version]
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Tian, L. Copper Active Sites in Biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar] [CrossRef] [Green Version]
- Laurie, S.H. Handbook of Metal-Ligand Interactions in Biological Fluids: Bioinorganic Chemistry; Berthon, G., Ed.; Marcel Dekker: New York, NY, USA, 1995; Volume 1, pp. 603–619. [Google Scholar]
- Khoury, R.R.; Sutton, G.J.; Ebrahimi, D.; Hibbert, D.B. Formation constants of copper(II) complexes with tripeptides containing Glu, Gly, and His: Potentiometric measurements and modeling by generalized multiplicative analysis of variance. Inorg. Chem. 2014, 53, 1278–1287. [Google Scholar] [CrossRef]
- Sóvágó, I.; Várnagy, K.; Lihi, N.; Grenács, Á. Coordinating properties of peptides containing histidyl residues. Coord. Chem. Rev. 2016, 327–328, 43–54. [Google Scholar] [CrossRef]
- Rimola, A.; Rodríguez-Santiago, L.; Sodupe, M. Cation-pi Interactions and oxidative effects on Cu+ and Cu2+ binding to Phe, Tyr, Trp, and His amino acids in the gas phase. Insights from first-principles calculations. J. Phys. Chem. B 2006, 110, 24189–24199. [Google Scholar] [CrossRef]
- Kozlowski, H.; Bal, W.; Dyba, M.; Kowalik-Jankowska, T. Specific structure-stability relations in metallopeptides. Coord. Chem. Rev. 1999, 184, 319–346. [Google Scholar] [CrossRef]
