Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning
Abstract
:1. Introduction
2. Related Work
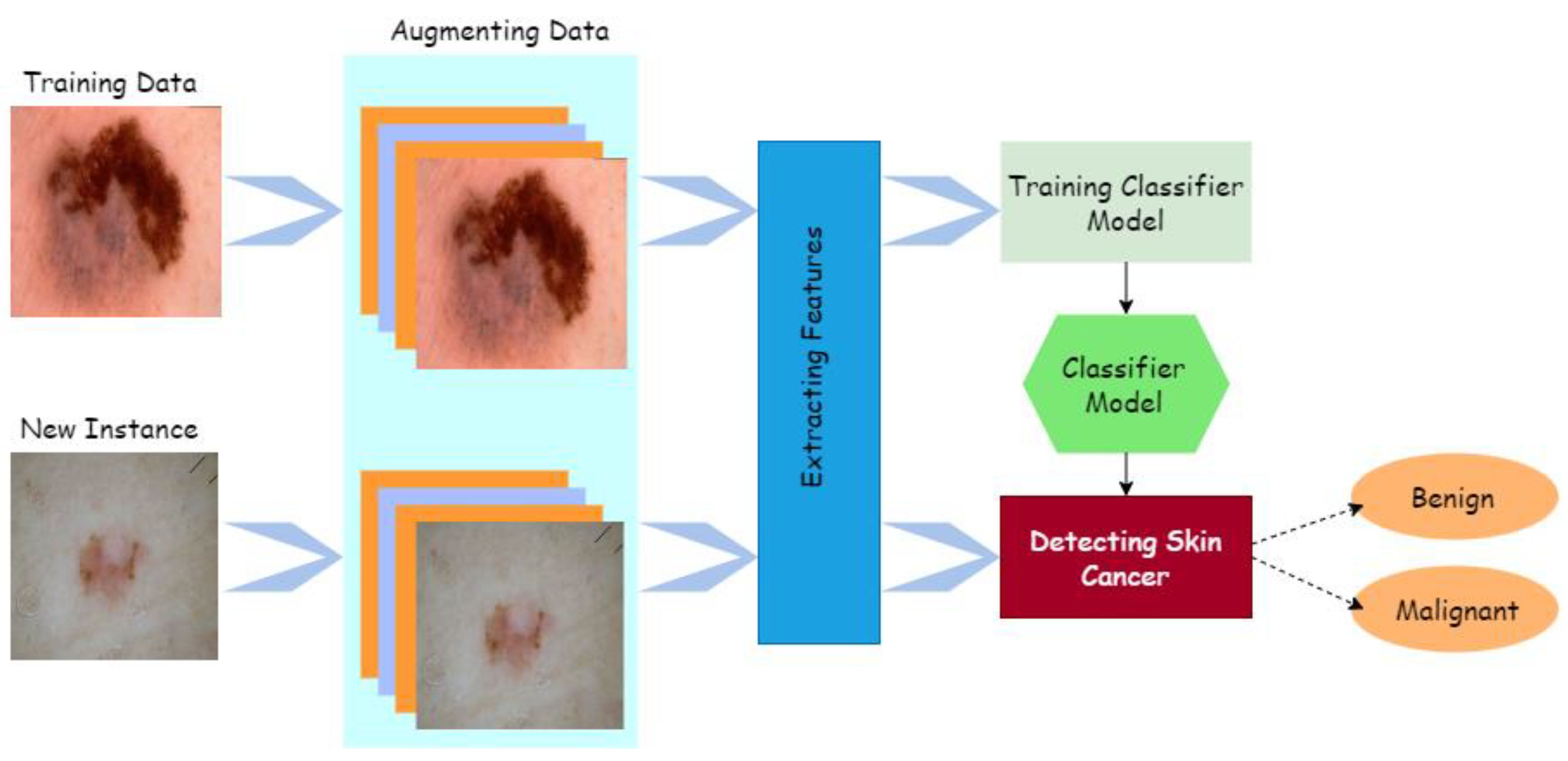
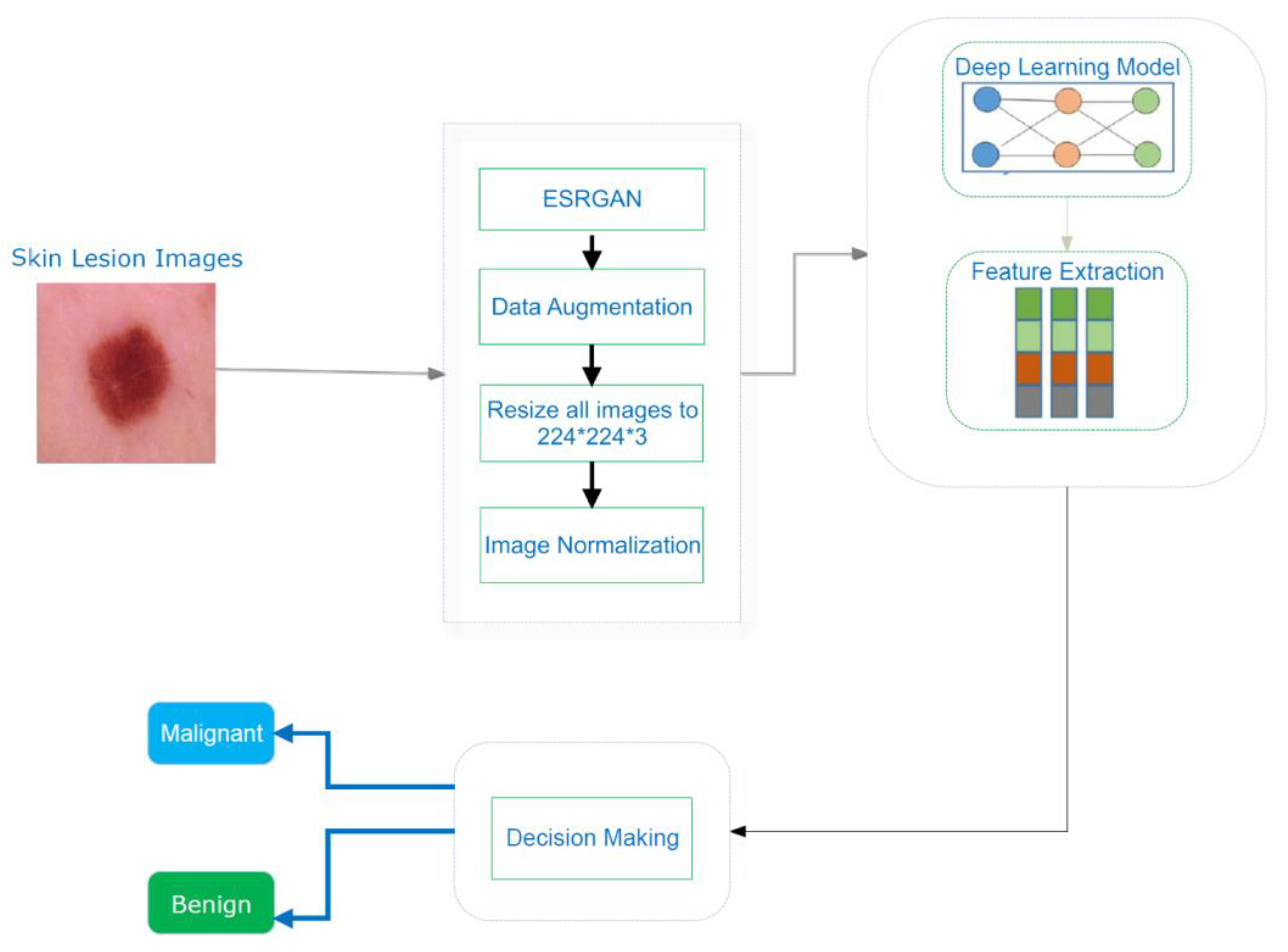
3. Proposed System
| Algorithm 1: Processes in the mechanism suggested |
| Let Ƀ = lesion image, aug = augmentation, ppr = preprocessing, ig = image, ig€ = image enhancement algorithm (ESRGAN), rt = rotation, sc = scaling, rl = reflection, and sh = shifting method Input: {Lesion image Ƀ} Output: {confusion matrix, accuracy, precision, ROC, F1, AUC, recall} Step 1: Browse(Ƀ) Step 2: Implement (ppr (ig)) 2.1. Operate (ig€)_ 2.2. aug(ig) w.r.t. rt, sc, rl, sh 2.2.1. perform rt 2.2.2. perform sc 2.2.3. perform rl 2.2.4. perform sh 2.3. Resize (ig)/224*24*3 2.4 Normalize pixelvalue (ig)/interval [0,1] Step 2: Split (dataset)/training, testing, and validating Step 3: Train CNN model Step 4: Train pretrained models (Resnet, Inception, Inception Resnet) 4.1 Fine-tune model parameters (freeze layers, learning rate, epochs, batch size) Step 5: Compute VPM (confusion matrix, accuracy, precision, ROC, F1, AUC, recall) Step 6: Evaluation (existing work) |
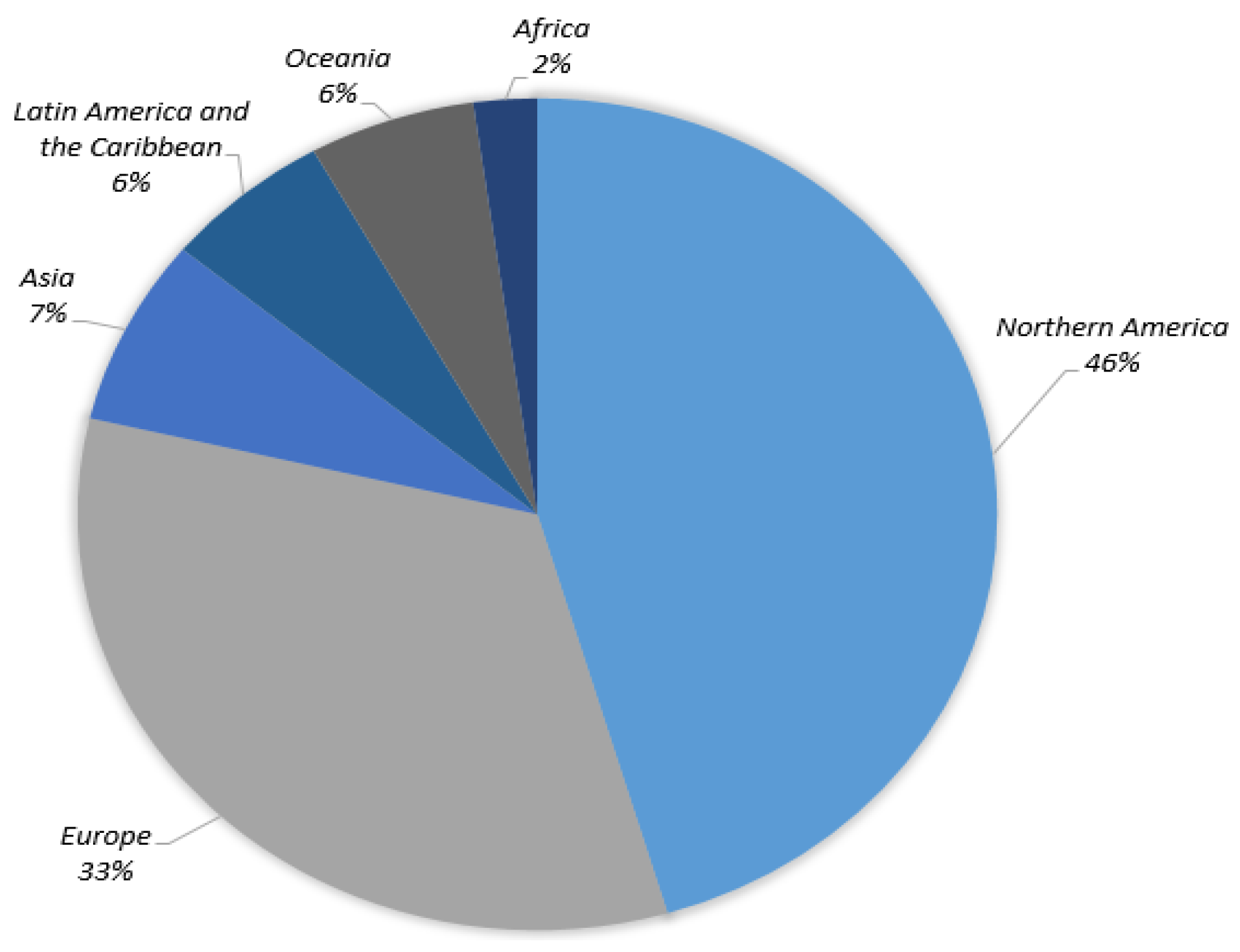
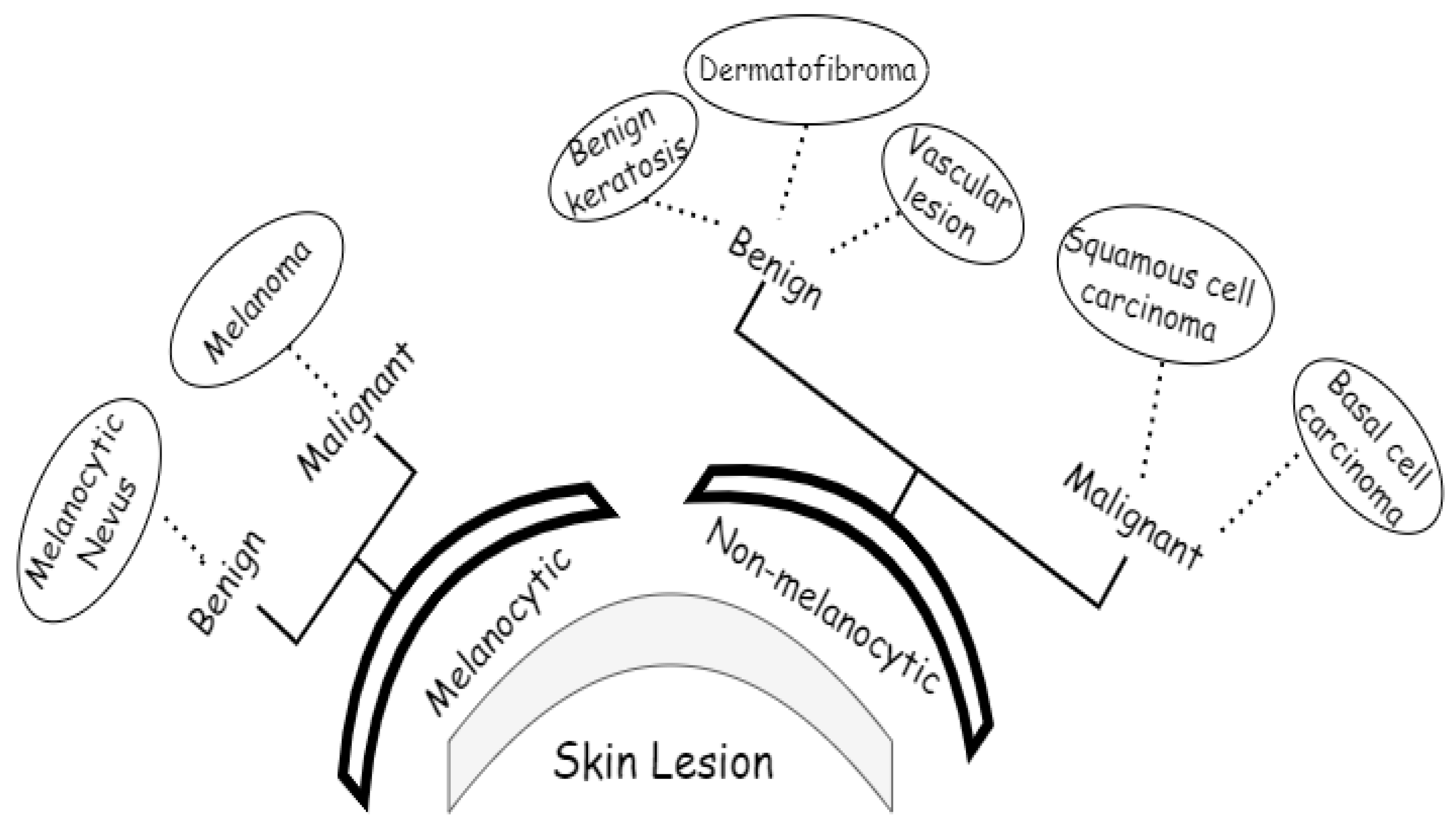
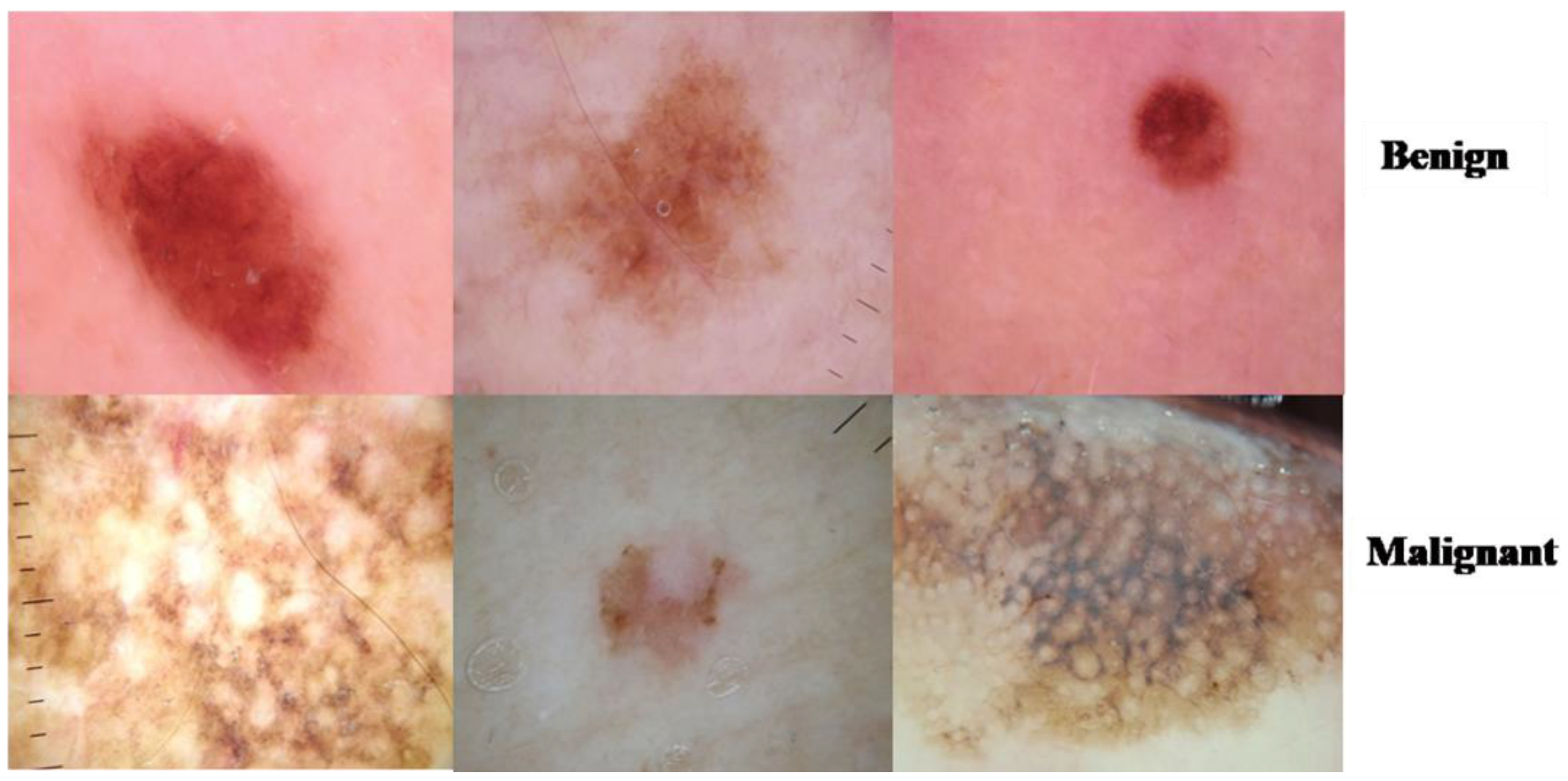
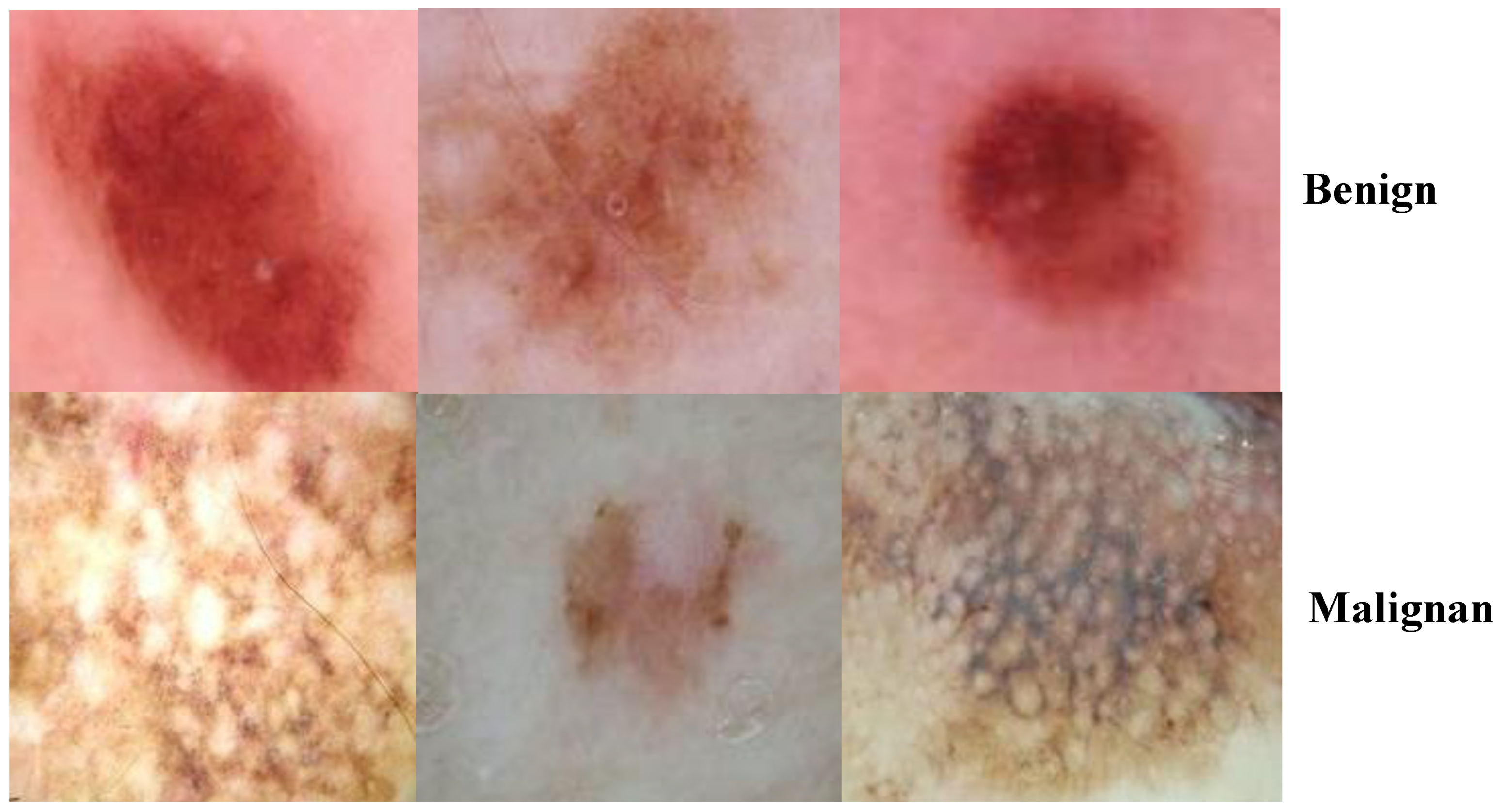
3.1. ISIC 2018 Image Dataset
3.2. Image Preprocessing
3.2.1. ESRGAN
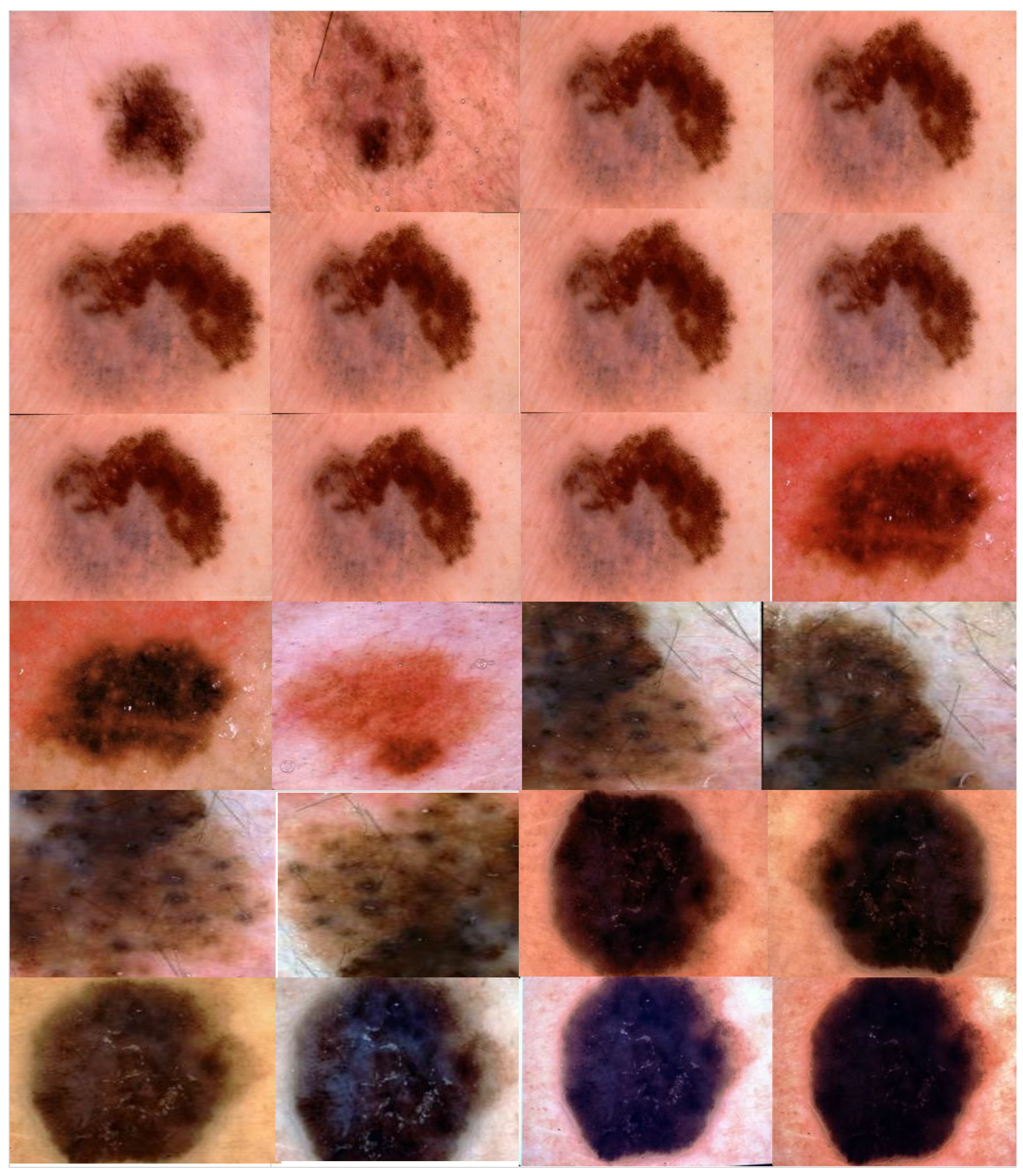
3.2.2. Augmentation
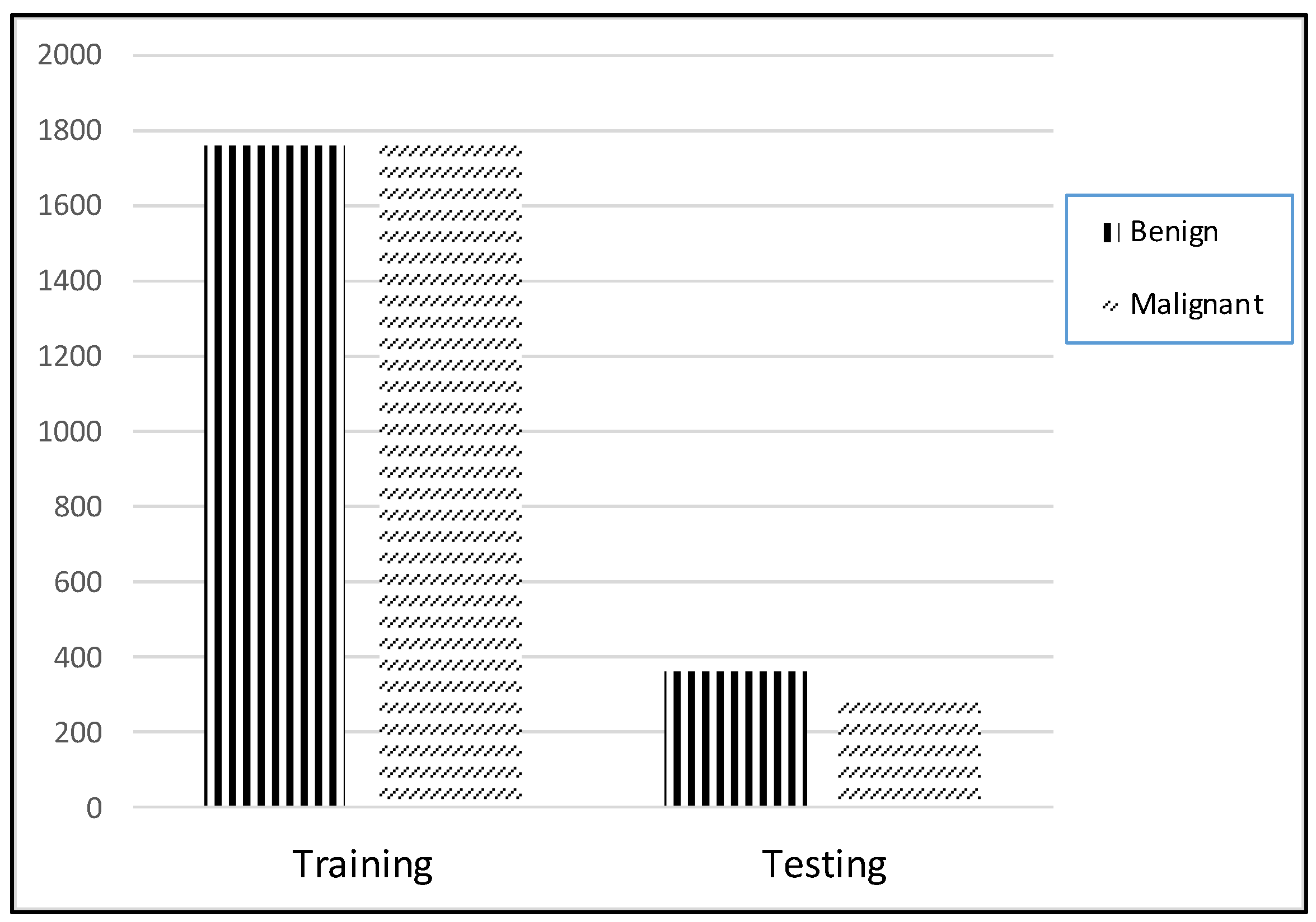
3.2.3. Data Preparation
3.3. Proposed CNN for ISIC2018 Detection
3.3.1. Resnet50
3.3.2. Inception V3
3.3.3. Inception Resnet
4. Experimental Results
4.1. Parameter Setting and Experimental Evaluation Index
