Deep Learning Techniques for the Effective Prediction of Alzheimer’s Disease: A Comprehensive Review
Abstract
:1. Introduction
2. Transformation from ML to DL Approaches for the Effective Prediction of AD
- “Free Surfer” is an application for cerebral localization with cortex-associated information.
- The “SPM5 (Statistical Parametric Mapping Tool)” is a device for the mapping of statistical parameters.
3. Diagnosis and Prognosis of AD Using DL Methods
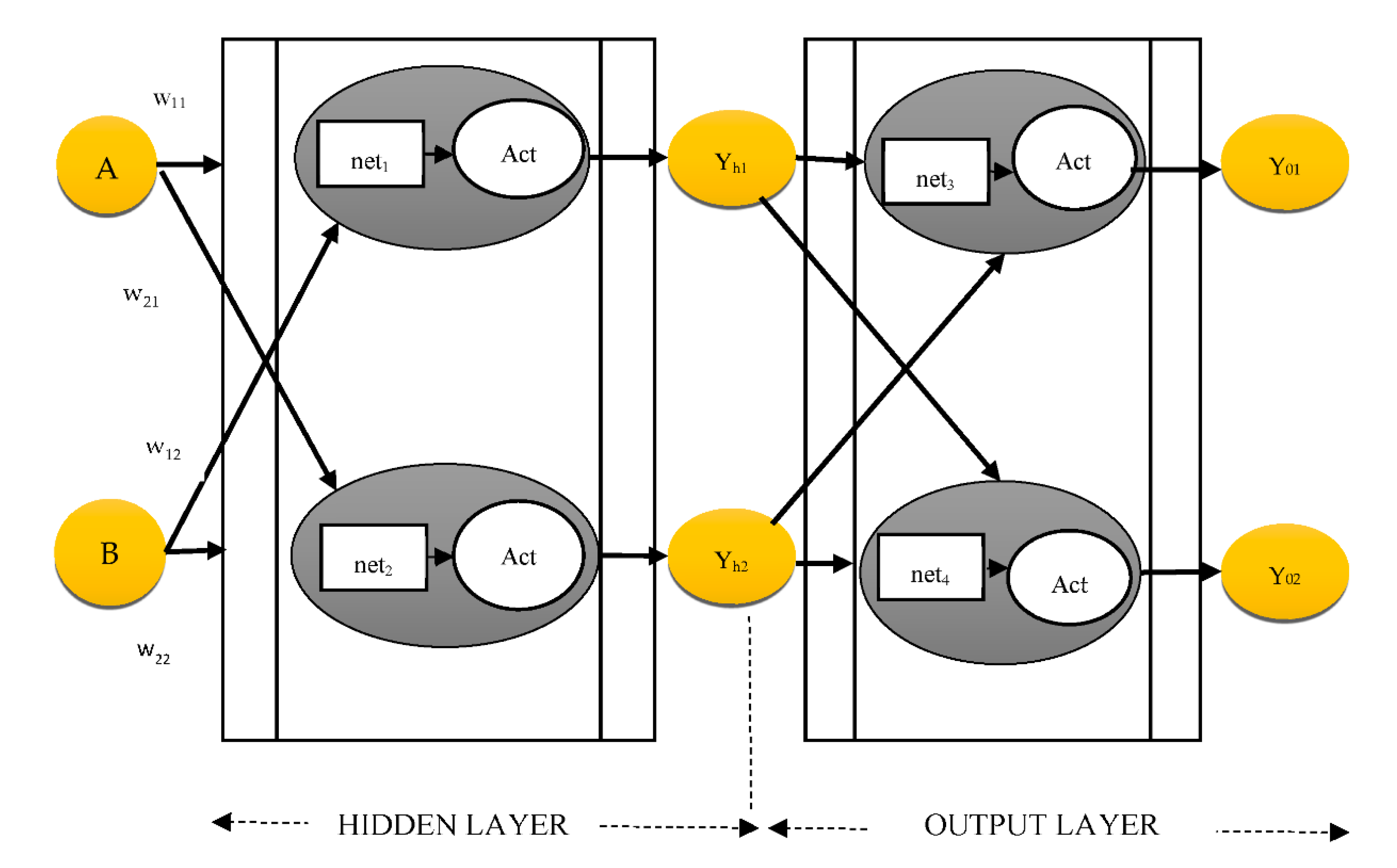
