Long-Term Glucose Forecasting for Open-Source Automated Insulin Delivery Systems: A Machine Learning Study with Real-World Variability Analysis
Abstract
1. Introduction
1.1. Overview of Data-Driven Automated Insulin Delivery Systems
1.2. Applications of Machine Learning and Deep Learning in AID Systems
1.3. Original Contributions
1.4. Organisation of the Paper
2. Related Work
2.1. Review of Machine Learning and Deep Learning Methods and Techniques for Glucose Forecasting
Limitations and Shortcomings
- The primary issue of all the reported methods is the evaluation of trained models for a limited prediction horizon of 30 min and 60 min, with the maximum being 120 min, i.e., the reported predictions for the trained models are in the range of 30, 60, or 120 min.
- The lack of consistency in the accuracies of the reported models makes it difficult to compare the existing work. This further affects the reliability of the trained models for further evaluation and reproducibility.
- Another drawback of the existing literature is the previous lack of large-scale and real-world datasets for individuals with diabetes that use automated insulin delivery systems. Therefore, the majority of the aforementioned models in the literature are trained on partial/fully simulated data or limited days of real-world CGM data.
- Multiple model performances and accuracy metrics have been used (including RMSE, specificity, MAE and F1 score) to evaluate the model predictions. However, to the best of our knowledge, none of the existing works has evaluated and studied the impact of glucose predictions by calculating the clinically validated glucose variability (GV) metrics.
- There is a lack of implementation details and open-source methods to reproduce the reported results which makes it difficult to independently evaluate them on additional datasets or to be able to evaluate their applicability for different modalities of insulin therapy, such as in sensor-augmented pump therapy as compared to automated insulin delivery therapy.
- Most of the existing works employed a limited number of machine learning models (one or two) for evaluation which certainly adds inconsistency. However, it is critical to evaluate model results for multiple machine learning and deep learning models along with tuned time series analysis frameworks like ARIMA. Evaluating the results of multiple model types would lay a foundation for benchmarking.
2.2. Clinically-Approved Statistical and Variability Metrics for Glucose Analysis
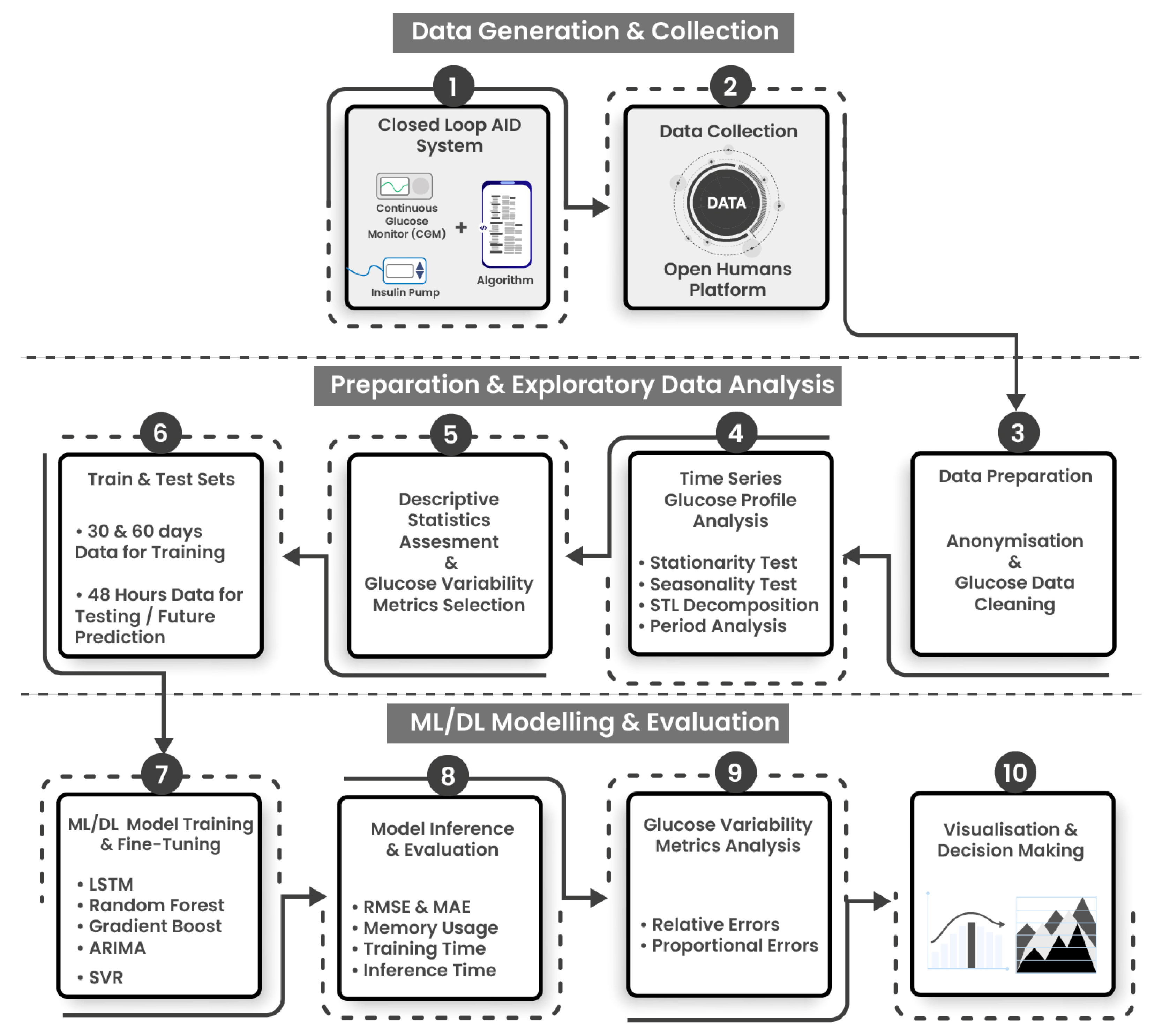
3. Materials and Methods
3.1. Experimental Workflow and ML Development Pipelines
- Stage 2: Data preparation and exploratory data analysis (EDA) is composed of four steps: Data is exported, prepared using anonymization and cleaning protocols (Step 3), a diverse subset of individuals are selected, and the glucose profiles are analyzed using descriptive statistics and clinically approved GV metrics (Steps 4 and 5). The data is then split into training and testing sets. Models have been trained on 30 and 60 days of glucose data and individually tested to predict upto 48 h of glucose data points. (Step 6).
- Stage 3: ML/DL modeling, evaluation and analysis consists of 4 steps. ML/DL algorithms are fine-tuned and evaluated for accuracy and resource consumption (step 7), and analyzed using statistical and glucose variability metrics from expected and predicted glucose profiles (Steps 8, 9, 10).