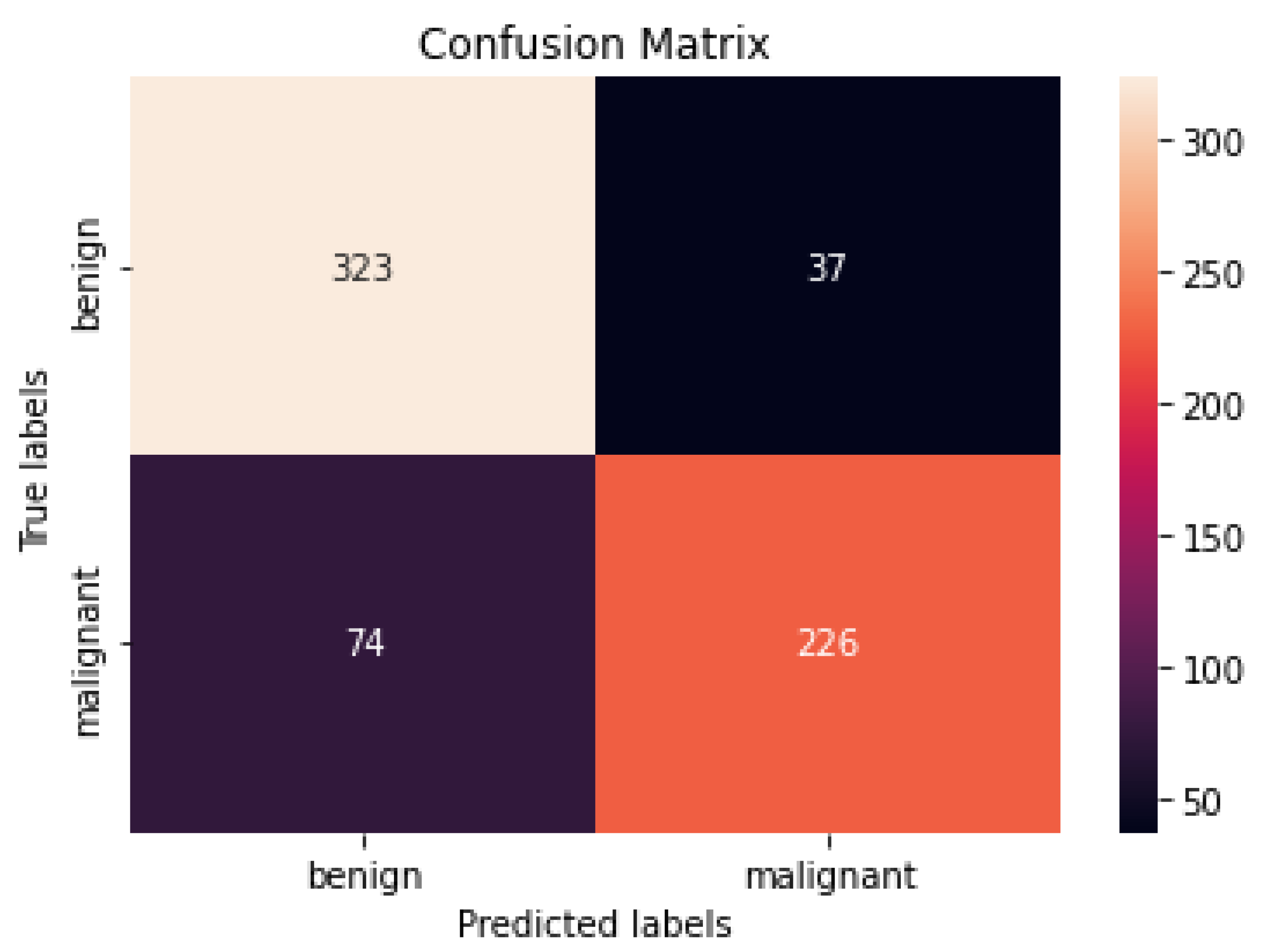
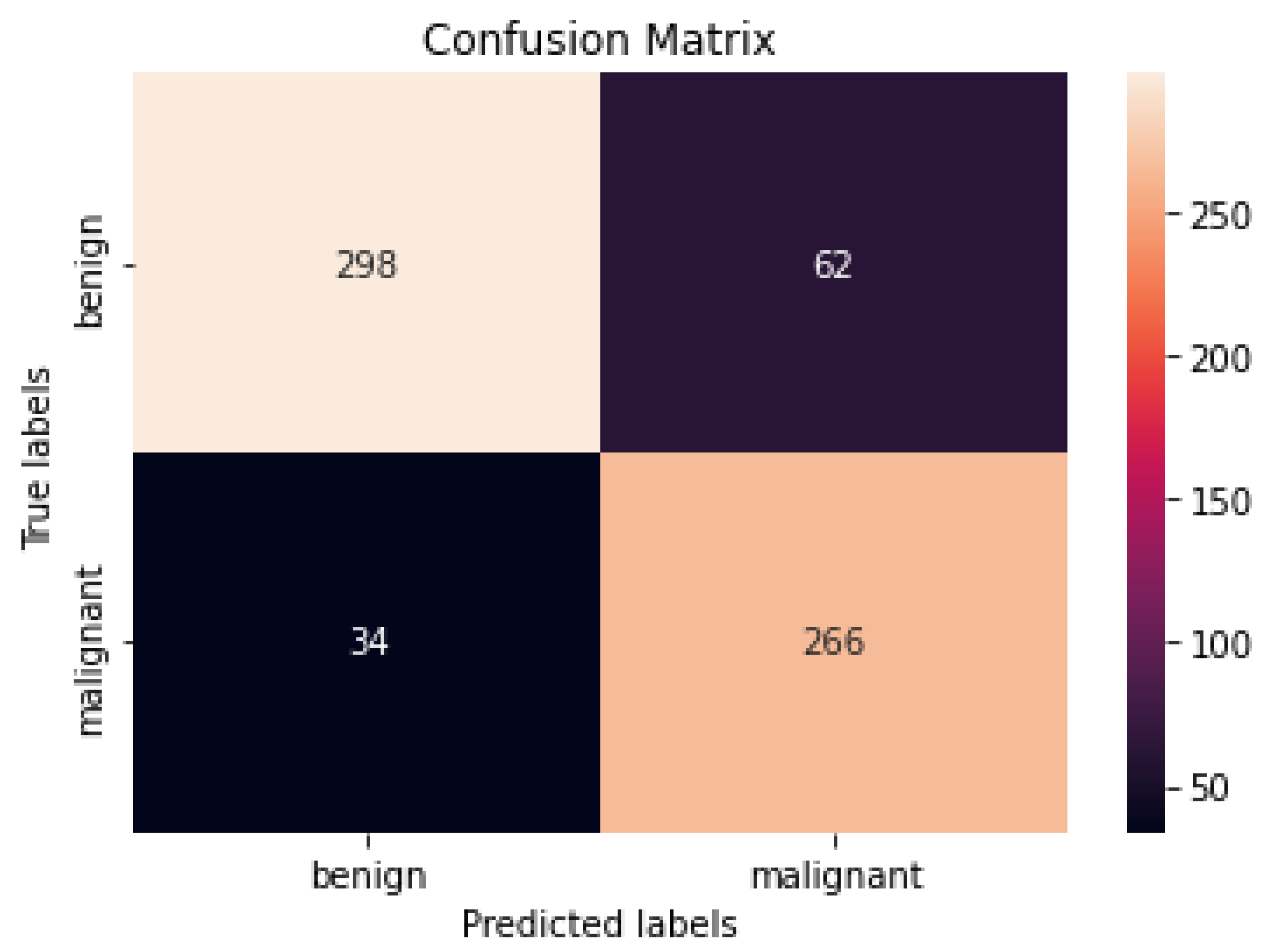
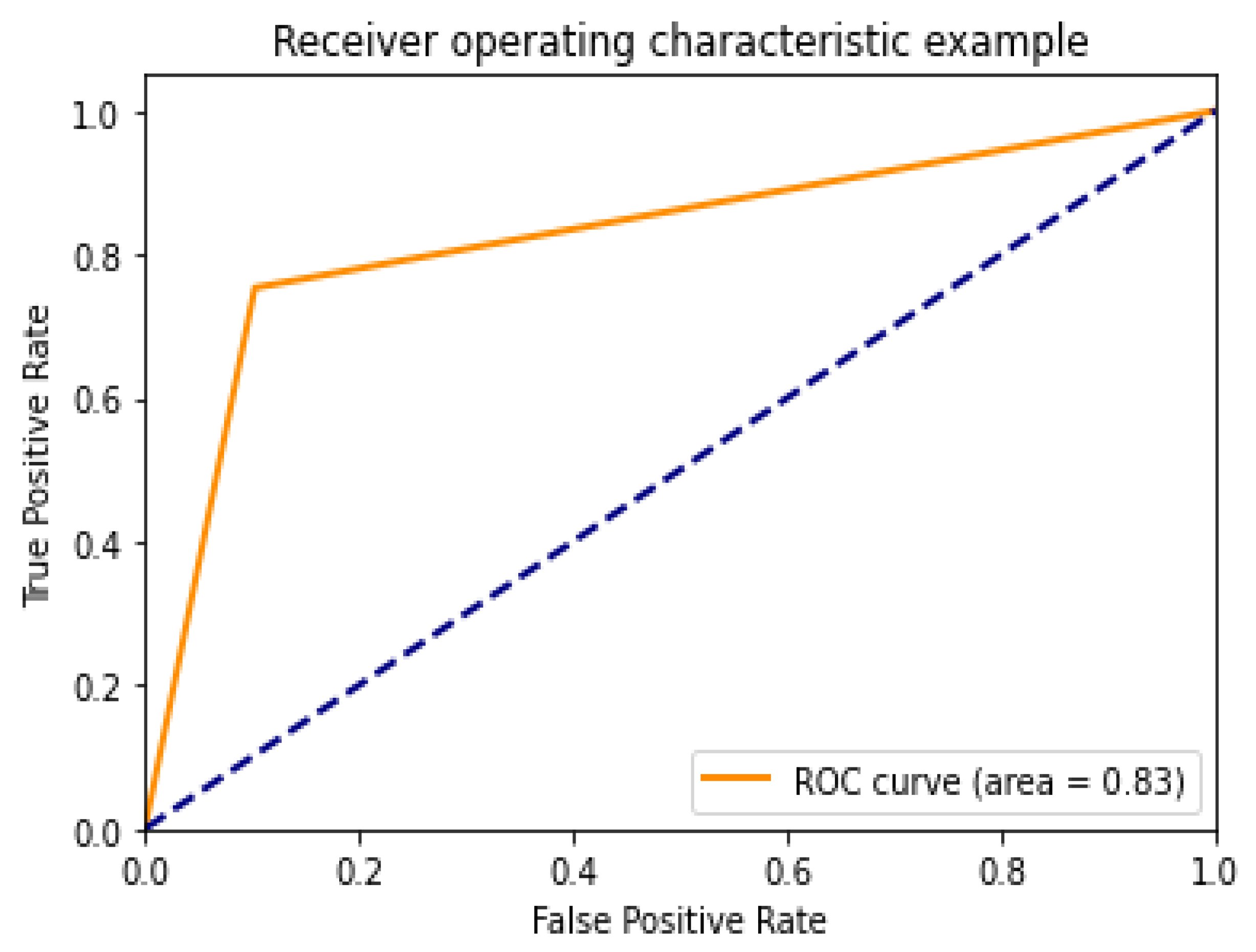
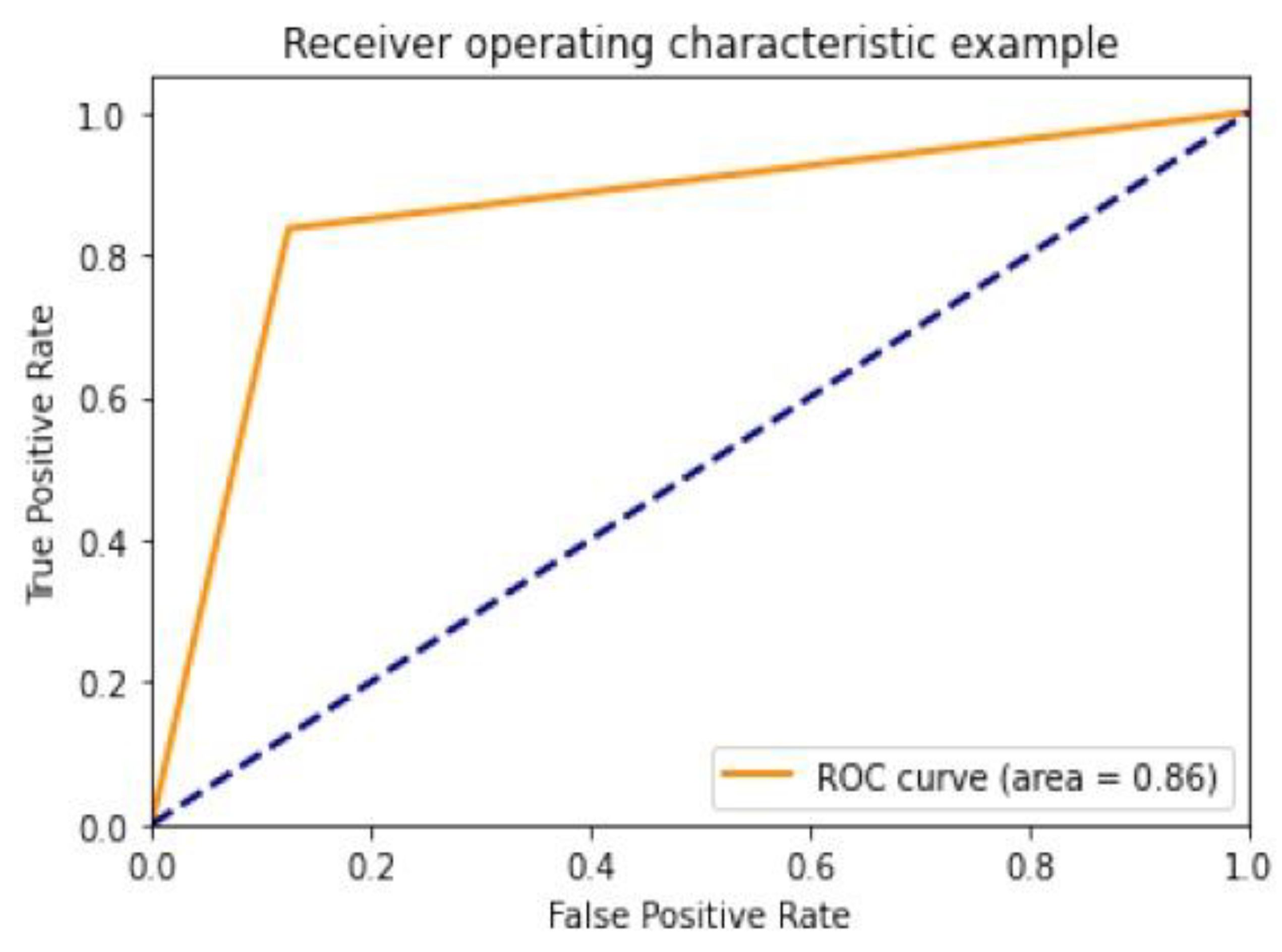
4.2. Performance Assessment
4.3. Performance of Different DCNN Models
4.4. Comparison with Other Methods
4.5. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Health Observatory; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Han, H.S.; Choi, K.Y. Advances in nanomaterial-mediated photothermal cancer therapies: Toward clinical applications. Biomedicines 2021, 9, 305. [Google Scholar] [CrossRef]
- Fuzzell, L.N.; Perkins, R.B.; Christy, S.M.; Lake, P.W.; Vadaparampil, S.T. Cervical cancer screening in the United States: Challenges and potential solutions for underscreened groups. Prev. Med. 2021, 144, 106400. [Google Scholar] [CrossRef] [PubMed]
- Ting, D.S.; Liu, Y.; Burlina, P.; Xu, X.; Bressler, N.M.; Wong, T.Y. AI for medical imaging goes deep. Nat. Med. 2018, 24, 539–540. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.; de Boer, A.; Sharma, K.; Boor, P.; Leiner, T.; Sunder-Plassmann, G.; Moser, E.; Caroli, A.; Jerome, N.P. Magnetic resonance imaging T1-and T2-mapping to assess renal structure and function: A systematic review and statement paper. Nephrol. Dial. Transplant. 2018, 33 (Suppl. S2), ii41–ii50. [Google Scholar] [CrossRef] [Green Version]
- Hooker, J.M.; Carson, R.E. Human positron emission tomography neuroimaging. Annu. Rev. Biomed. Eng. 2019, 21, 551–581. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.K.; Tiwari, P.; Kumar, S.; Gupta, D.; Khanna, A.; Rodrigues, J.J. Identifying pneumonia in chest X-rays: A deep learning approach. Measurement 2019, 145, 511–518. [Google Scholar] [CrossRef]