- Crowley, J.D.; McMorran, D.A. Topics in Heterocyclic 100. Chemistry 2012, 28, 31. [Google Scholar]
- Bedeche, S.; Daran, J.-C.; Ruiz, J.; Astruc, D. Synthesis and coordination chemistry of ferrocenyl-1, 2, 3-triazolyl ligands. Inorg. Chem. 2008, 47, 4903–4908. [Google Scholar] [CrossRef]
- Valverde, I.E.; Mindt, T.L. 1,2,3-Triazoles as Amide-bond Surrogates in Peptidomimetics. CHIMIA 2013, 67, 262–266. [Google Scholar] [CrossRef]
- Hamdan, F.; Tahoori, F.; Balalaie, S. Synthesis of novel cyclopeptides containing heterocyclic skeletons. RSC Adv. 2018, 8, 33893–33926. [Google Scholar] [CrossRef] [Green Version]
- Bonnamour, J.; Legros, J.; Crousse, B.; Bonnet-Delpon, D. Synthesis of new trifluoromethyl peptidomimetics with a triazole moiety. Tetrahedron Lett. 2007, 48, 8360–8362. [Google Scholar] [CrossRef]
- Agalave, S.G.; Maujan, S.R.; Pore, V.S. Click Chemistry: 1,2,3-Triazoles as Pharmacophores. Chem. Asian J. 2011, 6, 2696–2718. [Google Scholar] [CrossRef]
- Kharb, R.; Sharma, P.C.; Yar, M.S. Pharmacological significance of triazole scaffold. J. Enzym. Inhib. Med. Chem. 2010, 26, 1–21. [Google Scholar] [CrossRef]
- Sivakumar, K.; Xie, F.; Cash, B.M.; Long, S.; Barnhill, H.N.; Wang, Q. A Fluorogenic 1,3-Dipolar Cycloaddition Reaction of 3-Azidocoumarins and Acetylenes. Org. Lett. 2004, 6, 4603–4606. [Google Scholar] [CrossRef] [PubMed]