3.2. Highlights of Data Collection, Anonymisation, and Cleaning
3.3. Machine Learning and Deep Learning Algorithms Employed for Glucose Forecasting
3.4. Statistical and Variability Metrics for Glucose Analysis
4. Results
4.1. Descriptive Statistics and Glucose Variability Metrics for Selected AID Users
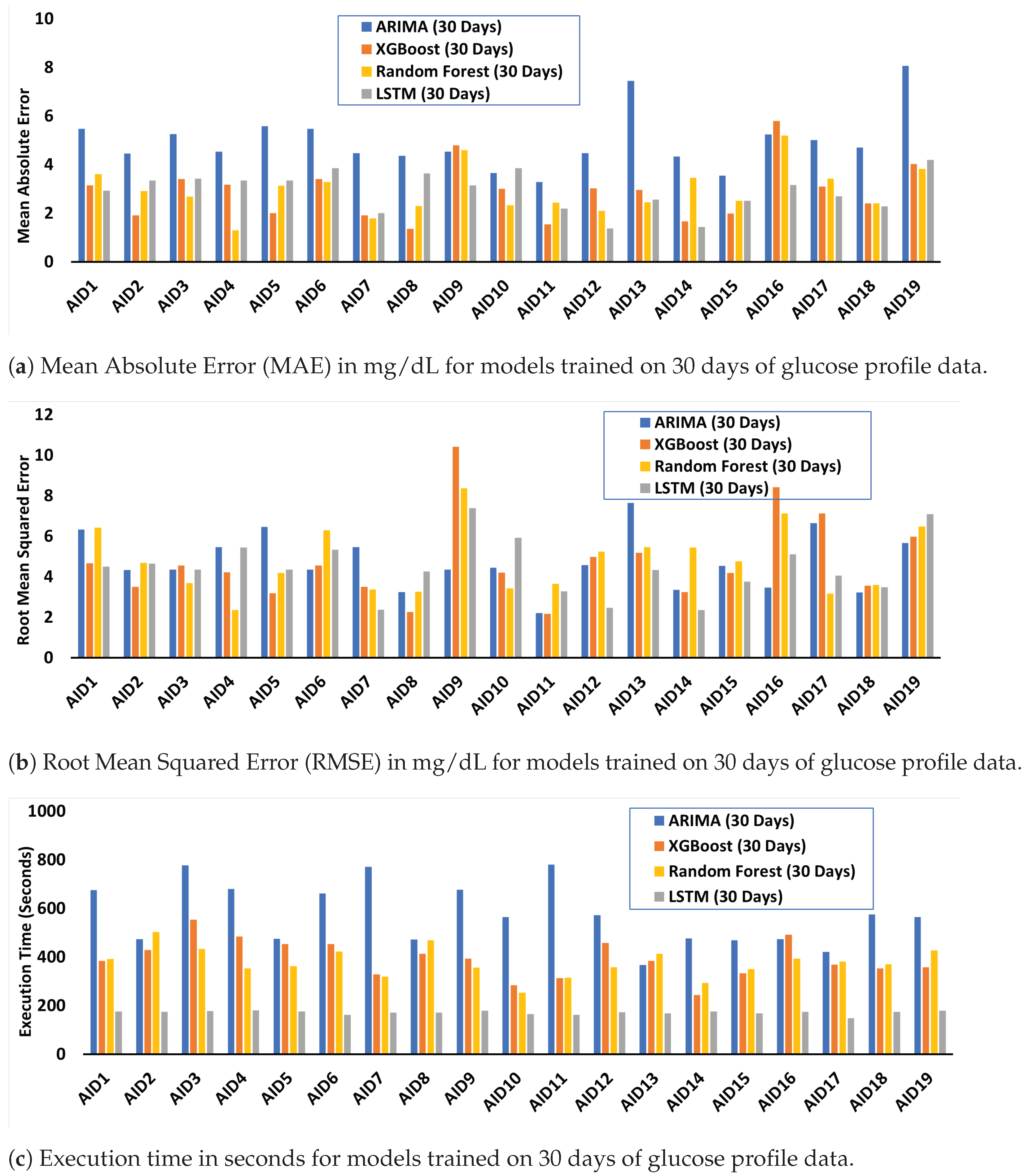
4.2. Performance and Resource Cost Evaluation and Analysis of Trained ML/DL Algorithms
4.3. Comparative Analysis of Glucose Variability for Predicted and Expected Glucose Profiles
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OPEN | Outcomes of Patients’ Evidence with Novel, Do-it-Yourself Artificial Pancreas Technology |
| OpenAPS | Open Source Artificial Pancreas System |
| AID | Automated Insulin Delivery |
| APS | Artificial Pancreas System |
| HCL | Hybrid Closed Loop |
| T1D | Type 1 Diabetes |
| CGM | Continuous Glucose Monitoring |
| PWD | People With Diabetes (any type) |
| HbA1c | Hemoglobin A1c |
| TIR | Time In Range |
| GV | Glucose Variability |
Appendix A. Model Evaluation Metrics for Performance and Resource Cost
Appendix B. Relative and Proportional Errors
Appendix C. Example Predicted and Expected Glucose Profiles for 48 h
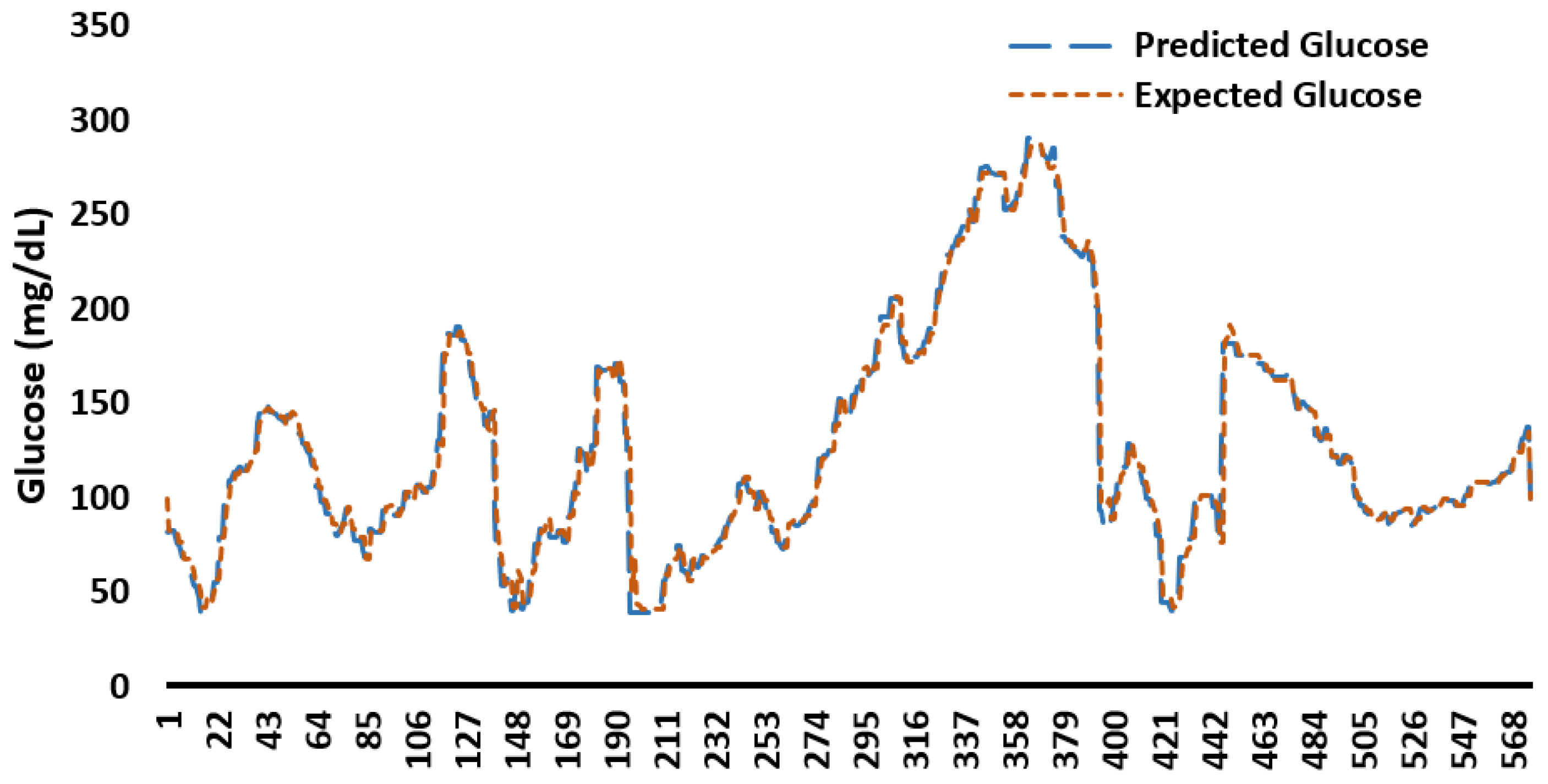
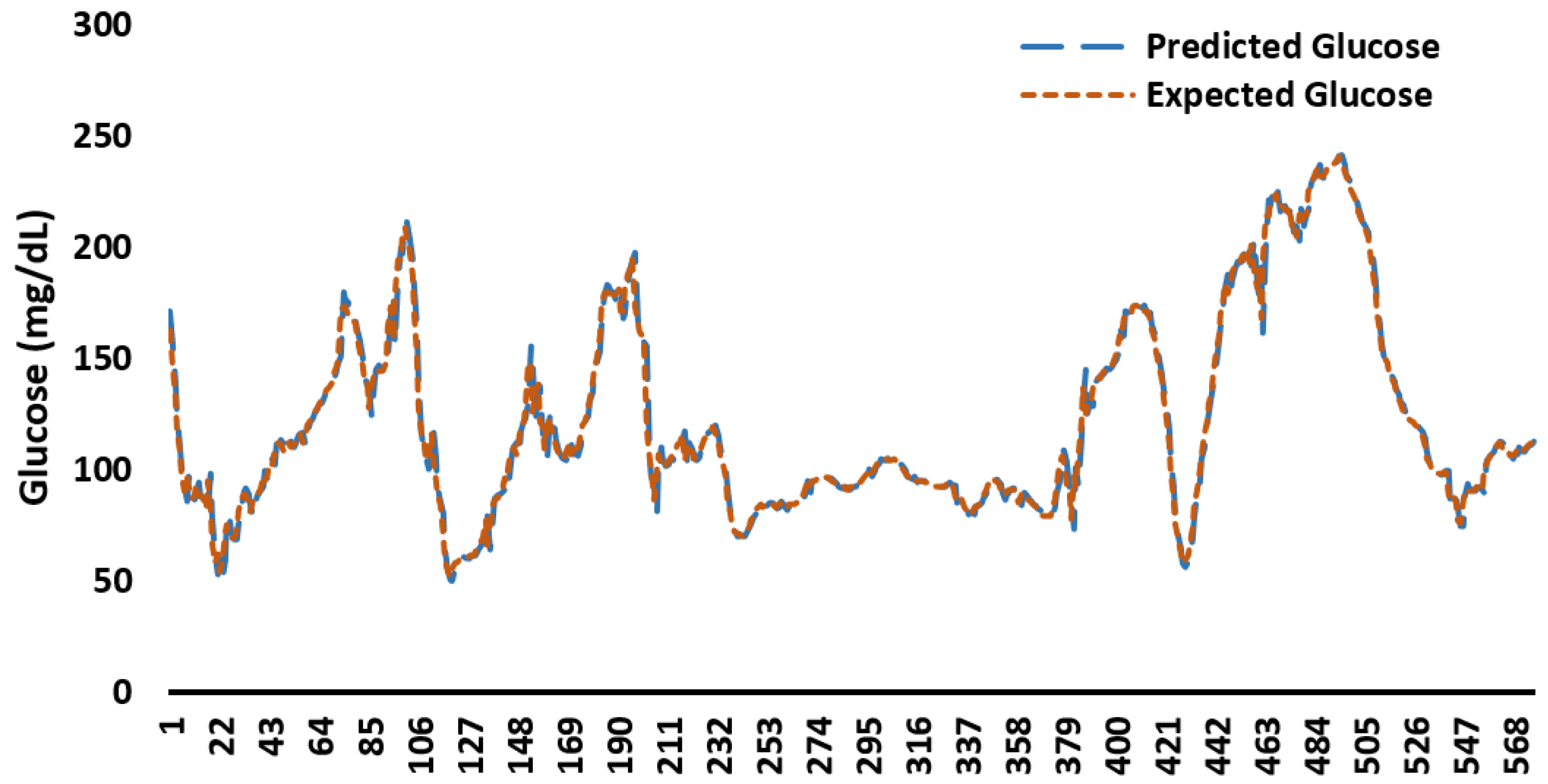
Appendix D. Memory Consumption by ML/DL Models
| Model | Memory Consumption Range (MB) |
|---|---|
| ARIMA | 994–1024 |
| XG Boost | 800–1024 |
| Random Forest | 750–1123 |
| LSTM | 1100–1993 |
| SVR | 1024–1345 |
Appendix E. MAE, RMSE, and Execution Time for Models Trained on 60 Days of Glucose Data
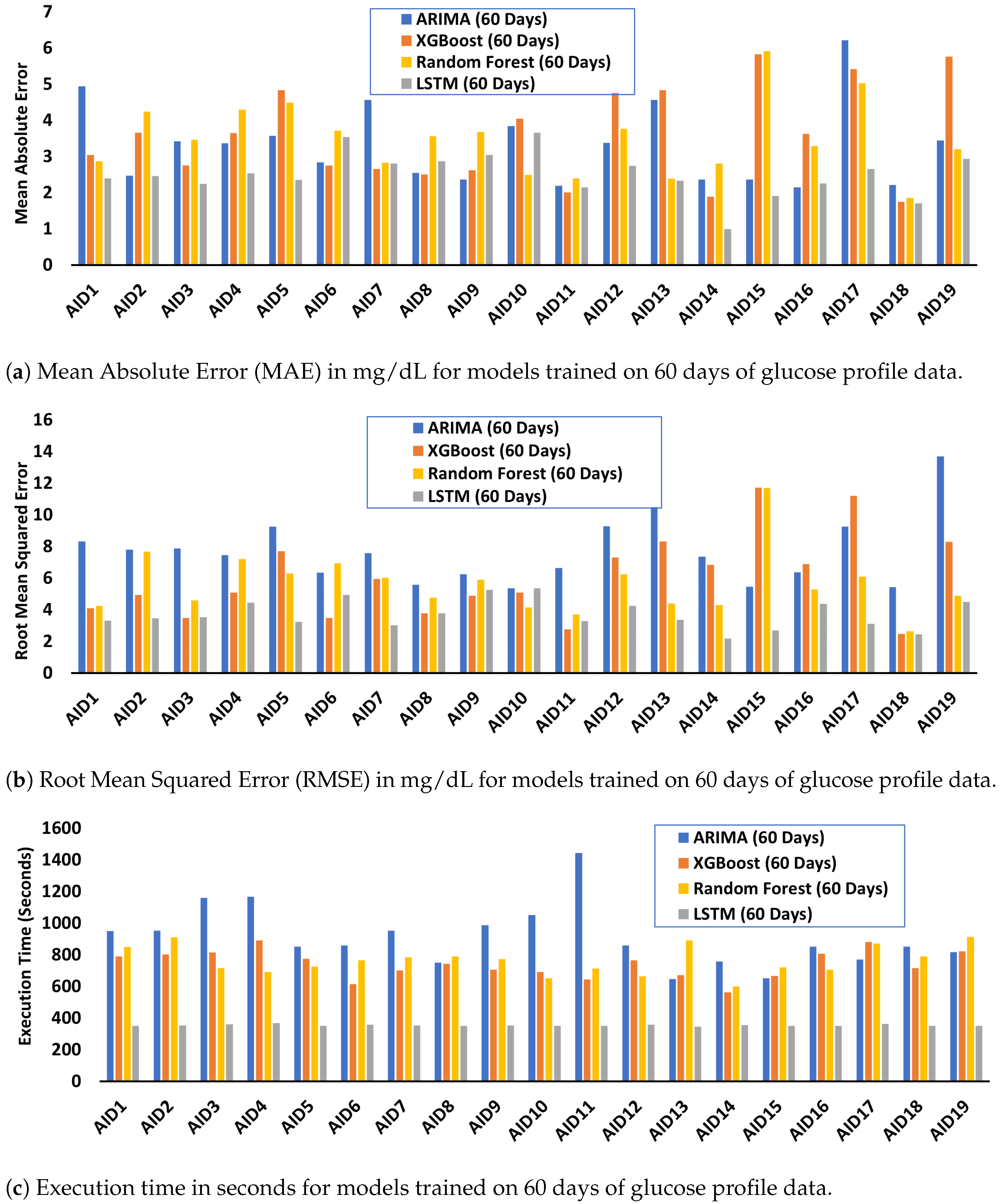
Appendix F. Relative and Proportional Errors for Models Trained on 60 Days of Glucose Data
| Relative Error (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARIMA60 | XGBoost60 | LSTM60 | RF60 | |||||||||
| Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | |
| CONGA | 0.08 | 3.85 | 47.28 | 0 | 0.5 | 1.04 | 0.1 | 0.72 | 2.21 | 0.13 | 0.64 | 1.7 |
| LI | 1.11 | 13.58 | 88.93 | 0.01 | 2.53 | 6.23 | 0.48 | 6.96 | 12.86 | 0.57 | 3.88 | 15.42 |
| JINDEX | 0.04 | 8.23 | 79.36 | 0.06 | 0.7 | 1.75 | 0.12 | 0.77 | 2.72 | 0.07 | 0.66 | 2.38 |
| LBGI | 0.21 | 12.27 | 70.35 | 2.22 | 6.05 | 12.17 | 0.02 | 4.52 | 15.09 | 0.87 | 4.39 | 8.87 |
| HBGI | 0.1 | 45.57 | 705.65 | 0.02 | 1.75 | 4.32 | 0.37 | 4.1 | 31.74 | 0.04 | 7.03 | 100 |
| GRADE | 0.1 | 20.42 | 262.81 | 0.27 | 4.68 | 10.2 | 0.04 | 2.86 | 6.45 | 0 | 4.59 | 8.63 |
| MODD | 0.06 | 11.92 | 152.79 | 0.31 | 1.57 | 2.79 | 0.33 | 1.4 | 5.58 | 0.03 | 1.19 | 2.43 |
| MAGE | 1.01 | 15.52 | 86.33 | 0.18 | 8.76 | 59.67 | 0.09 | 14.75 | 179.59 | 0.06 | 12.54 | 70.45 |
| ADDR | 0.08 | 43.29 | 621.83 | 0.17 | 1.67 | 5.35 | 0.06 | 8.91 | 67.25 | 1.01 | 12.83 | 100 |
| MVALUE | 0.41 | 11.6 | 76.71 | 1.53 | 6.38 | 11.28 | 0.17 | 4.07 | 9.01 | 0.58 | 4.15 | 8.32 |
| MAG | 3.52 | 20.59 | 100.51 | 0.14 | 5.47 | 11.3 | 8.63 | 36.25 | 60.68 | 18.18 | 29.44 | 45.99 |
| SD | 0.17 | 1.16 | 4.51 | 0.02 | 1.5 | 3 | 0.38 | 1.27 | 4.02 | 0.09 | 0.73 | 2.15 |
| MEAN | 0 | 0.03 | 0.13 | 0.04 | 0.26 | 0.82 | 0 | 0.4 | 1.29 | 0.03 | 0.34 | 1.05 |
| CV | 0.22 | 1.17 | 4.54 | 0.23 | 1.57 | 3.86 | 0.38 | 1.47 | 3.79 | 0.01 | 0.7 | 1.75 |
| TIR | 0 | 0.82 | 3.16 | 0 | 1.77 | 5.62 | 0 | 1.02 | 5.07 | 0 | 1.3 | 3.84 |
| TAR | 0 | 4.27 | 21.21 | 0 | 3.05 | 22.22 | 0 | 9.25 | 50 | 0 | 13.53 | 100 |
| TBR | 0 | 25.98 | 150 | 0 | 13.42 | 50 | 0 | 51.54 | 700 | 0 | 11.6 | 50 |
| Proportional Error | ||||||||||||
| ARIMA60 | XGBoost60 | LSTM60 | RF60 | |||||||||
| Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | |
| CONGA | 1 | 1.06 | 1.9 | 1 | 1 | 1.01 | 1 | 1.01 | 1.02 | 1 | 1.01 | 1.02 |
| LI | 1.01 | 1.4 | 6.83 | 1 | 1.03 | 1.06 | 1 | 1.07 | 1.14 | 1.01 | 1.04 | 1.18 |
| JINDEX | 1 | 1.24 | 4.84 | 1 | 1.01 | 1.02 | 1 | 1.01 | 1.03 | 1 | 1.01 | 1.02 |
| LBGI | 1 | 1.21 | 3.37 | 1.02 | 1.06 | 1.12 | 1 | 1.05 | 1.15 | 1.01 | 1.04 | 1.09 |
| HBGI | 1 | 3.5 | 40.78 | 1 | 1.02 | 1.04 | 1 | 1.04 | 1.32 | 1 | 1.02 | 1.07 |
| GRADE | 1 | 1.52 | 7.87 | 1 | 1.05 | 1.1 | 1 | 1.03 | 1.06 | 1 | 1.05 | 1.09 |
| MODD | 1 | 1.15 | 2.53 | 1 | 1.02 | 1.03 | 1 | 1.01 | 1.06 | 1 | 1.01 | 1.02 |
| MAGE | 1.01 | 1.23 | 2.86 | 1 | 1.13 | 2.48 | 1 | 1.15 | 2.8 | 1 | 1.22 | 3.38 |
| ADDR | 1 | 4 | 50.78 | 1 | 1.02 | 1.05 | 1 | 1.1 | 1.67 | 1.01 | 1.1 | 1.47 |
| MVALUE | 1 | 1.25 | 4.29 | 1.02 | 1.06 | 1.11 | 1 | 1.04 | 1.09 | 1.01 | 1.04 | 1.08 |
| MAG | 1.04 | 1.21 | 2.01 | 1 | 1.06 | 1.13 | 1.09 | 1.69 | 2.54 | 1.22 | 1.45 | 1.85 |
| SD | 1 | 1.01 | 1.05 | 1 | 1.02 | 1.03 | 1 | 1.01 | 1.04 | 1 | 1.01 | 1.02 |
| MEAN | 1 | 1 | 1 | 1 | 1 | 1.01 | 1 | 1 | 1.01 | 1 | 1 | 1.01 |
| CV | 1 | 1.01 | 1.05 | 1 | 1.02 | 1.04 | 1 | 1.01 | 1.04 | 1 | 1.01 | 1.02 |
| TIR | 1 | 1.01 | 1.03 | 1 | 1.02 | 1.06 | 1 | 1.01 | 1.05 | 1 | 1.01 | 1.04 |
| TAR | 1 | 1.04 | 1.21 | 1 | 1.04 | 1.29 | 1 | 1.09 | 1.5 | 1 | 1.08 | 1.5 |
| TBR | 1 | 1.26 | 2.5 | 1 | 1.16 | 1.86 | 1 | 1.52 | 8 | 1 | 1.15 | 2 |
References
- Benhamou, P.Y.; Reznik, Y. Closed-loop insulin delivery: Understanding when and how it is effective. Lancet Digit. Health 2020, 2, e50–e51. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.M. Quantifying input behaviors that influence clinical outcomes in diabetes and other chronic illnesses. J. Diabetes Sci. Technol. 2022, 16, 786–787. [Google Scholar] [CrossRef] [PubMed]
- Benhamou, P.Y.; Franc, S.; Reznik, Y.; Thivolet, C.; Schaepelynck, P.; Renard, E.; Guerci, B.; Chaillous, L.; Lukas-Croisier, C.; Jeandidier, N.; et al. Closed-loop insulin delivery in adults with type 1 diabetes in real-life conditions: A 12-week multicentre, open-label randomised controlled crossover trial. Lancet Digit. Health 2019, 1, e17–e25. [Google Scholar] [CrossRef] [PubMed]
- Mordvanyuk, N.; Torrent-Fontbona, F.; López, B. Prediction of Glucose Level Conditions from Sequential Data. In Proceedings of the CCIA, Terres de l’Ebre, Spain, 25–27 October 2017; pp. 227–232. [Google Scholar]
- Dave, D.; DeSalvo, D.J.; Haridas, B.; McKay, S.; Shenoy, A.; Koh, C.J.; Lawley, M.; Erraguntla, M. Feature-based machine learning model for real-time hypoglycemia prediction. J. Diabetes Sci. Technol. 2021, 15, 842–855. [Google Scholar] [CrossRef]
- Maritsch, M.; Foll, S.; Lehmann, V.; Bérubé, C.; Kraus, M.; Feuerriegel, S.; Kowatsch, T.; Zuger, T.; Stettler, C.; Fleisch, E.; et al. Towards wearable-based hypoglycemia detection and warning in diabetes. In Proceedings of the Extended Abstracts of the 2020 CHI Conference on Human Factors in Computing Systems, Honolulu, HI, USA, 25–30 April 2020; pp. 1–8. [Google Scholar]
- Zhu, T.; Kuang, L.; Li, K.; Zeng, J.; Herrero, P.; Georgiou, P. Blood Glucose Prediction in Type 1 Diabetes Using Deep Learning on the Edge. In Proceedings of the 2021 IEEE International Symposium on Circuits and Systems (ISCAS), Daegu, Republic of Korea, 22–28 May 2021; pp. 1–5. [Google Scholar]