3.1. Gradient Computation
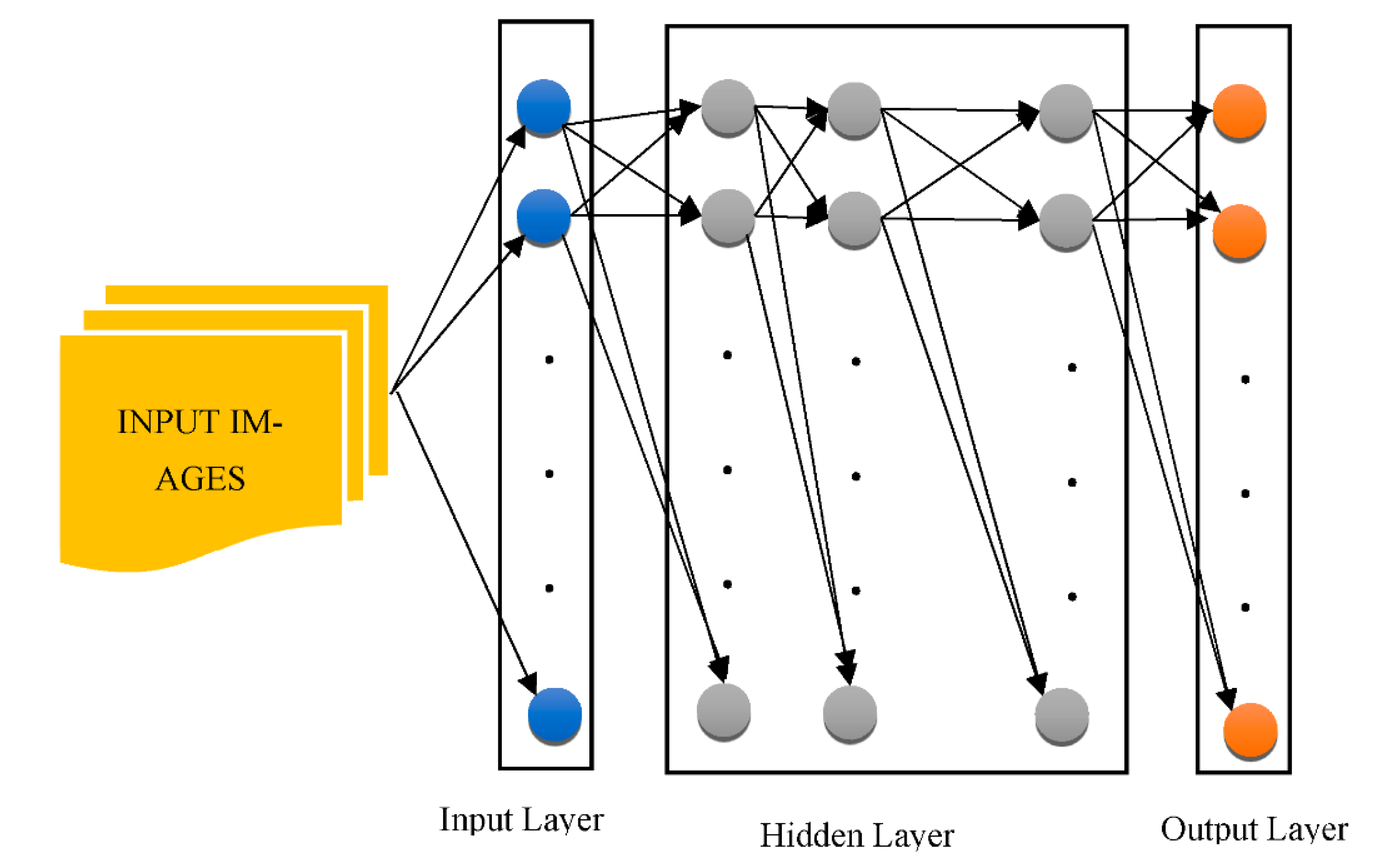
3.2. DNNs in the Real World
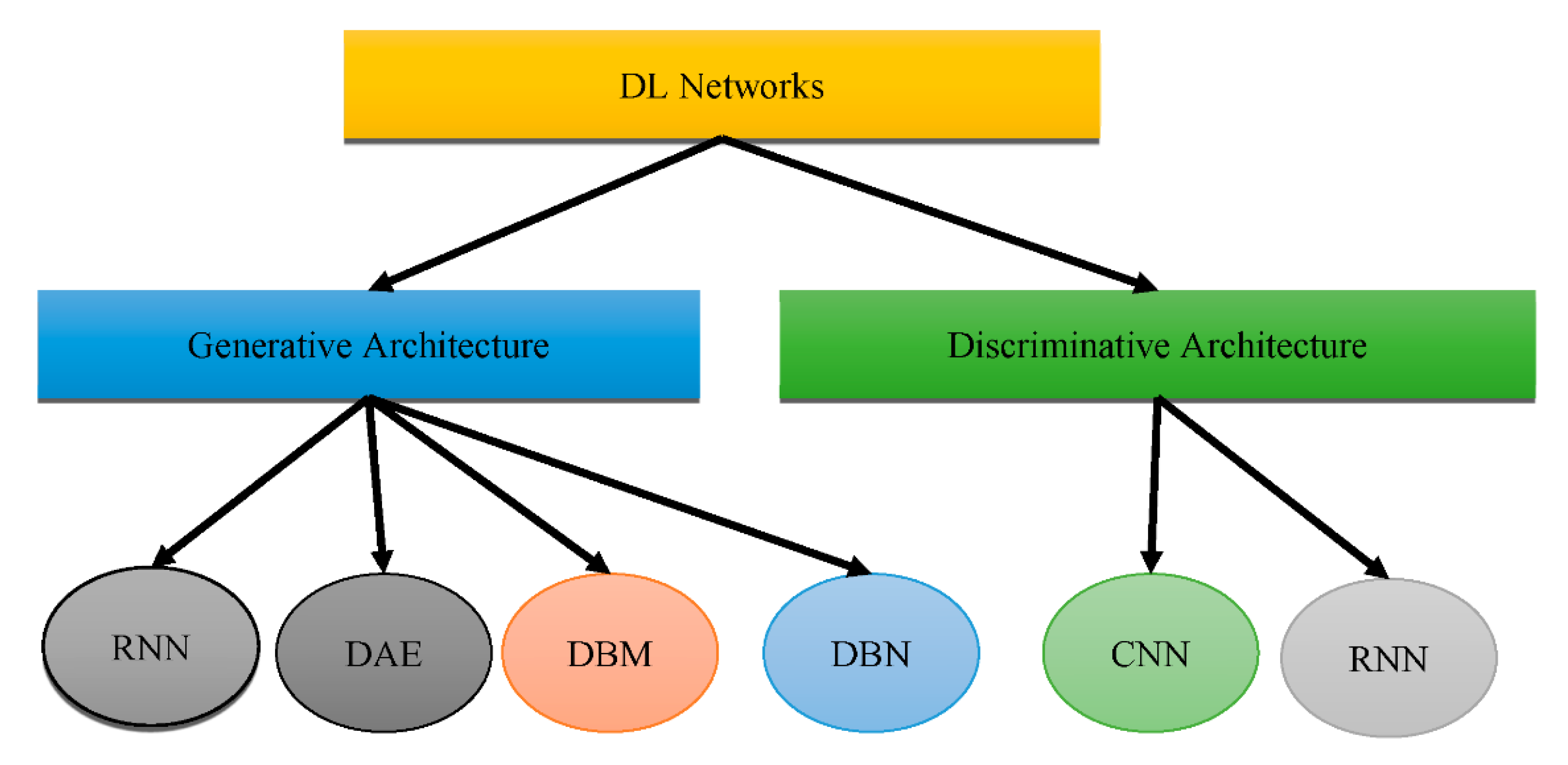
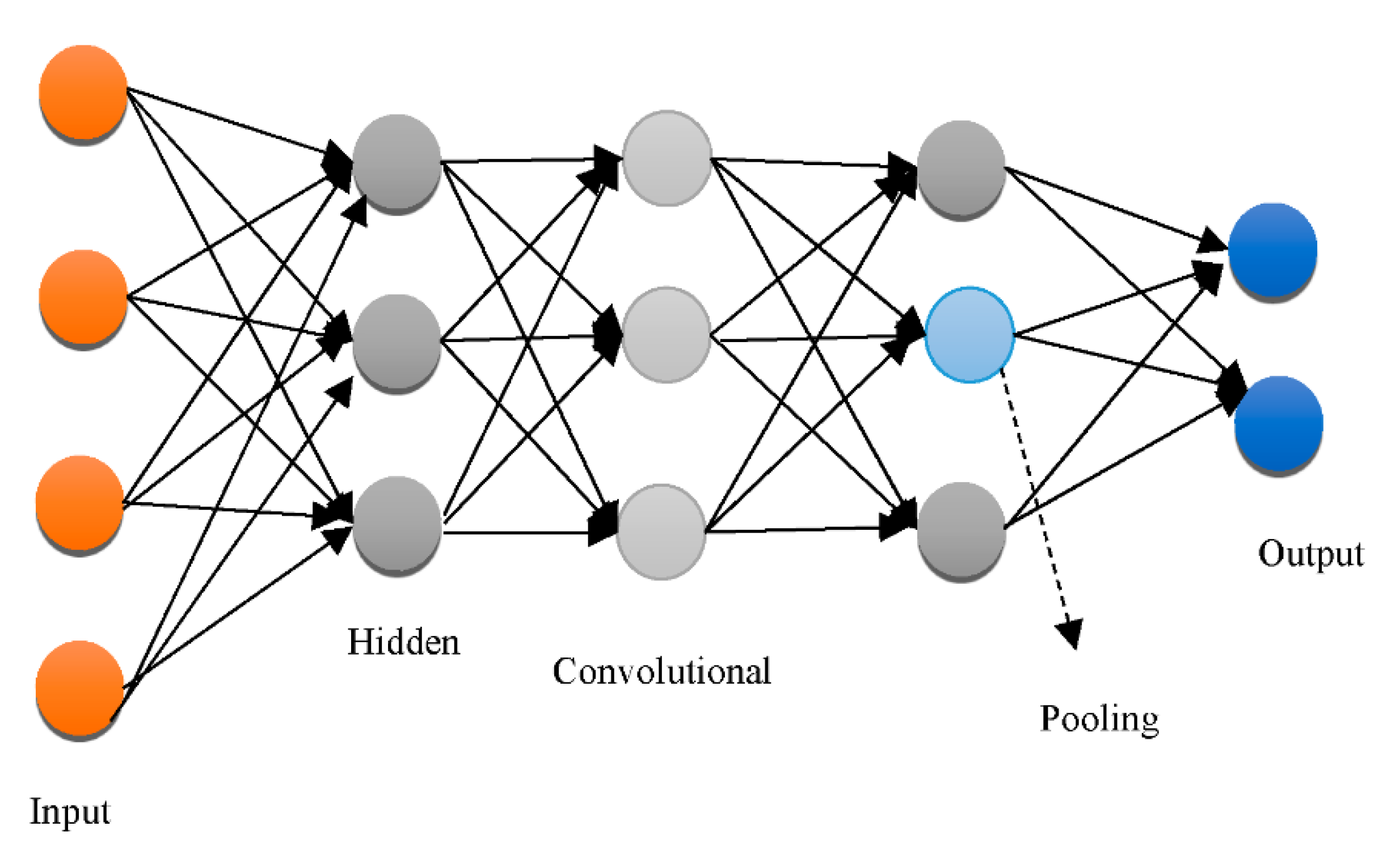
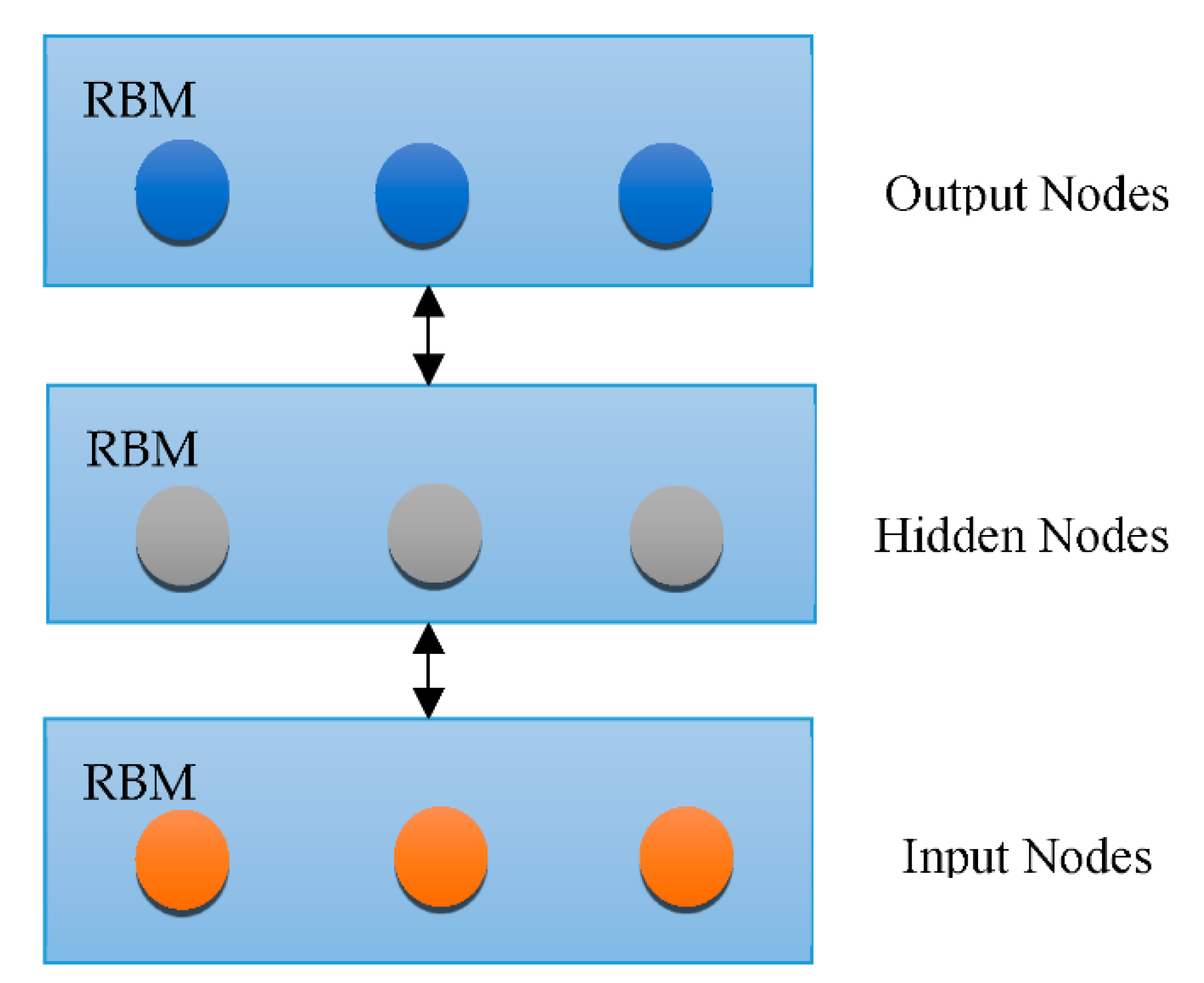
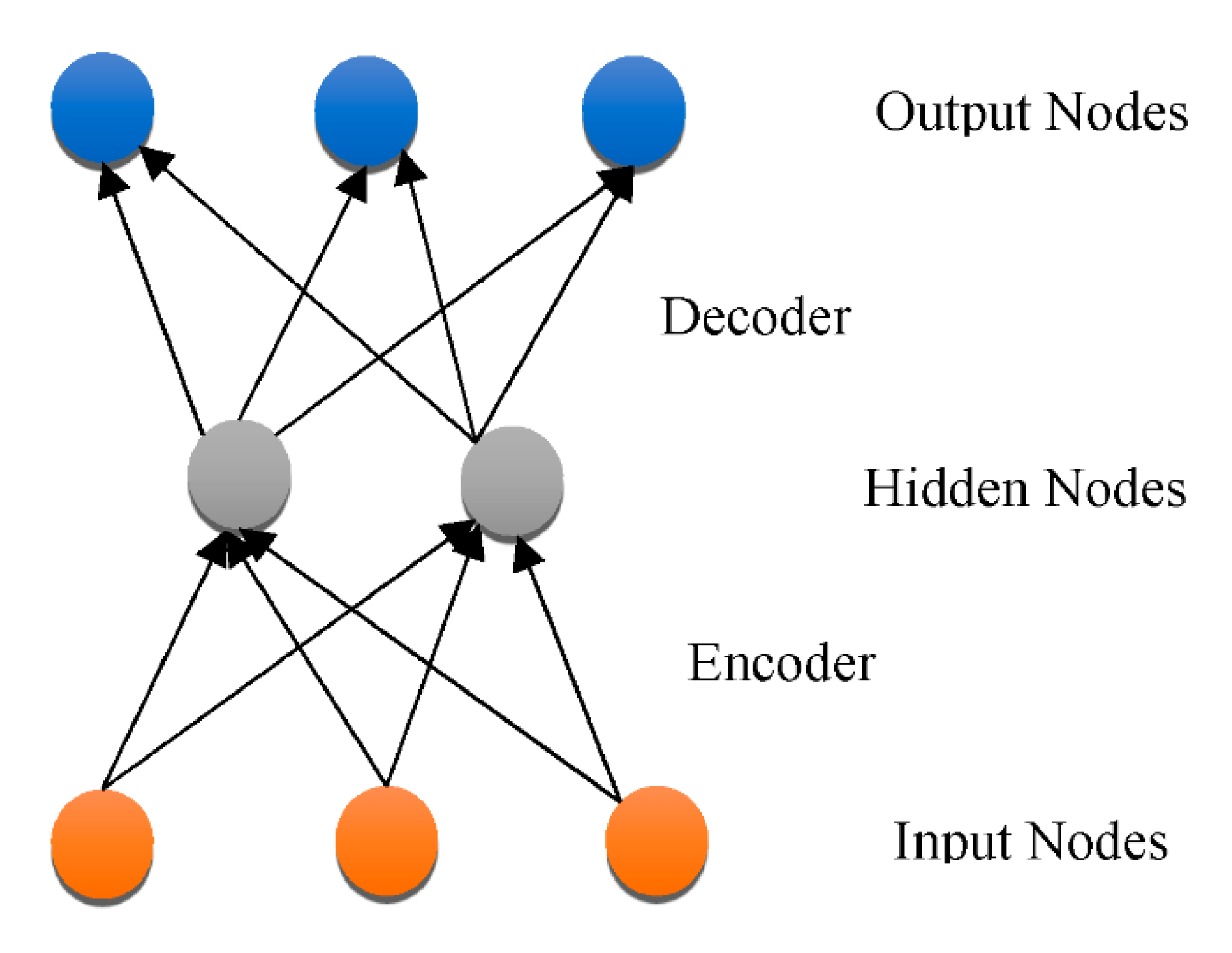
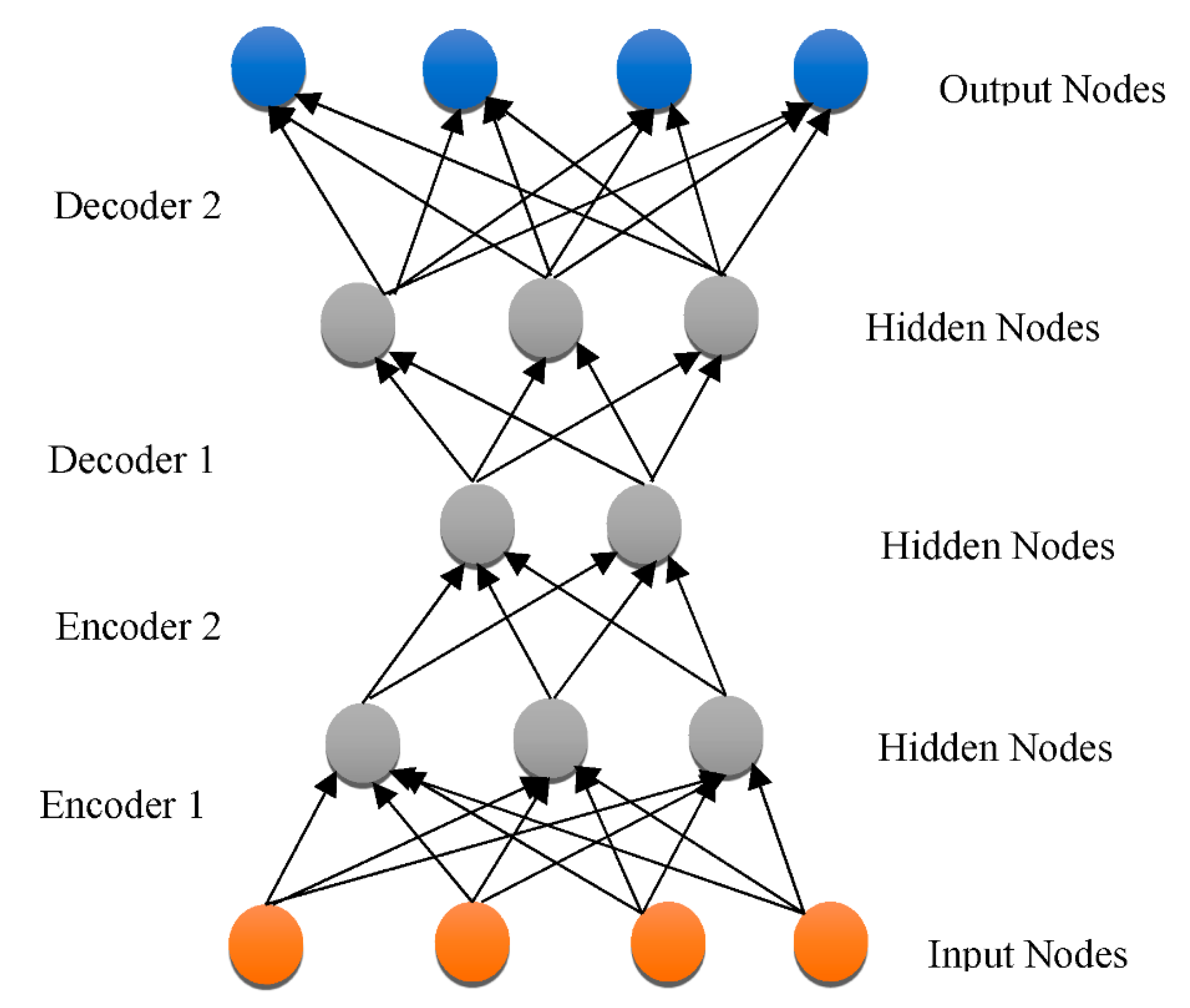
3.3. DNN Architectures
3.4. DL for Selection of Attributes from Neuroimaging Information
3.5. DL for Selection of Heterogeneous Neuroimaging Data
4. Case Studies on the Diagnosis of AD Using DL and Related Technologies
- During the last 15 years, the utilization of such techniques and AI technologies in medical applications has skyrocketed. There seem to be three important aspects to consider, e.g., information quantity and quality have both improved. In this sense, the discipline is approaching Big Data.
- In detection with the help of computers (CADe), certain components in the imagery, such as structures or neurons, can be identified. CADe can also be used to identify areas of focus for scientists, like malignancies.
- Segmentation is the separation of complete picture portions from the rest of the imaging.
- Computer-aided Diagnosis (CADx) denotes a diagnostic based on particular data that can be described as a categorization task in plain terms. Medical photos are employed in this scenario, which emphasizes the necessity of CNN. In the context of AD, there are three classifications: NC, MCI, and AD.