- Mama, N.; Battison, A. Synthesis and application of a fluorescent “turn-off” triazolyl-coumarin-based fluorescent chemosensor for the sensing of Fe3+ ions in aqueous solutions. Arkivoc 2020, 5, 59–84. [Google Scholar] [CrossRef]
- Singh, G.; Satija, P.; Singh, A.; Diksha, A.; Pawan, A.; Suman, A.; Sushma, A.; Mohit, A.; Soni, S. Azo dye featuring triazole appended organosilicon multifunctionalized sensor: Paradigm for detection of Cu+2 and Fe+2 ions. Mater. Chem. Phys. 2020, 249, 123005. [Google Scholar] [CrossRef]
- Musa, M.A.; Cooperwood, J.S.; Khan, M.O.F. A review of coumarin derivatives in pharmacotherapy of breast cancer. Curr. Med. Chem. 2008, 15, 2664–2679. [Google Scholar] [CrossRef] [Green Version]
- Canning, C.; Sun, S.; Ji, X.; Gupta, S.; Zhou, K. Antibacterial and cytotoxic activity of isoprenylated coumarin mammea A/AA isolated from Mammea Africana. J. Ethnopharmacol. 2013, 147, 259–262. [Google Scholar] [CrossRef]
- Nizamov, S.; Willig, K.I.; Sednev, M.V.; Belov, V.N.; Hell, S.W. Phosphorylated 3-Heteroarylcoumarins and Their Use in Fluorescence Microscopy and Nanoscopy. Chem. Eur. J. 2012, 18, 16339–16348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Lu, L.; Ye, W.; Zheng, O.; Qiu, B.; Lin, Z.; Guo, L.; Chen, G. Fluorescence sensor for Cu(II) in the serum sample based on click chemistry. Analyst 2014, 139, 656–659. [Google Scholar] [CrossRef]