- Zhu, T.; Li, K.; Chen, J.; Herrero, P.; Georgiou, P. Dilated recurrent neural networks for glucose forecasting in type 1 diabetes. J. Healthc. Informatics Res. 2020, 4, 308–324. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Yu, X.; Ma, N.; Wu, R.; Li, H. An autonomous channel deep learning framework for blood glucose prediction. Appl. Soft Comput. 2022, 120, 108636. [Google Scholar] [CrossRef]
- Berikov, V.B.; Kutnenko, O.A.; Semenova, J.F.; Klimontov, V.V. Machine Learning Models for Nocturnal Hypoglycemia Prediction in Hospitalized Patients with Type 1 Diabetes. J. Pers. Med. 2022, 12, 1262. [Google Scholar] [CrossRef]
- Duckworth, C.J.; Guy, M.J.; Kumaran, A.; O’Kane, A.; Ayobi, A.; Chapman, A.; Boniface, M. Explainable machine learning for real-time hypoglycaemia and hyperglycaemia prediction and personalised control recommendations. medRxiv 2022. [Google Scholar] [CrossRef]
- van Doorn, W.P.; Foreman, Y.D.; Schaper, N.C.; Savelberg, H.H.; Koster, A.; van der Kallen, C.J.; Wesselius, A.; Schram, M.T.; Henry, R.M.; Dagnelie, P.C.; et al. Machine learning-based glucose prediction with use of continuous glucose and physical activity monitoring data: The Maastricht Study. PLoS ONE 2021, 16, e0253125. [Google Scholar] [CrossRef]
- Iacono, F.; Magni, L.; Toffanin, C. Personalized LSTM models for glucose prediction in Type 1 diabetes subjects. In Proceedings of the 2022 30th Mediterranean Conference on Control and Automation (MED), Athens, Greece, 28 June–1 July 2022; pp. 324–329. [Google Scholar]
- Allam, F.; Nossai, Z.; Gomma, H.; Ibrahim, I.; Abdelsalam, M. A recurrent neural network approach for predicting glucose concentration in type-1 diabetic patients. In Engineering Applications of Neural Networks; Springer: Berlin/Heidelberg, Germany, 2011; pp. 254–259. [Google Scholar]
- Greshake Tzovaras, B.; Angrist, M.; Arvai, K.; Dulaney, M.; Estrada-Galiñanes, V.; Gunderson, B.; Head, T.; Lewis, D.; Nov, O.; Shaer, O.; et al. Open Humans: A platform for participant-centered research and personal data exploration. GigaScience 2019, 8, giz076. [Google Scholar] [CrossRef]
- Hameed, H.; Kleinberg, S. Comparing Machine Learning Techniques for Blood Glucose Forecasting Using Free-living and Patient Generated Data. In Proceedings of the 5th Machine Learning for Healthcare Conference; Doshi-Velez, F., Fackler, J., Jung, K., Kale, D., Ranganath, R., Wallace, B., Wiens, J., Eds.; Proceedings of Machine Learning Research, PMLR, MLResearchPress, 2020; Volume 126, pp. 871–894. Available online: http://proceedings.mlr.press/v126/hameed20a.html (accessed on 20 January 2023).
- Lal, R.A.; Maikawa, C.L.; Lewis, D.; Baker, S.W.; Smith, A.A.; Roth, G.A.; Gale, E.C.; Stapleton, L.M.; Mann, J.L.; Yu, A.C.; et al. Full closed loop open-source algorithm performance comparison in pigs with diabetes. Clin. Transl. Med. 2021, 11, e387. [Google Scholar] [CrossRef] [PubMed]
- Broome, D.T.; Hilton, C.B.; Mehta, N. Policy implications of artificial intelligence and machine learning in diabetes management. Curr. Diabetes Rep. 2020, 20, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Zafar, A. Machine Learning/Deep Learning Models and Statistical Analysis Scripts for the Analysis of Glucose Profiles. 2022. Available online: https://github.com/ahtshamzafar1/ML-and-DL-for-Diabetes-Datasets (accessed on 20 January 2023).
- Marling, C.; Bunescu, R. The OhioT1DM dataset for blood glucose level prediction: Update 2020. In Proceedings of the CEUR Workshop Proceedings; NIH Public Access: Bethesda, MD, USA, 2020; Volume 2675, p. 71. [Google Scholar]