- Morawitz, J.; Dietzel, F.; Ullrich, T.; Hoffmann, O.; Mohrmann, S.; Breuckmann, K.; Herrmann, K.; Buchbender, C.; Antoch, G.; Umultu, L.; et al. Comparison of nodal staging between CT, MRI, and [18F]-FDG PET/MRI in patients with newly diagnosed breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 992–1001. [Google Scholar] [CrossRef] [PubMed]
- Jinzaki, M.; Yamada, Y.; Nagura, T.; Nakahara, T.; Yokoyama, Y.; Narita, K.; Ogihara, N.; Yamada, M. Development of upright computed tomography with area detector for whole-body scans: Phantom study, efficacy on workflow, effect of gravity on human body, and potential clinical impact. Investig. Radiol. 2020, 55, 73. [Google Scholar] [CrossRef]
- Celebi, M.E.; Codella, N.; Halpern, A. Dermoscopy image analysis: Overview and future directions. IEEE J. Biomed. Health Inform. 2019, 23, 474–478. [Google Scholar] [CrossRef]
- Barata, C.; Celebi, M.E.; Marques, J.S. A survey of feature extraction in dermoscopy image analysis of skin cancer. IEEE J. Biomed. Health Inform. 2018, 23, 1096–1109. [Google Scholar] [CrossRef]
- Adeyinka, A.A.; Viriri, S. Skin lesion images segmentation: A survey of the state-of-the-art. In Proceedings of the International Conference on Mining Intelligence and Knowledge Exploration, Cluj-Napoca, Romania, 22 December 2018; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Al-Masni, M.A.; Al-Antari, M.A.; Choi, M.-T.; Han, S.-M.; Kim, T.-S. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput. Methods Programs Biomed. 