- Mital, M.; Wezynfeld, N.E.; Frączyk, T.; Wiloch, M.Z.; Wawrzyniak, U.E.; Bonna, A.; Tumpach, C.; Barnham, K.J.; Haigh, C.L.; Bal, W.; et al. A functional role for Aβ in metal homeostasis? N-truncation and high-affinity copper binding. Angew. Chem. 2015, 127, 10606–10610. [Google Scholar] [CrossRef]
- Mital, M.; Szutkowski, K.; Bossak-Ahmad, K.; Skrobecki, P.; Drew, S.C.; Poznański, J.; Zhukov, I.; Frączyk, T.; Bal, W. The Palladium(II) Complex of Aβ4−16 as Suitable Model for Structural Studies of Biorelevant Copper(II) Complexes of N-Truncated Beta-Amyloids. Int. J. Mol. Sci. 2020, 21, 9200. [Google Scholar] [CrossRef]
- Gonzalez, P.; Bossak, K.; Stefaniak, E.; Hureau, C.; Raibaut, L.; Bal, W.; Faller, P. N-Terminal Cu-Binding Motifs (Xxx-Zzz-His, Xxx-His) and Their Derivatives: Chemistry, Biology and Medicinal Applications. Chem.-Eur. J. 2018, 24, 8029–8041. [Google Scholar] [CrossRef]
- Li, L.B.; Vorobyov, I.; Allen, T.W. The Different Interactions of Lysine and Arginine Side Chains with Lipid Membranes. J. Phys. Chem. B 2013, 117, 11906–11920. [Google Scholar] [CrossRef]