- Man, C.D.; Micheletto, F.; Lv, D.; Breton, M.; Kovatchev, B.; Cobelli, C. The UVA/PADOVA type 1 diabetes simulator: New features. J. Diabetes Sci. Technol. 2014, 8, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Bunescu, R.; Struble, N.; Marling, C.; Shubrook, J.; Schwartz, F. Blood glucose level prediction using physiological models and support vector regression. In Proceedings of the 2013 12th International Conference on Machine Learning and Applications, Miami, FL, USA, 4–7 December 2013; Volume 1, pp. 135–140. [Google Scholar]
- Zecchin, C.; Facchinetti, A.; Sparacino, G.; Cobelli, C. Jump neural network for online short-time prediction of blood glucose from continuous monitoring sensors and meal information. Comput. Methods Programs Biomed. 2014, 113, 144–152. [Google Scholar] [CrossRef]
- Pustozerov, E.; Popova, P.; Tkachuk, A.; Bolotko, Y.; Yuldashev, Z.; Grineva, E. Development and evaluation of a mobile personalized blood glucose prediction system for patients with gestational diabetes mellitus. JMIR mHealth uHealth 2018, 6, e9236. [Google Scholar] [CrossRef]
- Tsai, C.W.; Li, C.H.; Lam, R.W.K.; Li, C.K.; Ho, S. Diabetes care in motion: Blood glucose estimation using wearable devices. IEEE Consum. Electron. Mag. 2019, 9, 30–34. [Google Scholar] [CrossRef]
- Georga, E.I.; Protopappas, V.C.; Ardigo, D.; Marina, M.; Zavaroni, I.; Polyzos, D.; Fotiadis, D.I. Multivariate prediction of subcutaneous glucose concentration in type 1 diabetes patients based on support vector regression. IEEE J. Biomed. Health Inform. 2012, 17, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Gandía, C.; Facchinetti, A.; Sparacino, G.; Cobelli, C.; Gómez, E.; Rigla, M.; de Leiva, A.; Hernando, M. Artificial neural network algorithm for online glucose prediction from continuous glucose monitoring. Diabetes Technol. Ther. 2010, 12, 81–88. [Google Scholar] [CrossRef]
- Bent, B.; Henriquez, M.; Dunn, J.P. Cgmquantify: Python and R Software Packages for Comprehensive Analysis of Interstitial Glucose and Glycemic Variability from Continuous Glucose Monitor Data. IEEE Open J. Eng. Med. Biol. 2021, 2, 263–266. [Google Scholar] [CrossRef]
- Rawlings, R.A.; Shi, H.; Yuan, L.H.; Brehm, W.; Pop-Busui, R.; Nelson, P.W. Translating Glucose Variability Metrics into the Clinic via C ontinuous G lucose M onitoring: AG raphical U ser I nterface for D iabetes E valuation (CGM-GUIDE©). Diabetes Technol. Ther. 2011, 13, 1241–1248. [Google Scholar] [CrossRef]
- Attaye, I.; van der Vossen, E.W.; Mendes Bastos, D.N.; Nieuwdorp, M.; Levin, E. Introducing the Continuous Glucose Data Analysis (CGDA) R Package: An Intuitive Package to Analyze Continuous Glucose Monitoring Data. J. Diabetes Sci. Technol. 2022, 16, 783–785. [Google Scholar] [CrossRef] [PubMed]
- Moscardó, V.; Giménez, M.; Oliver, N.; Hill, N.R. Updated software for automated assessment of glucose variability and quality of glycemic control in diabetes. Diabetes Technol. Ther. 2020, 22, 701–708. [Google Scholar] [CrossRef]
- Vigers, T.; Chan, C.L.; Snell-Bergeon, J.; Bjornstad, P.; Zeitler, P.S.; Forlenza, G.; Pyle, L. cgmanalysis: An R package for descriptive analysis of continuous glucose monitor data. PLoS ONE 2019, 14, e0216851. [Google Scholar] [CrossRef] [PubMed]
- Czerwoniuk, D.; Fendler, W.; Walenciak, L.; Mlynarski, W. GlyCulator: A glycemic variability calculation tool for continuous glucose monitoring data. J. Diabetes Sci. Technol. 2011, 5, 447–451. [Google Scholar] [CrossRef] [PubMed]
- OpenAPS Data Commons. Available online: https://openaps.org/outcomes/data-commons/ (accessed on 20 January 2023).