2018, 162, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Ünver, H.M.; Ayan, E. Skin lesion segmentation in dermoscopic images with combination of YOLO and grabcut algorithm. Diagnostics 2019, 9, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Tang, J.; Wang, Z.; Zhang, K.; Zhang, L.; Sun, Q. Deep learning for image-based cancer detection and Diagnosis—A survey. Pattern Recognit. 2018, 83, 134–149. [Google Scholar] [CrossRef]
- Fujisawa, Y.; Otomo, Y.; Ogata, Y.; Nakamura, Y.; Okiyama, N.; Ohara, K.; Fujimoto, M.; Fujita, R.; Ishitsuka, Y.; Watanabe, R. Deep-learning-based, computer-aided classifier developed with a small dataset of clinical images surpasses board-certified dermatologists in skin tumour diagnosis. Br. J. Dermatol. 2019, 180, 373–381. [Google Scholar] [CrossRef]
- Adegun, A.; Viriri, S. Deep learning techniques for skin lesion analysis and melanoma cancer detection: A survey of state-of-the-art. Artif. Intell. Rev. 2021, 54, 811–841. [Google Scholar] [CrossRef]
- Iqbal, S.; Siddiqui, G.F.; Rehman, A.; Hussain, L.; Saba, T.; Tariq, U.; Abbasi, A.A. Prostate cancer detection using deep learning and traditional techniques. IEEE Access 2021, 9, 27085–27100. [Google Scholar] [CrossRef]
- Dildar, M.; Akram, S.; Mahmood, A.R.; Mahnashi, M.H.; Alsaiari, S.A.; Irfan, M.; Khan, H.U.; Saeed, A.H.M.; Ramzan, M.; Alraddadi, M.O. Skin cancer detection: A review using deep learning techniques. Int. J. Environ. Res. Public Health 2021, 18, 5479. [Google Scholar] [CrossRef]
- Vaishnavi, K.; Ramadas, M.A.; Chanalya, N.; Manoj, A.; Nair, J.J. Deep learning approaches for detection of COVID-19 using chest X-ray images. In Proceedings of the 2021 Fourth International Conference on Electrical, Computer and Communication Technologies (ICECCT), Piscataway, NJ, USA, 15–17 September 2021. [Google Scholar]