- Mergny, J.L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Feldman, A.K.; Colasson, B.; Fokin, V.V. One-Pot Synthesis of 1,4-Disubstituted 1,2,3-Triazoles from In Situ Generated Azides. Org. Lett. 2004, 6, 3897–3899. [Google Scholar] [CrossRef]
- Hassan, S.; Müller, T.J.J. Multicomponent syntheses based upon copper-catalyzed alkyne-azide cycloaddition. Adv. Synth. Catal. 2015, 357, 617–666. [Google Scholar] [CrossRef]
- Aioub, A.G.; Dahora, L.; Gamble, K.; Finn, M.G. Selection of Natural Peptide Ligands for Copper-Catalyzed Azide–Alkyne Cycloaddition Catalysis. Bioconjug. Chem. 2017, 28, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- Hoshiyama, M.; Kubo, K.; Igarashi, T.; Sakurai, T. Complexation and proton dissociation behavior of 7-hydroxy-4-methylcoumarin and related compounds in the presence of beta-cyclodextrin. J. Photochem. Photobiol. A Chem. 2001, 138, 227–233. [Google Scholar] [CrossRef]
- Gampp, H.; Maeder, M.; Meyer, C.J.; Zuberbuhler, A.D. Calculation of Equilibrium-Constants from Multiwavelength Spectroscopic Data.2. Specfit-2 User-Friendly Programs in Basic and Standard Fortran-77. Talanta 1985, 32, 257–264. [Google Scholar] [CrossRef]
- Maeder, M.; Zuberbuehler, A.D. Nonlinear Least-Squares Fitting of Multivariate Absorption Data. Anal. Chem. 1990, 62, 2220–2224. [Google Scholar] [CrossRef]