- Shahid, A.; Lewis, D.M. Large-Scale Data Analysis for Glucose Variability Outcomes with Open-Source Automated Insulin Delivery Systems. Nutrients 2022, 14, 1906. [Google Scholar] [CrossRef]
- Shahid, A. Programming Scripts for Demographics and Glucose Variability Analysis for OpenAPS Data Commons Dataset. 2022. Available online: https://github.com/danamlewis/OpenHumansDataTools/tree/master/bin/GV-demographics-scripts (accessed on 20 January 2023).
- Newbold, P. ARIMA model building and the time series analysis approach to forecasting. J. Forecast. 1983, 2, 23–35. [Google Scholar] [CrossRef]
- Taieb, S.B.; Hyndman, R.J. A gradient boosting approach to the Kaggle load forecasting competition. Int. J. Forecast. 2014, 30, 382–394. [Google Scholar] [CrossRef]
- Masini, R.P.; Medeiros, M.C.; Mendes, E.F. Machine learning advances for time series forecasting. J. Econ. Surv. 2021, 37, 76–111. [Google Scholar] [CrossRef]
- Siami-Namini, S.; Tavakoli, N.; Namin, A.S. A comparison of ARIMA and LSTM in forecasting time series. In Proceedings of the 2018 17th IEEE international conference on machine learning and applications (ICMLA), Orlando, FL, USA, 17–20 December 2018; pp. 1394–1401. [Google Scholar]
- Lin, K.; Lin, Q.; Zhou, C.; Yao, J. Time series prediction based on linear regression and SVR. In Proceedings of the Third International Conference on Natural Computation (ICNC 2007), Haikou, China, 24–27 August 2007; Volume 1, pp. 688–691. [Google Scholar]
- Burnside, M.J.; Lewis, D.M.; Crocket, H.R.; Meier, R.A.; Williman, J.A.; Sanders, O.J.; Jefferies, C.A.; Faherty, A.M.; Paul, R.G.; Lever, C.S.; et al. Open-source automated insulin delivery in type 1 diabetes. N. Engl. J. Med. 2022, 387, 869–881. [Google Scholar] [CrossRef]
- Lewis, D.M.; Leibrand, S. Automatic estimation of Basals, ISF, and CARB ratio for sensor-augmented pump and hybrid closed-loop therapy. In Proceedings of the Diabetes; American Diabetes Association: Alexandria, VA, USA, 2017; Volume 66, p. LB33. [Google Scholar]
| Acronym | Description |
|---|---|
| ADRR | The average daily risk range (ADRR) measures the overall daily variation of glucose, within a specific risk range meanwhile the risk is defined based on the target. |
| CONGA | Continuous overall net glycemic action (CONGA) is applicably close to standard deviation (SD) and measures the possible changes in glucose for a defined period. |
| CV | Coefficient of Variation (CV) is a statistical metric to evaluate the diversity in glucose data and is commonly subdivided into inter-day and intra-day CV metrics. |
| GRADE | The glycemic risk assessment diabetes equation (GRADE) score evaluates the risk correlated with a particular glucose profile comprehensively. |
| HBGI | High blood glucose index (HBGI) is a metric that quantifies the possible risk of hyperglycemia and it can be calculated using self-monitoring of blood glucose (SMBG) or continuous glucose monitor (CGM) data. |
| LBGI | Low blood glucose index (LBGI) is used for hypoglycemic risk management. |
| MAG | Mean absolute glucose (MAG) represents the difference of summation between sequential glucose profiles over 24 h, which is divided by the time (in hours) between the starting and ending glucose values. |
| MAGE | The Mean Amplitude of Glycemic Excursion (MAGE) is defined as the mean of glucose values that exceed the 24-h mean blood glucose value, by one standard deviation. |
| MODD | Mean of daily differences (MODD) evaluates the inter-day variability; the average difference between glucose values is calculated over multiple days at the same time. |
| SD | Standard deviation (SD) determines the deviation of values in a group from the mean value of the same group of values. |
| TIR | Time In Range (TIR) quantifies the percentage of time spent within the target sensor glucose range (between 70 mg/dL and 180 mg/dL). |
| TAR | Time Above Range (TAR) quantifies the percentage of time spent above (>180 mg/dL) the target sensor glucose range. |
| TBR | Time Below Range (TBR) quantifies the percentage of time spent below (<70 mg/dL) the target sensor glucose range. |
| ID | Age | Daily Insulin (units) | Daily Basal Insulin (units) | Height (cm) | Weight (lb) | Gender | Country | AID Technology |
|---|---|---|---|---|---|---|---|---|
| AID1 | 51–60 | 52 | 16.7 | 195.07 | 198 | Male | Netherlands | OpenAPS |
| AID2 | 11–20 | 66.6 | 21 | 155.45 | 158 | Male | Canada | OpenAPS |
| AID3 | 21–30 | 30.97 | 11.09 | 170.69 | 132 | Female | Hungary | OpenAPS |
| AID4 | 11–20 | 36.61 | 9.14 | 155.45 | 105 | Female | USA | OpenAPS |
| AID5 | 11–20 | 78.63 | 16.75 | 155.45 | 131 | Male | USA | OpenAPS |
| AID6 | 31–40 | 86.98 | 38.72 | 173.74 | 277 | Female | Canada | OpenAPS |
| AID7 | 41–50 | 45 | 26 | 182.88 | 196 | Male | UK | AndroidAPS |
| AID8 | 31–40 | 35 | 12 | 164.59 | 132 | Female | Australia | AndroidAPS |
| AID9 | 31–40 | 32 | 15 | 167.64 | 160 | Trans Male | USA | OpenAPS |
| AID10 | 51–60 | 44 | 18 | 179.83 | 191 | Male | Norway | Loop |