5. Research Challenges in DL for AD
6. Discussion
7. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mathew, A.; Amudha, P.; Sivakumari, S. Deep Learning Techniques: An Overview. In Advanced Machine Learning Technologies and Applications. AMLTA 2020. Advances in Intelligent Systems and Computing; Hassanien, A., Bhatnagar, R., Darwish, A., Eds.; Springer: Singapore, 2020; Volume 1141. [Google Scholar] [CrossRef]
- Jiang, T. Deep Learning Application in Alzheimer Disease Diagnoses and Prediction. In Proceedings of the 2020 4th International Conference on Artificial Intelligence and Virtual Reality, Kumamoto, Japan, 23–25 October 2020. [Google Scholar] [CrossRef]
- Kraemer, H.C.; Taylor, J.L.; Tinklenberg, J.R.; Yesavage, J.A. The Stages of Alzheimer’s Disease: A Reappraisal. Dement. Geriatr. Cogn. Disord. 1998, 9, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Pais, M.; Martinez, L.; Ribeiro, O.; Loureiro, J.; Fernandez, R.; Valiengo, L.; Canineu, P.; Stella, F.; Talib, L.; Radanovic, M.; et al. Early diagnosis and treatment of Alzheimer’s disease: New definitions and challenges. Braz. J. Psychiatry 2020, 42, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Al-Shoukry, S.; Rassem, T.H.; Makbol, N.M. Alzheimer’s Diseases Detection by Using Deep Learning Algorithms: A Mini-Review. IEEE Access 2020, 8, 77131–77141. [Google Scholar] [CrossRef]
- Haller, S.; Nguyen, D.; Rodriguez, C.; Emch, J.; Gold, G.; Bartsch, A.; Lovblad, K.O.; Giannakopoulos, P. Individual prediction of cognitive decline in mild cognitive impairment using support vector machine-based analysis of diffusion tensor imaging data. J. Alzheimer’s Dis. 2010, 22, 315–327. [Google Scholar] [CrossRef]
- Gamarra, M.; Mitre-Ortiz, A.; Escalante, H. Automatic cell image segmentation using genetic algorithms. In Proceedings of the 2019 XXII Symposium on Image, Signal Processing and Artificial Vision (STSIVA), Bucaramanga, Colombia, 24–26 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Fogel, I.; Sagi, D. Gabor filters as texture discriminator. Biol. Cybern. 1989, 61, 103–113. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Deng, L.; Yu, D. Deep Learning: Methods and Applications. Found. Trends Signal Processing 2014, 7, 197–387. [Google Scholar] [CrossRef]
- Nanni, L.; Brahnam, S.; Ghidoni, S.; Menegatti, E.; Barrier, T. A comparison of methods for extracting information from the co-occurrence matrix for subcellular classification. Expert Syst. Appl. 2013, 40, 7457–7467. [Google Scholar] [CrossRef]
- Xu, Y.; Zhu, J.Y.; Eric, I.; Chang, C.; Lai, M.; Tu, Z. Weakly supervised histopathology cancer image segmentation and classification. Med. Image Anal. 2014, 18, 591–604. [Google Scholar] [CrossRef]
- Barker, J.; Hoogi, A.; Depeursinge, A.; Rubin, D.L. Automated classification of brain tumor type in whole-slide digital pathology images using local representative tiles. Med. Image Anal. 2015, 30, 60–71. [Google Scholar] [CrossRef]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Feng, C.; Elazab, A.; Yang, P.; Wang, T.; Zhou, F.; Hu, H.; Xiao, X.; Lei, B. Deep learning framework for alzheimer’s disease diagnosis via 3d-cnn and fsbi-lstm. IEEE Access 2019, 7, 63605–63618. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Proceedings of the 27th International Conference on Neural Information Processing Systems, Ser. NIPS’14, Montreal, QC, Canada, 8–13 December 2014; MIT Press: Cambridge, MA, USA, 2014; Volume 2, pp. 3320–3328. [Google Scholar]
- Sarraf, S.; Tofighi, G. Classification of alzheimer’s disease using fmri data and deep learning convolutional neural networks. arXiv 2016, arXiv:1603.08631. [Google Scholar]
- Li, Y.; Huang, C.; Ding, L.; Li, Z.; Pan, Y.; Gao, X. Deep learning in bioinformatics: Introduction, application, and perspective in the big data era. Methods 2019, 166, 4–21. [Google Scholar] [CrossRef]
- Mussap, M.; Noto, A.; Cibecchini, F.; Fanos, V. The importance of biomarkers in neonatology. Semin. Fetal Neonatal Med. 2013, 18, 56–64. [Google Scholar] [CrossRef]
- Cedazo-Minguez, A.; Winblad, B. Biomarkers for Alzheimer’s disease and other forms of dementia: Clinical needs, limitations and future aspects. Exp. Gerontol. 2010, 45, 5–14. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Shijie, J.; Ping, W.; Peiyi, J.; Siping, H. Research on data augmentation for image classification based on convolution neural networks. In Proceedings of the 2017 Chinese Automation Congress (CAC), Jinan, China, 20–22 October 2017; pp. 4165–4170. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. Gan-based synthetic medical image augmentation for increased cnn performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Zhu, D.; Lu, J.; Luo, Y.; Zhang, G. Synthetic medical images using f&bgan for improved lung nodules classification by multi-scale vgg16. Symmetry 2018, 10, 519. [Google Scholar]
- Long, M.; Cao, Y.; Cao, Z.; Wnag, J.; Jordan, M.I. Transferable Representation Learning with Deep Adaptation Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 3071–3085. [Google Scholar] [CrossRef]
- Pichler, B.J.; Kolb, A.; Nägele, T.; Schlemmer, H.-P. PET/MRI: Paving the Way for the Next Generation of Clinical Multimodality Imaging Applications. J. Nucl. Med. 2010, 51, 333–336. [Google Scholar] [CrossRef]
- Ding, J.; Chen, B.; Liu, H.; Huang, M. Convolutional neural network with data augmentation for sar target recognition. IEEE Geosci. Remote Sens. Lett. 2016, 13, 364–368. [Google Scholar] [CrossRef]
- Castro, E.; Cardoso, J.S.; Pereira, J.C. Elastic deformations for data augmentation in breast cancer mass detection. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), Las Vegas, NV, USA, 4–7 March 2018; pp. 230–234. [Google Scholar]
- Nordberg, A.; Rinne, J.O.; Kadir, A.; Långström, B. The use of PET in Alzheimer disease. Nat. Rev. Neurol. 2010, 6, 78–87. [Google Scholar] [CrossRef]
- Shen, T.; Jiang, J.; Li, Y.; Wu, P.; Zuo, C.; Yan, Z. Decision supporting model for one-year conversion probability from mci to ad using cnn and svm. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 738–741. [Google Scholar]
- Shmulev, Y.; Belyaev, M. Predicting conversion of mild cognitive impairments to alzheimer’s disease and exploring impact of neuroimaging. In Graphs in Biomedical Image Analysis and Integrating Medical Imaging and Non-Imaging Modalities; Stoyanov, D., Taylor, Z., Ferrante, E., Dalca, A.V., Martel, A., Maier-Hein, L., Parisot, S., Sotiras, A., Papiez, B., Sabuncu, M.R., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 83–91. [Google Scholar]