- Alies, B.; Renaglia, E.; Rozga, M.; Bal, W.; Faller, P.; Hureau, C. Cu(II) Affinity for the Alzheimer’s Peptide: Tyrosine Fluorescence Studies Revisited. Anal. Chem. 2013, 85, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Pescitelli, G.; Di Bari, L.; Berova, N. Conformational aspects in the studies of organic compounds by electronic circular dichroism. Chem. Soc. Rev. 2011, 40, 4603–4625. [Google Scholar] [CrossRef]
- Saha, B.; Ikbal, S.A.; Petrovic, A.G.; Berova, N.; Rath, S.P. Complexation of Chiral Zinc-Porphyrin Tweezer with Achiral Diamines: Induction and Two-Step Inversion of Interporphyrin Helicity Monitored by ECD. Inorg. Chem. 2017, 56, 3849–3860. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Petrovic, A.G.; Roje, M.; Pescitelli, G.; Kayser, M.M.; Yang, Y.; Berova, N.; Proni, G. CD-Sensitive Zn-Porphyrin Tweezer Host-Guest Complexes, Part 2: Cis- and Trans-3-Hydroxy-4-aryl/alkyl-beta-Lactams. A Case Study. Chirality 2010, 22, 140–152. [Google Scholar] [CrossRef] [PubMed]
- McGhee, J.D.; von Hippel, P.H. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J. Mol. Biol. 1974, 86, 469–489. [Google Scholar] [CrossRef]
- Sedgwick, A.C.; Brewster, J.T.; Wu, T.H.; Feng, X.; Bull, S.D.; Qian, X.H.; Sessler, J.L.; James, T.D.; Anslyn, E.V.; Sun, X.L. Indicator displacement assays (IDAs): The past, present and future. Chem. Soc. Rev. 2021, 50, 9–38. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, M.; Norden, B. Linear and Circular Dichroism of Drug-Nucleic Acid Complexes. Methods Enzymol. 2001, 340, 68–98. [Google Scholar] [PubMed]