- Duc, N.T.; Lee, Y.-M.; Park, J.H.; Lee, B. An ensemble deep learning for automatic prediction of papillary thyroid carcinoma using fine needle aspiration cytology. Expert Syst. Appl. 2022, 188, 115927. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Damaševičius, R.; Eltoukhy, M.M. Machine learning and deep learning methods for skin lesion classification and Diagnosis: A systematic review. Diagnostics 2021, 11, 1390. [Google Scholar] [CrossRef]
- Abayomi-Alli, O.O.; Damasevicius, R.; Misra, S.; Maskeliunas, R.; Abayomi-Alli, A. Malignant skin melanoma detection using image augmentation by oversamplingin nonlinear lower-dimensional embedding manifold. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 2600–2614. [Google Scholar] [CrossRef]
- Kadry, S.; Taniar, D.; Damaševičius, R.; Rajinikanth, V.; Lawal, I.A. Extraction of abnormal skin lesion from dermoscopy image using VGG-SegNet. In Proceedings of the 2021 Seventh International Conference on Bio Signals, Images, and Instrumentation (ICBSII), Chennai, India, 25 March 2021. [Google Scholar]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Hauschild, A.; Weichenthal, M.; Maron, R.C.; Berking, C.; Haferkamp, S.; Klode, J.; Schadendorf, D.; et al. Superior skin cancer classification by the combination of human and artificial intelligence. Eur. J. Cancer 2019, 120, 114–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reis, H.C.; Turk, V.; Khoshelham, K.; Kaya, S. InSiNet: A deep convolutional approach to skin cancer detection and segmentation. Med. Biol. Eng. Comput. 