| AID11 | 11–20 | 35 | 11 | 167.64 | 111 | Male | Australia | Loop |
| AID12 | 41–50 | 25 | 14.65 | 188.98 | 180 | Male | Sweden | AndroidAPS |
| AID13 | 11–20 | 36.31 | 15.36 | 146.3 | 49 | Female | Australia | AndroidAPS |
| AID14 | 31–40 | 30 | 10 | 167.64 | 130 | Female | USA | OpenAPS |
| AID15 | 11–20 | 35 | 18 | 173.74 | 160 | Female | UK | OpenAPS |
| AID16 | 21–30 | 36.85 | 23.9 | 179.83 | 150 | Female | Italy | AndroidAPS |
| AID17 | 51–60 | 40 | 19.5 | 173.74 | 224 | Female | Germany | OpenAPS |
| AID18 | 21–30 | 90 | 49 | 155.75 | 264 | Male | Australia | Loop |
| AID19 | 41–50 | 43.21 | 26.65 | 188.98 | 180 | Male | Germany | AndroidAPS |
| Model | Description | Category | Parameters | Python Library | Optimizing Function |
|---|---|---|---|---|---|
| ARIMA | A modelling technique for estimating or foreseeing future results in light of previous time series data. Since constant variance and normal distribution are observed between actual and predicted glucose data, fine-tuned hyperparameters have been reported. | Auto Regressor | P = 7, Q = 0, D = 1, Lags = 7 | Statsmodels | ACF, PACF, Stationarity |
| XGBoost | An additive model is generated by this estimator in a forward fashion which incorporates multiple stages. Further, it adds optimization for differential loss functions. In each stage, a tree is on a negative gradient for a provided loss function. | Regressor | learning rate = 0.1, estimators = 100, sub-sample = 1, max depth = 3 | Scikit Learn | Squared Error |
| Random Forest | A meta assessor that fits various characterizing decision trees on different sub-samples of the dataset and utilizes averaging to work on the exactness and avoid over-fitting. | Regressor | max depth = none, estimators = 100, min sample split = 2 | Scikit Learn | Squared Error |
| LSTM | The models use a progression of ’gates’ to control and manage the data in a string of information as input and output to the framework. There are three gates in a usual LSTM; forget gate, input gate and output gate. These gates can be considered as channels each having its own cognitive framework. | Deep Learning | lags = 1, epochs = 15, batch size = 1, neurons = 50 | Keras | Mean Squared Error |
| SVR | The model implementation is based on libsvm library with high training time complexity, i.e., proportionally more than quadratic with the number of samples. The implementation becomes challenging with large datasets. | Regressor | Kernel = RBF, Gamma = Scale, Epsilon = 0.1, C(regularization param) = 1 | Scikit Learn | Epsilon Value |
| ID | Count | Mean | SD | Q1 | Q2 | Q3 | CV | Skewness | Distribution |
|---|---|---|---|---|---|---|---|---|---|
| AID1 | 312,212 | 147.94 | 60.27 | 101 | 139 | 185 | 40.74 | 0.37 | Symmetrical |
| AID2 | 357,587 | 144.51 | 47.51 | 110 | 135 | 171 | 32.88 | 0.93 | Moderately Skewed |
| AID3 | 486,197 | 133.09 | 50.68 | 96 | 123 | 160 | 38.08 | 1.08 | Highly Skewed |
| AID4 | 282,441 | 140.11 | 48.22 | 102 | 131 | 169 | 34.41 | 0.89 | Moderately Skewed |
| AID5 | 242,279 | 145.53 | 56 | 106 | 135 | 174 | 38.48 | −0.12 | Symmetrical |
| AID6 | 276,622 | 140.05 | 58.85 | 99 | 126 | 167 | 42.02 | 0.89 | Moderately Skewed |
| AID7 | 280,822 | 127.89 | 43.35 | 97 | 120 | 151 | 33.9 | 1.04 | Highly Skewed |
| AID8 | 206,778 | 158.42 | 57.6 | 115 | 153 | 195 | 36.36 | 0.47 | Symmetrical |
| AID9 | 201,712 | 116.22 | 51.57 | 82 | 104 | 137 | 44.37 | 1.46 | Highly Skewed |
| AID10 | 168,848 | 147.35 | 55.88 | 107 | 135 | 177 | 37.92 | 1.08 | Highly Skewed |
| AID11 | 163,267 | 117.18 | 30.68 | 95 | 112 | 134 | 26.18 | 0.30 | Symmetrical |
| AID12 | 145,692 | 147.67 | 53.23 | 108 | 138 | 178 | 36.04 | 0.90 | Moderately Skewed |
| AID13 | 122,557 | 148.45 | 55.32 | 107 | 138 | 178 | 37.27 | 0.99 | Moderately Skewed |
| AID14 | 102,673 | 152.71 | 56.36 | 112 | 138 | 184 | 36.91 | 0.73 | Moderately Skewed |
| AID15 | 104,669 | 138.08 | 40.84 | 109 | 130 | 160 | 29.58 | 1.03 | Highly Skewed |
| AID16 | 96,270 | 143.14 | 59.29 | 101 | 131 | 175 | 41.42 | 0.98 | Moderately Skewed |
| AID17 | 77,946 | 134.24 | 46.37 | 100 | 126 | 162 | 34.54 | 0.13 | Symmetrical |
| AID18 | 78,798 | 98.42 | 33.3 | 76 | 91 | 114 | 33.84 | 1.43 | Highly Skewed |
| AID19 | 27,786 | 132.75 | 47.66 | 97 | 126 | 160 | 35.9 | 0.45 | Symmetrical |
| ID | SD ROC | TBR (%) | TIR (%) | TAR (%) | LBGI | HBGI | GMI | J_Index |
|---|---|---|---|---|---|---|---|---|
| AID1 | 1.45 | 6.24 | 67.13 | 26.63 | 1.48 | 5.85 | 6.85 | 43.35 |
| AID2 | 1.07 | 1.7 | 77.77 | 20.52 | 0.54 | 4.42 | 6.77 | 36.87 |
| AID3 | 1.68 | 5.78 | 78.05 | 16.17 | 1.42 | 3.64 | 6.49 | 33.77 |
| AID4 | 1.15 | 1.42 | 79.1 | 19.48 | 0.68 | 4.12 | 6.66 | 35.47 |
| AID5 | 1.53 | 4.14 | 74.33 | 21.54 | 0.99 | 5.1 | 6.79 | 40.61 |
| AID6 | 1.74 | 4.68 | 76.29 | 19.03 | 1.2 | 4.77 | 6.66 | 39.56 |
| AID7 | 1.35 | 4.56 | 83.76 | 11.68 | 1.25 | 2.71 | 6.37 | 29.32 |
| AID8 | 1.73 | 3.97 | 63.6 | 32.43 | 0.93 | 6.84 | 7.1 | 46.66 |
| AID9 | 1.72 | 14.51 | 75.06 | 10.43 | 3.26 | 2.56 | 6.09 | 28.15 |