- Šmidlehner, T.; Piantanida, I.; Pescitelli, G. Polarization Spectroscopy Methods in the Determination of Interactions of Small Molecules with Nucleic Acids-Tutorial. Beilstein J. Org. Chem. 2018, 14, 84–105. [Google Scholar] [CrossRef]
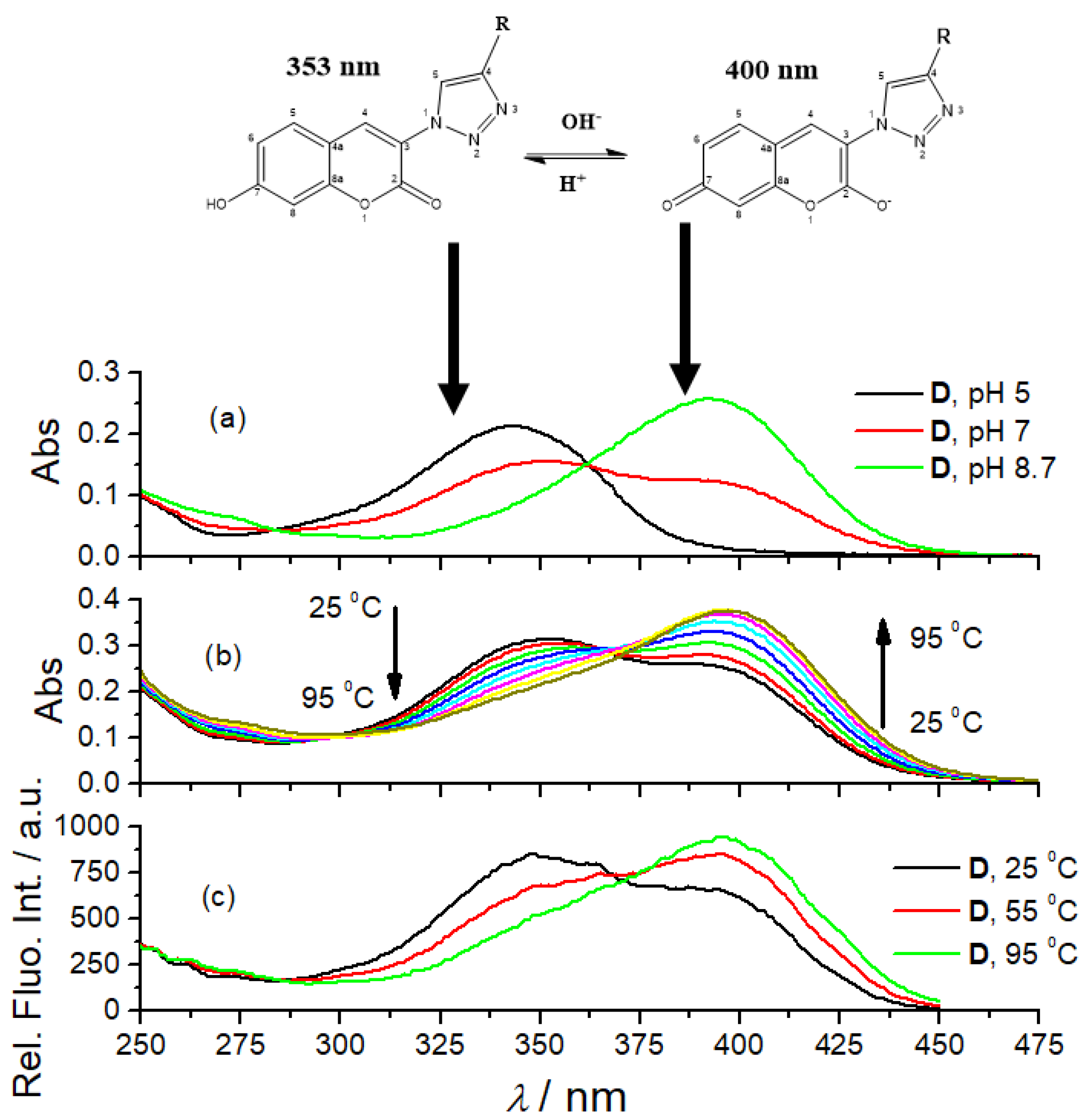
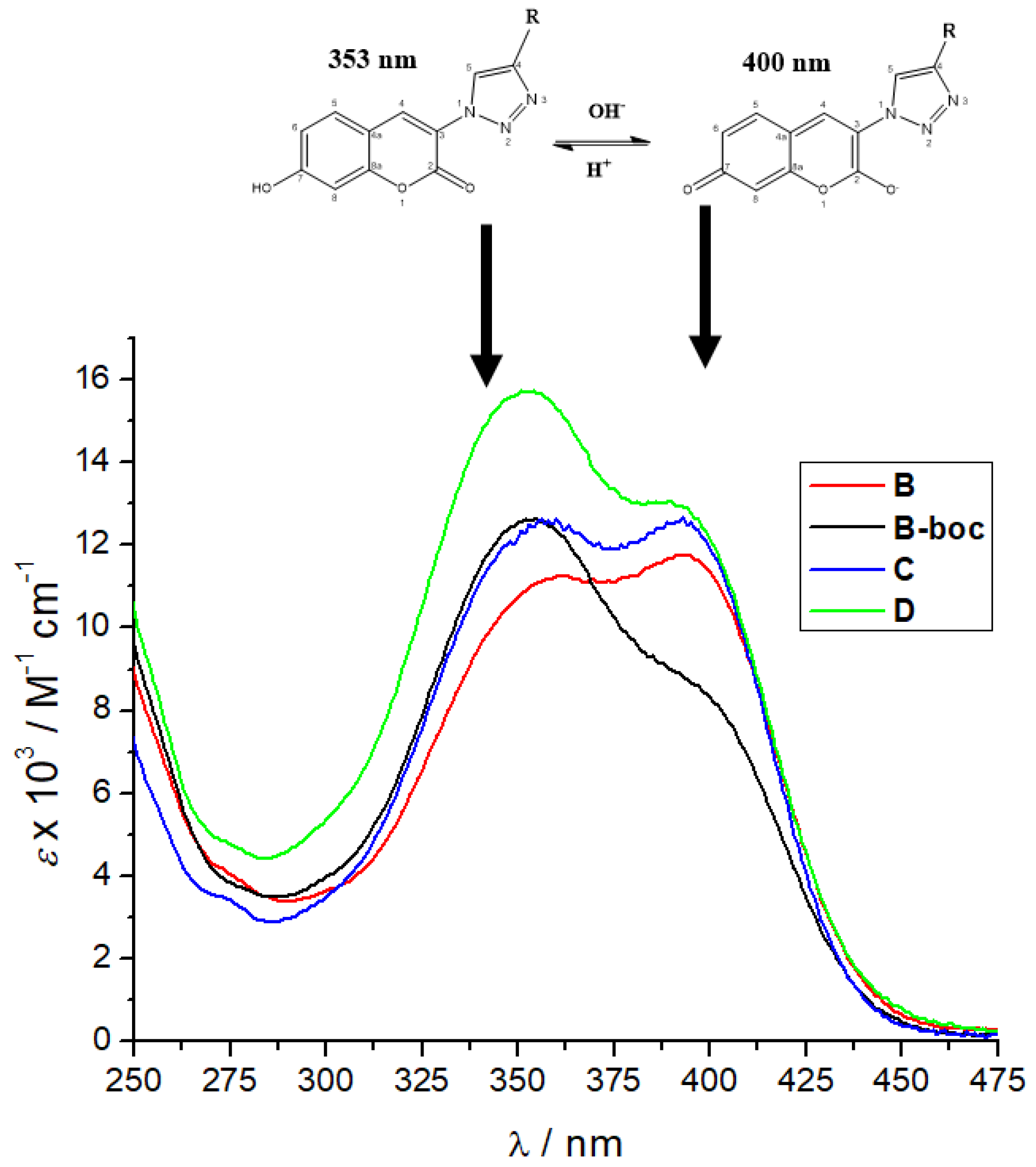
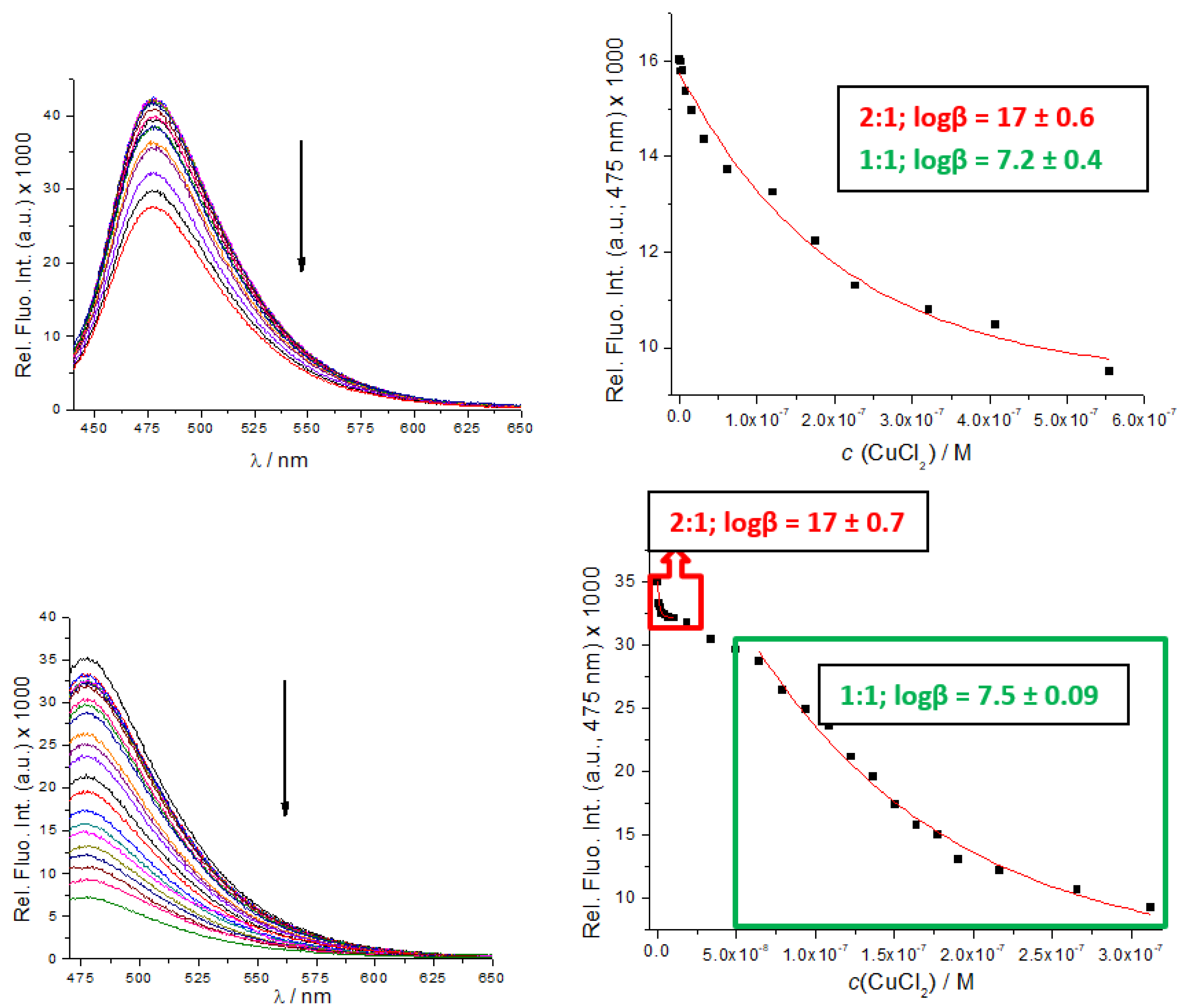
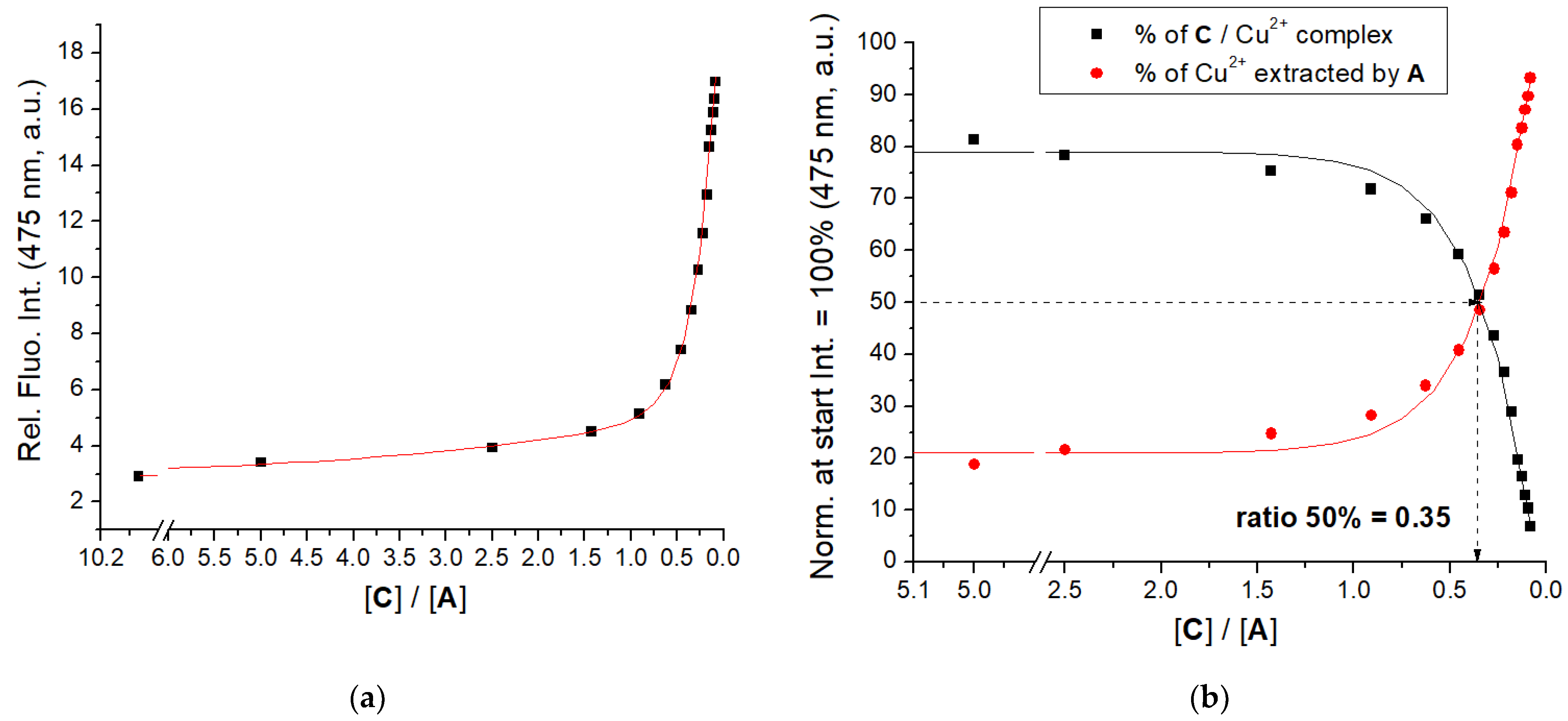
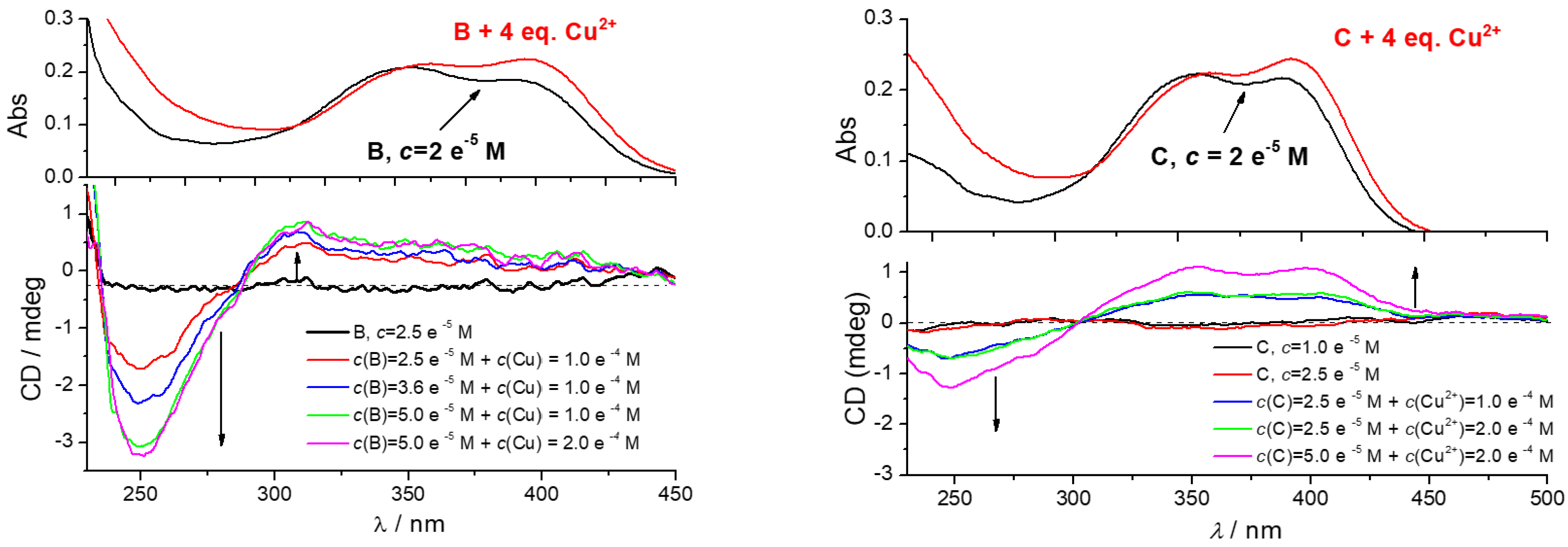
| Compd. | λmax/nm | ε/M−1cm−1 | b Φf | a λexc/nm | a λem/nm | /ns a (purged) | χ2 |
|---|---|---|---|---|---|---|---|
| A | 266 | 5826 ± 60 | c | c | c | c | c |
| B | 392 | 11,633 ± 167 | 0.77 ± 0.05 | 340 | 477 | 4.36 (100%) | 1.062 |
| 405 | 4.32 (100%) | 1.040 | |||||
| B-boc | 353 | 12,620 ± 60 | 0.36 ± 0.05 | 340 | 476 | 4.47 (100%) | 1.055 |
| 405 | 0.49 (5.84%) 4.40 (94.16%) | 0.933 | |||||
| C | 393 | 11,262 ± 496 | 0.58 ± 0.05 | 340 | 475 | 4.22 (100%) | 1.082 |
| 405 | 4.21 (100%) | 1.079 | |||||
| D | 353 | 15,479 ± 106 | 0.45 ± 0.05 | 340 | 472 | 4.20 (100%) | 1.009 |
| 405 | 4.19 (100%) | 1.032 |
| Compound | ctDNA | Poly A–Poly U |
|---|---|---|