- Suk, H.-I.; Lee, S.W.; Shen, D. Hierarchical feature representation and multimodal fusion with deep learning for ad/mci diagnosis. NeuroImage 2014, 101, 569–582. [Google Scholar] [CrossRef]
- Nho, K.; Shen, L.; Kim, S.; Risacher, S.L.; West, J.D.; Foroud, T.; Jack, C.R., Jr.; Weiner, M.W.; Saykin, A.J. Automatic prediction of conversion from mild cognitive impairment to probable alzheimer’s disease using structural magnetic resonance imaging. In Annual Symposium Proceedings/AMIA Symposium; American Medical Informatics Association: Bethesda, MD, USA, 2010; Volume 2010, pp. 542–546. [Google Scholar]
- Costafreda, S.G.; Dinov, I.D.; Tu, Z.; Shi, Y.; Liu, C.Y.; Kloszewska, I.; Mecocci, P.; Soininen, H.; Tsolaki, M.; Vellas, B.; et al. Automated hippocampal shape analysis predicts the onset of dementia in mild cognitive impairment. NeuroImage 2011, 56, 212–219. [Google Scholar] [CrossRef]
- Saraiva, C.; Praça, C.; Ferreira, R.; Santos, T.; Ferreira, L.; Bernardino, L. Nanoparticle-mediated brain drug delivery: Overcoming blood–brain barrier to treat neurodegenerative diseases. J. Control. Release 2016, 235, 34–47. [Google Scholar] [CrossRef]
- Coupé, P.; Eskildsen, S.F.; Manjón, J.V.; Fonov, V.S.; Collins, D.L. Simultaneous segmentation and grading of anatomical structures for patient’s classification: Application to Alzheimer’s disease. Neuroimage 2011, 59, 3736–3747. [Google Scholar] [CrossRef]
- Wolz, R.; Julkunen, V.; Koikkalainen, J.; Niskanen, E.; Zhang, D.P.; Rueckert, D.; Soininen, H.; Lötjönen, J.; The Alzheimer’s Disease Neuroimaging Initiative. Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. PLoS ONE 2011, 6, e25446. [Google Scholar] [CrossRef]
- Liu, M.; Cheng, D.; Wang, K.; Wang, Y.; The Alzheimer’s Disease Neuroimaging Initiative. Multi-modality cascaded convolutional neural networks for Alzheimer’s disease diagnosis. Neuroinformatics 2018, 16, 295–308. [Google Scholar] [CrossRef]
- Kruthika, K.R.; Maheshappa, H.D. Multistage classifier-based approach for Alzheimer’s disease prediction and retrieval. Inform. Med. Unlocked 2019, 14, 34–42. [Google Scholar] [CrossRef]
- Basaia, S.; Agosta, F.; Wagner, L.; Canu, E.; Magnani, G.; Santangelo, R.; Filippi, M. Automated classification of Alzheimer’s disease and mild cognitive impairment using a single MRI and deep neural networks. NeuroImage Clin. 2018, 21, 101645. [Google Scholar] [CrossRef] [PubMed]
- Payan, A.; Montana, G. Predicting Alzheimer’s disease: A neuroimaging study with 3d convolutional neural networks. In Proceedings of the ICPRAM 2015 4th International Conference on Pattern Recognition Applications and Methods, Lisbon, Portugal, 10–12 January 2015; Volume 2. [Google Scholar]
- Asl, E.H.; Ghazal, M.; Mahmoud, A.; Aslantas, A.; Shalaby, A.; Casanova, M.; Barnes, G.; Gimel’farb, G.; Keynton, R.; El Baz, A. Alzheimer’s disease diagnostics by a 3d deeply supervised adaptable convolutional network. Front. Biosci. 2018, 23, 584–596. [Google Scholar]
- Feldman, M.D. Positron Emission Tomography (PET) for the Evaluation of Alzheimer’s Disease/Dementia. In Proceedings of the California Technology Assessment Forum, New York, NY, USA, June 2010. [Google Scholar]
- Bengio, Y. Learning deep architectures for AI. Found. Trends Mach. Learn. 2009, 2, 1–127. [Google Scholar] [CrossRef]
- Ciregan, D.; Meier, U.; Schmidhuber, J. Multi-column deep neural networks for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 3642–3649. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; ACM: Stateline, NV, USA, 2012; pp. 1097–1105. [Google Scholar]
- Farabet, C.; Couprie, C.; Najman, L.; Lecun, Y. Learning hierarchical features for scene labeling. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1915–1929. [Google Scholar] [CrossRef]
- Hinton, G.; Deng, L.; Yu, D.; Dahl, G.; Mohamed, A.-R.; Jaitly, N.; Senior, A.; Vanhoucke, V.; Nguyen, P.; Sainath, T.N.; et al. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal Process. Mag. 2012, 29, 82–97. [Google Scholar] [CrossRef]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Advances in Neural Information Processing Systems 26. Burges, C.J.C., Bottou, L., Welling, M., Ghahramani, Z., Weinberger, K.Q., Eds.; ACM: Stateline, NV, USA, 2013; pp. 3111–3119. [Google Scholar]
- Jo, T.; Nho, K.; Saykin, A.J. Deep Learning in Alzheimer’s Disease: Diagnostic Classification and Prognostic Prediction Using Neuroimaging Data. Front. Aging Neurosci. 2019, 11, 220. [Google Scholar] [CrossRef]
- Boureau, Y.-L.; Ponce, J.; Lecun, Y. A theoretical analysis of feature pooling in visual recognition. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 111–118. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comp. Vision 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Bengio, Y. Deep learning of representations: Looking forward. In Proceedings of the International Conference on Statistical Language and Speech Processing, First International Conference, SLSP 2013, Tarragona, Spain, 29–31 July 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 1–37. [Google Scholar]
- Bengio, Y.; Courville, A.; Vincent, P. Representation learning: A review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 1798–1828. [Google Scholar] [CrossRef]
- Yi, H.; Sun, S.; Duan, X.; Chen, Z. A study on Deep Neural Networks framework. In Proceedings of the 2016 IEEE Advanced Information Management, Communicates, Electronic and Automation Control Conference (IMCEC), Xi’an, China, 3–5 October 2016. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Werbos, P.J. Backwards differentiation in AD and neural nets: Past links and new opportunities. In Automatic Differentiation: Applications, Theory, and Implementations; Bücker, H.M., Corliss, G., Hovland, P., Naumann, U., Norris, B., Eds.; Springer: New York, NY, USA, 2006; pp. 15–34. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted Boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Glorot, X.; Bordes, A.; Bengio, Y. Deep sparse rectifier neural networks. In Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics, Fort Lauderdale, FL, USA, 11–13 April 2011; pp. 315–323. [Google Scholar]