| AID10 | 1.6 | 2.42 | 74.36 | 23.23 | 0.73 | 5.31 | 6.83 | 41.3 |
| AID11 | 0.89 | 2.33 | 93.9 | 3.77 | 0.98 | 1.23 | 6.11 | 21.86 |
| AID12 | 1.58 | 2.6 | 73.55 | 23.86 | 0.74 | 5.19 | 6.84 | 40.36 |
| AID13 | 0.79 | 2.37 | 74 | 23.63 | 0.73 | 5.41 | 6.86 | 41.52 |
| AID14 | 2.05 | 0.78 | 73.45 | 25.76 | 0.43 | 5.9 | 6.96 | 43.71 |
| AID15 | 1.7 | 0.83 | 84.51 | 14.66 | 0.41 | 3.32 | 6.61 | 32.01 |
| AID16 | 1.94 | 6.13 | 71.66 | 22.21 | 1.48 | 5.17 | 6.73 | 40.98 |
| AID17 | 1.52 | 4.3 | 79.7 | 16 | 1.14 | 3.45 | 6.52 | 32.62 |
| AID18 | 0.95 | 16.97 | 80.42 | 2.6 | 3.82 | 0.74 | 5.66 | 17.35 |
| AID19 | 1.59 | 5.21 | 79.55 | 15.24 | 1.34 | 3.4 | 6.49 | 32.55 |
| Relative Error (%) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARIMA30 | XGBoost30 | LSTM30 | RF30 | |||||||||
| Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | |
| CONGA | 0.12 | 0.33 | 0.85 | 0.02 | 0.45 | 1.21 | 0.01 | 2.66 | 44.29 | 0.06 | 0.5 | 1.18 |
| LI | 1.86 | 4.53 | 10.54 | 0.24 | 3.48 | 11.2 | 0.06 | 15.57 | 207.66 | 0.02 | 3.8 | 15.63 |
| JINDEX | 0.02 | 0.37 | 0.86 | 0.03 | 1.01 | 3.27 | 0.03 | 9.16 | 163.53 | 0.06 | 0.75 | 2.78 |
| LBGI | 0.42 | 2.85 | 6.28 | 0.17 | 9.84 | 31.76 | 0.23 | 5.91 | 40.19 | 0.38 | 6.07 | 29.45 |
| HBGI | 0.02 | 1.38 | 4.52 | 0.15 | 3.74 | 14 | 0.45 | 35.39 | 626.52 | 0.07 | 2.87 | 13.03 |
| GRADE | 0.44 | 2.77 | 9.78 | 0.59 | 4.96 | 18.24 | 0.08 | 20.95 | 327.02 | 0.14 | 4.32 | 15.47 |
| MODD | 0.03 | 0.68 | 1.84 | 0.46 | 2.18 | 4.8 | 0.04 | 10.59 | 184.19 | 0.17 | 1.46 | 4.36 |
| MAGE | 0.01 | 6.9 | 19.88 | 1.05 | 10.51 | 36.59 | 0.16 | 9.29 | 43.58 | 0 | 18.2 | 182.65 |
| ADDR | 0.19 | 3.63 | 15.73 | 0.78 | 4.51 | 39.98 | 0.26 | 47.47 | 753.86 | 0.54 | 5.69 | 35.9 |
| MVALUE | 0.44 | 3.31 | 8.65 | 1.67 | 12.18 | 64.69 | 0.24 | 8.33 | 77.16 | 0.13 | 7.97 | 58.22 |
| MAG | 6.85 | 13.68 | 25.51 | 0.2 | 5.29 | 10.2 | 12.54 | 37.11 | 110.75 | 12.62 | 26.48 | 46 |
| SD | 0.14 | 0.82 | 1.89 | 0.53 | 2.4 | 5.77 | 0.28 | 0.91 | 2.82 | 0.07 | 1.49 | 4.75 |
| MEAN | 0 | 0.03 | 0.11 | 0 | 0.34 | 1.06 | 0.02 | 0.25 | 0.77 | 0 | 0.27 | 0.78 |
| CV | 0.13 | 0.81 | 1.89 | 0.53 | 2.55 | 5.85 | 0.04 | 0.91 | 2.69 | 0.08 | 1.59 | 4.62 |
| TIR | 0 | 0.33 | 0.91 | 0 | 1.04 | 4.56 | 0 | 0.51 | 2.19 | 0 | 1.15 | 4.12 |
| TAR | 0 | 2.6 | 14.29 | 0 | 1.24 | 4.44 | 0 | 3.68 | 13.63 | 0 | 4.03 | 13.04 |
| TBR | 0 | 11.78 | 54.55 | 0 | 7.7 | 40 | 0 | 15.76 | 150 | 0 | 11.34 | 71.43 |
| Proportional Error | ||||||||||||
| ARIMA30 | XGBoost30 | LSTM30 | RF30 | |||||||||
| Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | Min | Avg | Max | |
| CONGA | 1 | 1 | 1.01 | 1 | 1 | 1.01 | 1 | 1.03 | 1.44 | 1 | 1.01 | 1.01 |
| LI | 1.02 | 1.05 | 1.11 | 1 | 1.04 | 1.11 | 1 | 1.16 | 3.08 | 1 | 1.04 | 1.19 |
| JINDEX | 1 | 1 | 1.01 | 1 | 1.01 | 1.03 | 1 | 1.09 | 2.64 | 1 | 1.01 | 1.03 |
| LBGI | 1 | 1.03 | 1.06 | 1 | 1.1 | 1.32 | 1 | 1.07 | 1.67 | 1 | 1.06 | 1.29 |
| HBGI | 1 | 1.01 | 1.05 | 1 | 1.04 | 1.14 | 1 | 1.35 | 7.27 | 1 | 1.03 | 1.13 |
| GRADE | 1 | 1.03 | 1.1 | 1.01 | 1.05 | 1.18 | 1 | 1.21 | 4.27 | 1 | 1.04 | 1.15 |
| MODD | 1 | 1.01 | 1.02 | 1 | 1.02 | 1.05 | 1 | 1.11 | 2.84 | 1 | 1.01 | 1.04 |
| MAGE | 1 | 1.07 | 1.25 | 1 | 1.1 | 1.37 | 1 | 1.09 | 1.44 | 1 | 1.17 | 2.83 |
| ADDR | 1 | 1.04 | 1.16 | 1.01 | 1.05 | 1.4 | 1 | 1.52 | 8.54 | 1.01 | 1.06 | 1.36 |
| MVALUE | 1 | 1.03 | 1.09 | 1.02 | 1.12 | 1.65 | 1 | 1.09 | 1.77 | 1 | 1.08 | 1.58 |
| MAG | 1.07 | 1.14 | 1.26 | 1 | 1.06 | 1.11 | 1.14 | 1.63 | 2.57 | 1.14 | 1.38 | 1.85 |
| SD | 1 | 1.01 | 1.02 | 1.01 | 1.02 | 1.06 | 1 | 1 | 1.02 | 1 | 1.01 | 1.05 |
| MEAN | 1 | 1 | 1 | 1 | 1 | 1.01 | 1 | 1 | 1 | 1 | 1 | 1.01 |
| CV | 1 | 1.01 | 1.02 | 1.01 | 1.03 | 1.06 | 1 | 1 | 1.02 | 1 | 1.02 | 1.05 |
| TIR | 1 | 1 | 1.01 | 1 | 1.01 | 1.05 | 1 | 1 | 1.02 | 1 | 1.01 | 1.04 |
| TAR | 1 | 1.03 | 1.14 | 1 | 1.01 | 1.05 | 1 | 1.03 | 1.15 | 1 | 1.04 | 1.15 |
| TBR | 1 | 1.12 | 1.55 | 1 | 1.63 | 11 | 1 | 1.16 | 2.5 | 1 | 1.22 | 3.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafar, A.; Lewis, D.M.; Shahid, A. Long-Term Glucose Forecasting for Open-Source Automated Insulin Delivery Systems: A Machine Learning Study with Real-World Variability Analysis. Healthcare 2023, 11, 779. https://doi.org/10.3390/healthcare11060779
Zafar A, Lewis DM, Shahid A. Long-Term Glucose Forecasting for Open-Source Automated Insulin Delivery Systems: A Machine Learning Study with Real-World Variability Analysis. Healthcare. 2023; 11(6):779. https://doi.org/10.3390/healthcare11060779
Chicago/Turabian StyleZafar, Ahtsham, Dana M. Lewis, and Arsalan Shahid. 2023. "Long-Term Glucose Forecasting for Open-Source Automated Insulin Delivery Systems: A Machine Learning Study with Real-World Variability Analysis" Healthcare 11, no. 6: 779. https://doi.org/10.3390/healthcare11060779
APA StyleZafar, A., Lewis, D. M., & Shahid, A. (2023). Long-Term Glucose Forecasting for Open-Source Automated Insulin Delivery Systems: A Machine Learning Study with Real-World Variability Analysis. Healthcare, 11(6), 779. https://doi.org/10.3390/healthcare11060779