| A bA + 4Cu2+ | 0 +3.7 | 0 c 0; +9.5 |
| B-boc bB-boc + 4Cu2+ | 0 +2.7 | 0 c 0; +10.3 |
| B bB + 4Cu2+ | 0 +2.0 | 0 c 0; +11.6 |
| C bC + 4Cu2+ | 0 +2.0 | −2.5 c 0; +11.0 |
| D bD + 4Cu2+ | 0 +2.5 | 0 c 0; +8.5 |
| Compound | ct-DNA | poly A–poly U |
|---|---|---|
| B-boc | 5.4 | 5.7 |
| B | 4.3 | 3.8 |
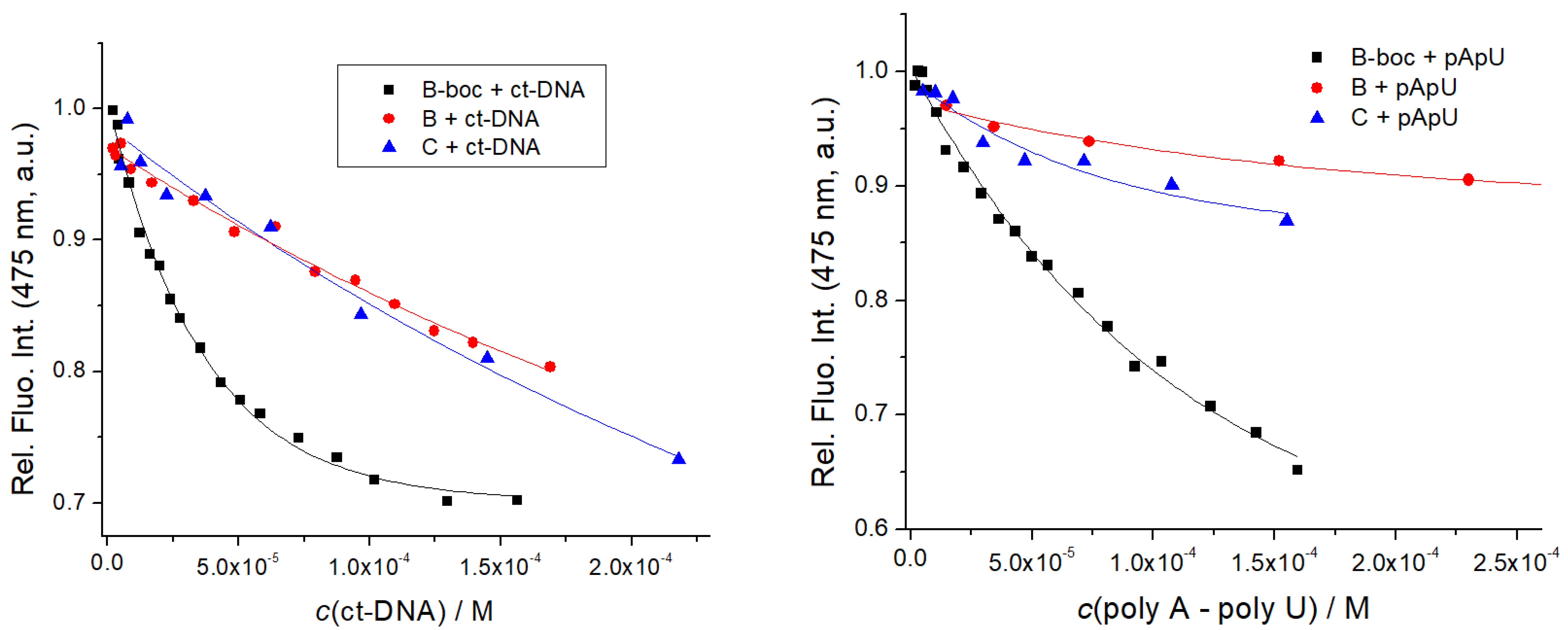
| C | <4 b | 4.8 |
| D | b | b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Košćak, M.; Krošl, I.; Žinić, B.; Piantanida, I. Fluorescent Analogues of FRH Peptide: Cu(II) Binding and Interactions with ds-DNA/RNA. Chemosensors 2022, 10, 34. https://doi.org/10.3390/chemosensors10010034
Košćak M, Krošl I, Žinić B, Piantanida I. Fluorescent Analogues of FRH Peptide: Cu(II) Binding and Interactions with ds-DNA/RNA. Chemosensors. 2022; 10(1):34. https://doi.org/10.3390/chemosensors10010034
Chicago/Turabian StyleKošćak, Marta, Ivona Krošl, Biserka Žinić, and Ivo Piantanida. 2022. "Fluorescent Analogues of FRH Peptide: Cu(II) Binding and Interactions with ds-DNA/RNA" Chemosensors 10, no. 1: 34. https://doi.org/10.3390/chemosensors10010034
APA StyleKošćak, M., Krošl, I., Žinić, B., & Piantanida, I. (2022). Fluorescent Analogues of FRH Peptide: Cu(II) Binding and Interactions with ds-DNA/RNA. Chemosensors, 10(1), 34. https://doi.org/10.3390/chemosensors10010034