2022, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.; Mehmood, Z.; Nazir, T.; Naqvi, R.A.; Rehman, A.; Iqbal, M.; Saba, T. Skin cancer detection from dermoscopic images using deep learning and fuzzy k-means clustering. Microsc. Res. Tech. 2022, 85, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Humayun, M.; Sujatha, R.; Almuayqil, S.N.; Jhanjhi, N.Z. A Transfer Learning Approach with a Convolutional Neural Network for the Classification of Lung Carcinoma. Healthcare 2022, 10, 1058. [Google Scholar] [CrossRef]
- Adegun, A.A.; Viriri, S. Deep learning-based system for automatic melanoma detection. IEEE Access 2019, 8, 7160–7172. [Google Scholar] [CrossRef]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018, a challenge hosted by the international skin imaging collaboration (isic). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Kinyanjui, N.M.; Odonga, T.; Cintas, C.; Codella, N.C.; Panda, R.; Sattigeri, P.; Varshney, K.R. Fairness of classifiers across skin tones in dermatology. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4 October 2020; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Ech-Cherif, A.; Misbhauddin, M.; Ech-Cherif, M. Deep neural network based mobile dermoscopy application for triaging skin cancer detection. In Proceedings of the 2019 2nd International Conference on Computer Applications & Information Security (ICCAIS), Riyadh, Saudi Arabia, 1 May 2019. [Google Scholar]
- Le, D.N.; Le, H.X.; Ngo, L.T.; Ngo, H.T. Transfer learning with class-weighted and focal loss function for automatic skin cancer classification. arXiv 2020, arXiv:2009.05977. [Google Scholar]