- Bottou, L. Large-scale machine learning with stochastic gradient descent. In Proceedings of the COMPSTAT’2010, 19th International Conference on Computational Statistics, Paris, France, 22–27 August 2010; Springer: Berlin, Germany, 2010; pp. 177–186. [Google Scholar]
- Sutskever, I.; Martens, J.; Dahl, G.; Hinton, G. On the importance of initialization and momentum in deep learning. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA USA, 16–21 June 2013; pp. 1139–1147. [Google Scholar]
- Shrestha, A.; Mahmood, A. Review of Deep Learning Algorithms and Architectures. IEEE Access 2019, 7, 53040–53065. [Google Scholar] [CrossRef]
- Salakhutdinov, R.; Larochelle, H. Efficient learning of deep Boltzmann machines. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; pp. 693–700. [Google Scholar]
- Makhzani, A.; Frey, B. k-sparse autoencoders. In Advances in Neural Information Processing Systems 28; ICLR: Montreal, QC, USA, 2015; pp. 2791–2799. [Google Scholar]
- Liu, S.; Liu, S.; Cai, W.; Pujol, S.; Kikinis, R.; Feng, D. Early diagnosis of Alzheimer’s disease with deep learning. In Proceedings of the 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI), Beijing, China, 29 April–2 May 2014; pp. 1015–1018. [Google Scholar]
- Cheng, D.; Liu, M.; Fu, J.; Wang, Y. Classification of MR brain images by combination of multi-CNNs for AD diagnosis. In Proceedings of the Ninth International Conference on Digital Image Processing (ICDIP 2017), Hong Kong, China, 19–22 May 2017; pp. 875–879. [Google Scholar]
- Ngiam, J.; Khosla, A.; Kim, M.; Nam, J.; Lee, H.; Ng, A.Y. Multimodal deep learning. In Proceedings of the 28th International Conference on Machine Learning (ICML-11), Bellevue, WA, USA, 18 June–2 July 2011; pp. 689–696. [Google Scholar]
- Liu, S.; Liu, S.; Cai, W.; Che, H.; Pujol, S.; Kikinis, R.; Feng, D.; Fulham, M.J. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s Disease. IEEE Trans. Biomed. Eng. 2015, 62, 1132–1140. [Google Scholar] [CrossRef]
- Lu, D.; Popuri, K.; Ding, G.W.; Balachandar, R.; Beg, M.F. Multimodal and multiscale deep neural networks for the early diagnosis of Alzheimer’s disease using structural structuralMR and FDG-PET images. Sci. Rep. 2018, 8, 5697. [Google Scholar] [CrossRef]
- De strooper, B.; Karran, E. The cellular phase of Alzheimer’s disease. Cell 2016, 164, 603–615. [Google Scholar] [CrossRef]
- Cheng, D.; Liu, M. CNNs based multi-modality classification for AD diagnosis. In Proceedings of the 2017 10th International Congress on Image and Signal Processing, BioMedical Engineering, and Informatics (CISP-BMEI), Shanghai, China, 14–16 October 2017; pp. 1–5. [Google Scholar]
- Korolev, S.; Safiullin, A.; Belyaev, M.; Dodonova, Y. Residual and plain convolutional neural networks for 3D brain MRI classification. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017; pp. 835–838. [Google Scholar]
- Aderghal, K.; Benois-Pineau, J.; Afdel, K.; Catheline, G. FuseMe: Classification of sMRI images by fusion of deep CNNs in 2D+ǫ projections. In Proceedings of the 15th International Workshop on Content-Based Multimedia Indexing, New York, NY, USA, 19–21 June 2017. [Google Scholar]
- Liu, M.; Zhang, J.; Adeli, E.; Shen, D. Landmark-based deep multiinstance learning for brain disease diagnosis. Med. Image Anal. 2018, 43, 157–168. [Google Scholar] [CrossRef]
- Li, R.; Zhang, W.; Suk, H.-I.; Wang, L.; Li, J.; Shen, D.; Ji, S. Deep learning-based imaging data completion for improved brain disease diagnosis. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Boston, MA, USA, 14–18 September 2014; Volume 17, pp. 305–312. [Google Scholar]
- Vu, T.D.; Yang, H.-J.; Nguyen, V.Q.; Oh, A.R.; Kim, M.-S. Multimodal learning using convolution neural network and Sparse Autoencoder. In Proceedings of the 2017 IEEE International Conference on Big Data and Smart Computing (BigComp), Jeju, Korea, 13–16 February 2017; pp. 309–312. [Google Scholar]
- Liu, M.; Cheng, D.; Yan, W.; Alzheimer’s Disease Neuroimaging Initiative. Classification of Alzheimer’s disease by combination of convolutional and recurrent neural networks using FDG-PET images. Front. Neuroinform. 2018, 12, 35. [Google Scholar] [CrossRef]
- Venugopalan, J.; Tong, L.; Hassanzadeh, H.R.; Wang, M. Multimodal deep learning models for early detection of Alzheimer’s disease stage. Sci. Rep. 2021, 11, 3254. [Google Scholar] [CrossRef]
- Choi, H.; Jin, K.H. Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav. Brain Res. 2018, 344, 103–109. [Google Scholar] [CrossRef]
- O’Connor, J.P.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef]
- Ker, J.; Wang, L.; Rao, J.; Lim, T. Deep Learning Applications in Medical Image Analysis. IEEE Access 2018, 6, 9375–9389. [Google Scholar] [CrossRef]
- Klöppel, S.; Stonnington, C.M.; Barnes, J.; Chen, F.; Chu, C.; Good, C.D.; Mader, I.; Mitchell, L.A.; Patel, A.C.; Roberts, C.C.; et al. Accuracy of dementia diagnosis—A direct comparison between radiologists and a computerized method. Brain 2008, 131, 2969–2974. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Processing Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Yang, W.; Lui, R.L.; Gao, J.H.; Chan, T.F.; Yau, S.T.; Sperling, R.A.; Huang, X. Independent component analysis-based classification of Alzheimer’s disease MRI data. J. Alzheimer’s Dis. 2011, 24, 775–783. [Google Scholar] [CrossRef]
- Xiaoqing, L.; Kunlun, G.; Bo, L.; Chengwei, P.; Kongming, L.; Lifeng, Y.; Jiechao, M.; Fujin, H.; Shu, Z.; Siyuan, P.; et al. Advances in Deep Learning-Based Medical Image Analysis. Health Data Sci. 2021, 14, 8786793. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Gupta, A.; Ayhan, M.; Maida, A. Natural image bases to represent neuroimaging data. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013; pp. 987–994. [Google Scholar]
- Evans, A.C. An MRI-based stereotactic atlas from 250 young normal subjects. Soc. Neurosci. Abstr. 1992, 18, 408. [Google Scholar]
- Hosseini-Asl, E.; Keynton, R.; El-Baz, A. Alzheimer’s disease diagnostics by adaptation of 3D convolutievaonal network. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), Phoenix, AZ, USA, 25–28 September 2016; pp. 126–130. [Google Scholar]
- Hellström, E. Feature Learning with Deep Neural Networks for Keystroke Biometrics: A Study of Supervised Pre-Training and Autoencoders. 2018. Available online: https://www.diva-portal.org/smash/get/diva2:1172405/FULLTEXT01.pdf (accessed on 12 March 2021).