- Dorj, U.-O.; Lee, K.-K.; Choi, J.-Y.; Lee, M. The skin cancer classification using deep convolutional neural network. Multimed. Tools Appl. 2018, 77, 9909–9924. [Google Scholar] [CrossRef]
- Rahman, M.M.; Nasir, M.K.; Nur, A.; Khan, S.I.; Band, S.; Dehzangi, I.; Beheshti, A.; Rokny, H.A. Hybrid Feature Fusion and Machine Learning Approaches for Melanoma Skin Cancer Detection; UNSW: Sydney, Australia, 2022. [Google Scholar]
- Murugan, A.; Nair, S.A.H.; Preethi, A.A.P.; Kumar, K.S. Diagnosis of skin cancer using machine learning techniques. Microprocess. Microsyst. 2021, 81, 103727. [Google Scholar] [CrossRef]
- Ali, M.S.; Miah, M.S.; Haque, J.; Rahman, M.M.; Islam, M.K. An enhanced technique of skin cancer classification using deep convolutional neural network with transfer learning models. Mach. Learn. Appl. 2021, 5, 100036. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, Y.; Ping, B.; Li, D.; Du, J.; Qin, Y.; Lu, H.; Wan, X.; Xiang, J. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: A pilot study. J. Cancer 2019, 10, 4876. [Google Scholar] [CrossRef] [PubMed]
- Rajput, G.; Agrawal, S.; Raut, G.; Vishvakarma, S.K. An accurate and noninvasive skin cancer screening based on imaging technique. Int. J. Imaging Syst. Technol. 2022, 32, 354–368. [Google Scholar] [CrossRef]
- Kanani, P.; Padole, M. Deep learning to detect skin cancer using google colab. Int. J. Eng. Adv. Technol. Regul. Issue 2019, 8, 2176–2183. [Google Scholar] [CrossRef]
- MNOWAK061. Skin Lesion Dataset. ISIC2018 Kaggle Repository. 2021. Available online: https://www.kaggle.com/datasets/mnowak061/isic2018-and-ph2-384x384-jpg (accessed on 10 April 2022).