- Woods, R.P.; Mazziotta, J.C.; Cherry, S.R. MRI-PET registration with automated algorithm. J. Comput. Assist. Tomogr. 1993, 17, 536. [Google Scholar] [CrossRef]
- Klein, A.; Andersson, J.; Ardekani, B.A.; Ashburner, J.; Avants, B.; Chiang, M.C.; Christensen, G.E.; Collins, D.L.; Gee, J.; Hellier, P.; et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 2009, 46, 786–802. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Welsh-Bohmer, K.A.; Hanson, M.; Crain, B.; Hulette, C.; Earl, N.; Coleman, R.E. FDG PET imaging in patients with pathologically verified dementia. J. Nucl. Med. 2000, 41, 1920–1928. [Google Scholar] [PubMed]
- Evans, A.C.; Collins, D.L.; Mills, S.R.; Brown, E.D.; Kelly, R.L.; Peters, T.M. 3D statistical neuroanatomical models from 305 MRI volumes. In Proceedings of the 1993 IEEE Conference Record Nuclear Science Symposium and Medical Imaging Conference, San Francisco, CA, USA, 31 October–6 November 1993; pp. 1813–1817. [Google Scholar]
- Suk, H.I.; Lee, S.W.; Shen, D.; The Alzheimer’s Disease Neuroimaging Initiative. Latent feature representation with stacked auto-encoder for AD/MCI diagnosis. Brain Struct. Funct. 2015, 220, 841–859. [Google Scholar] [CrossRef]
- Rueckert, D.; Sonoda, L.I.; Hayes, C.; Hill, D.L.; Leach, M.O.; Hawkes, D.J. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Trans. Med. Imaging 1999, 18, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Rajchl, M.; Ktena, S.I.; Pawlowski, N. An Introduction to Biomedical Image Analysis with TensorFlow and DLTK. Medium.com. 2018. Available online: https://medium.com/tensorflow/an-introduction-to-biomedical-image-analysis-with-tensorflow-and-dltk-2c25304e7c13 (accessed on 19 April 2021).
- Ding, Y.; Sohn, J.H.; Kawczynski, M.G.; Trivedi, H.; Harnish, R.; Jenkins, N.W.; Lituiev, D.; Copeland, T.P.; Aboian, M.S.; Mari Aparici, C.; et al. A Deep learning model to predict a diagnosis of Alzheimer disease by using 18F-FDG PET of the brain. Radiology 2018, 290, 456–464. [Google Scholar] [CrossRef]
- Vesal, S.; Ravikumar, N.; Davari, A.A.; Ellmann, S.; Maier, A. Classification of Breast Cancer Histology Images Using Transfer Learning. In Image Analysis and Recognition, Proceedings of the 5th International Conference, ICIAR 2018, Póvoa de Varzim, Portugal, 27–29 June 2018; Springer: Cham, Switzerland, 2017; pp. 812–819. [Google Scholar]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- OASIS Brains Datasets. Available online: https://www.oasis-brains.org/#data (accessed on 7 October 2021).
- Tong, T.; Wolz, R.; Gao, Q.; Guerrero, R.; Hajnal, J.V.; Rueckert, D.; Alzheimer’s Disease Neuroimaging Initiative. Multiple instance learning for classification of dementia in brain MRI. Med. Image Anal. 2014, 18, 808–818. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Habas, P.A.; Zurada, J.M.; Lo, J.Y.; Baker, J.A.; Tourassi, G.D. Training neural network classifiers for medical decision making: The effects of imbalanced datasets on classification performance. Neural Netw. 2008, 21, 427–436. [Google Scholar] [CrossRef]
- Montavon, G.; Lapuschkin, S.; Binder, A.; Samek, W.; Müller, K.R. Explaining nonlinear classification decisions with deep taylor decomposition. Pattern Recognit. 2017, 65, 211–222. [Google Scholar] [CrossRef]
- Hutson, M. Artificial intelligence faces reproducibility crisis. Science 2018, 359, 725–726. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, A.; Bengio, S.; Brevdo, E.; Chollet, F.; Gomez, A.N.; Gouws, S.; Jones, L.; Kaiser, Ł.; Kalchbrenner, N.; Parmar, N.; et al. Tensor2tensor for neural machine translation. In Proceedings of the 13th Conference of the Association for Machine Translation in the Americas, Boston, MA, USA, 17–21 March 2018; pp. 193–199. [Google Scholar]
- Smialowski, P.; Frishman, D.; Kramer, S. Pitfalls of supervised feature selection. Bioinformatics 2009, 26, 440–443. [Google Scholar] [CrossRef] [PubMed]
- König, I.R. Validation in genetic association studies. Brief. Bioinform. 2011, 12, 253–258. [Google Scholar] [CrossRef] [PubMed]


| Reference | DL Technique | Accuracy |
|---|---|---|
| [69] | SAEsoftmax” regression layer | >86% |
| [70] | 3D-CNN | >87% |
| [72] | SAE SoftMax” regression layer | >90% |
| [73] | SAE DNN | >84% for AD/CN classification >82% for MCI to AD classification |
| [74] | 3D CNN | >92% for AD/CN classification >72% for MCI to AD conversion |
| [75] | VoxCNN ResNet | >79% |
| [77] | 2D CNN | >85% |
| [78] | 3D CNN | >75% for MCI to AD conversion |
| [80] | SAE 3D CNN | >90% |
| [81] | Ensemble of 2D CNN and RNN | >91% |
| [83] | 3D CNN | >95% for AD/CN classification >84% for MCI to AD conversion |
| Reference | Illness | CNN Model | Accuracy |
|---|---|---|---|
| [19] | AD | “Inception + LeNet5” | 96.85% |
| [76] | AD | VoxCNN + VoxResNet | 80% |
| [103] | AD | “Inception V3” | 92% |
| [104] | Breast Cancer | “Inception V3 + ResNet50” | 85% |
| [105] | Skin Cancer | “Inception V3” | 93.33% |
| [106] | Diabetic Retinopathy | “Inception V3” | 90.3% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shastry, K.A.; Vijayakumar, V.; V, M.K.M.; B A, M.; B N, C. Deep Learning Techniques for the Effective Prediction of Alzheimer’s Disease: A Comprehensive Review. Healthcare 2022, 10, 1842. https://doi.org/10.3390/healthcare10101842
Shastry KA, Vijayakumar V, V MKM, B A M, B N C. Deep Learning Techniques for the Effective Prediction of Alzheimer’s Disease: A Comprehensive Review. Healthcare. 2022; 10(10):1842. https://doi.org/10.3390/healthcare10101842
Chicago/Turabian StyleShastry, K Aditya, V Vijayakumar, Manoj Kumar M V, Manjunatha B A, and Chandrashekhar B N. 2022. "Deep Learning Techniques for the Effective Prediction of Alzheimer’s Disease: A Comprehensive Review" Healthcare 10, no. 10: 1842. https://doi.org/10.3390/healthcare10101842
APA StyleShastry, K. A., Vijayakumar, V., V, M. K. M., B A, M., & B N, C. (2022). Deep Learning Techniques for the Effective Prediction of Alzheimer’s Disease: A Comprehensive Review. Healthcare, 10(10), 1842. https://doi.org/10.3390/healthcare10101842