- Takano, N.; Alaghband, G. Srgan: Training dataset matters. arXiv 2019, arXiv:1903.09922. [Google Scholar]
- Ledig, C.; Theis, L.; Huszar, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21 July 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 30 June 2016. [Google Scholar]
- Wang, C.; Chen, D.; Hao, L.; Liu, X.; Zeng, Y.; Chen, J.; Zhang, G. Pulmonary image classification based on inception-v3 transfer learning model. IEEE Access 2019, 7, 146533–146541. [Google Scholar] [CrossRef]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, inception-ResNet and the impact of residual connections on learning. arXiv 2018, arXiv:1602.07261. [Google Scholar]
- Foahom Gouabou, A.C.; Damoiseaux, J.-L.; Monnier, J.; Iguernaissi, R.; Moudafi, A.; Merad, D. Ensemble Method of Convolutional Neural Networks with Directed Acyclic Graph Using Dermoscopic Images: Melanoma Detection Application. Sensors 2021, 21, 3999. [Google Scholar] [CrossRef]
- Lopez, A.R.; Giro-i-Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20 February 2017. [Google Scholar]
- Harangi, B. Skin lesion classification with ensembles of deep convolutional neural networks. J. Biomed. Inform. 2018, 86, 25–32. [Google Scholar] [CrossRef]
- Kim, C.-I.; Hwang, S.-M.; Park, E.-B.; Won, C.-H.; Lee, J.-H. Computer-Aided Diagnosis Algorithm for Classification of Malignant Melanoma Using Deep Neural Networks. Sensors 2021, 21, 5551. [Google Scholar] [CrossRef]
- Alnowami, M. Very Deep Convolutional Networks for Skin Lesion Classification. J. King Abdulaziz Univ. Eng. Sci. 2019, 30, 43–54. [Google Scholar]
- Ameri, A. A deep learning approach to skin cancer detection in dermoscopy images. J. Biomed. Phys. Eng. 2020, 10, 801. [Google Scholar] [CrossRef] [PubMed]
- Sae-Lim, W.; Wettayaprasit, W.; Aiyarak, P. Convolutional neural networks using MobileNet for skin lesion classification. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chonburi, Thailand, 10 July 2019. [Google Scholar]
| Recent Work | Data Size | Data Set | Techniques Used | Number of Classes |
|---|---|---|---|---|
| [25] | 300 | HAM10000 | CNN with XGBoost | Five |
| [26] | 1323 | HAM10000 | InSiNet | Two |
| [27] | 1280 | ISIC-2016 | Region-based CNN (RCNN) | Two |
| 2000 | ISIC-2017 | |||
| 200 | PH2 | |||
| [29] | 2000 | ISBI2017 | Deep convolutional encoder–decoder network (DCNN) | Two |
| [32] | 48,373 | DermNet, ISIC Archive, Dermofit image library | MobileNetV2 | Two |
| [33] | 7470 | HAM10000 | ResNet50 | Seven |
| [34] | 3753 | ImageNet | ECOC SVM | Two |
| [35] | 16,170 | HAM10000 | Anisotropic diffusion filtering | Two |
| [36] | 1000 | ISIC | SVM + RF | Eight |
| [37] | 6705 | HAM10000 | DCNN | Two |
| [38] | 279 | ImageNet | DCNN VGG-16 | Two |
| [39] | 10,015 | HAM10000 | AlexNet | Seven |
| [40] | 10,015 | HAM10000 | CNN | Seven |
| Parameter | Value |
|---|---|
| Batch size | 2–32 |
| Loss function | categorical cross-entropy |
| Momentum | 0.95 |
| Batch Size | Ensemble Using Several Runs | ||
|---|---|---|---|
| Run 1 | Run 2 | Run 3 | |
| 2 | 0.7818 | 0.7606 | 0.7011 |
| 4 | 0.7636 | 0.7833 | 0.7363 |
| 8 | 0.7363 | 0.75 | 0.7439 |
| 16 | 0.7939 | 0.7727 | 0.7636 |
| 32 | 0.7651 | 0.7363 | 0.7363 |
| Batch Size | Ensemble Using Several Runs | ||
|---|---|---|---|
| Run 1 | Run 2 | Run 3 | |
| 2 | 0.8212 | 0.8196 | 0.8136 |
| 4 | 0.8121 | 0.8227 | 0.7924 |
| 8 | 0.8227 | 0.8227 | 0.8167 |
| 16 | 0.8000 | 0.7651 | 0.7985 |
| 32 | 0.8045 | 0.8136 | 0.8152 |
| Batch Size | Ensemble Using Several Runs | ||
|---|---|---|---|
| Run 1 | Run 2 | Run 3 | |
| 2 | 0.8182 | 0.8000 | 0.8136 |
| 4 | 0.8318 | 0.8257 | 0.8121 |
| 8 | 0.8061 | 0.7909 | 0.8091 |
| 16 | 0.7879 | 0.7879 | 0.7985 |
| 32 | 0.7864 | 0.7969 | 0.7985 |
| CNN | Resnet50 | InceptionV3 | Inception Resnet |
|---|---|---|---|
| 0.8318 | 0.8364 | 0. 8576 | 0.8409 |
| Reference | Dataset | Model | Accuracy |
|---|---|---|---|
| [47] | ISIC2018 | VGG19_2 | 76.6% |
| [48] | ISIC2016 | VGGNet | 78.6% |
| [49] | ISBI2017 | AlexNet + VGGNet | 79.9% |
| [50] | ISIC2017 | U-Net | 80.0% |
| [51] | 2-ary, 3-ary, 9-ary | DenseNet | 82% |
| [52] | HAM10000 | AlexNet | 84% |
| [53] | HAM10000 | MobileNet | 83.9% |
| Proposed | ISIC2018 | CNN | 83.1% |
| Proposed | ISIC2018 | Resnet50 | 83.6% |
| Proposed | ISIC2018 | Resnet50-Inception | 84.1% |
| Proposed | ISIC2018 | Inception V3 | 85.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183. https://doi.org/10.3390/healthcare10071183
Gouda W, Sama NU, Al-Waakid G, Humayun M, Jhanjhi NZ. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare. 2022; 10(7):1183. https://doi.org/10.3390/healthcare10071183
Chicago/Turabian StyleGouda, Walaa, Najm Us Sama, Ghada Al-Waakid, Mamoona Humayun, and Noor Zaman Jhanjhi. 2022. "Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning" Healthcare 10, no. 7: 1183. https://doi.org/10.3390/healthcare10071183
APA StyleGouda, W., Sama, N. U., Al-Waakid, G., Humayun, M., & Jhanjhi, N. Z. (2022). Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare, 10(7), 1183. https://doi.org/10.3390/healthcare10071183







