Rapid and Easy High-Molecular-Weight Glutenin Subunit Identification System by Lab-on-a-Chip in Wheat (Triticum aestivum L.)
Abstract
1. Introduction
2. Results
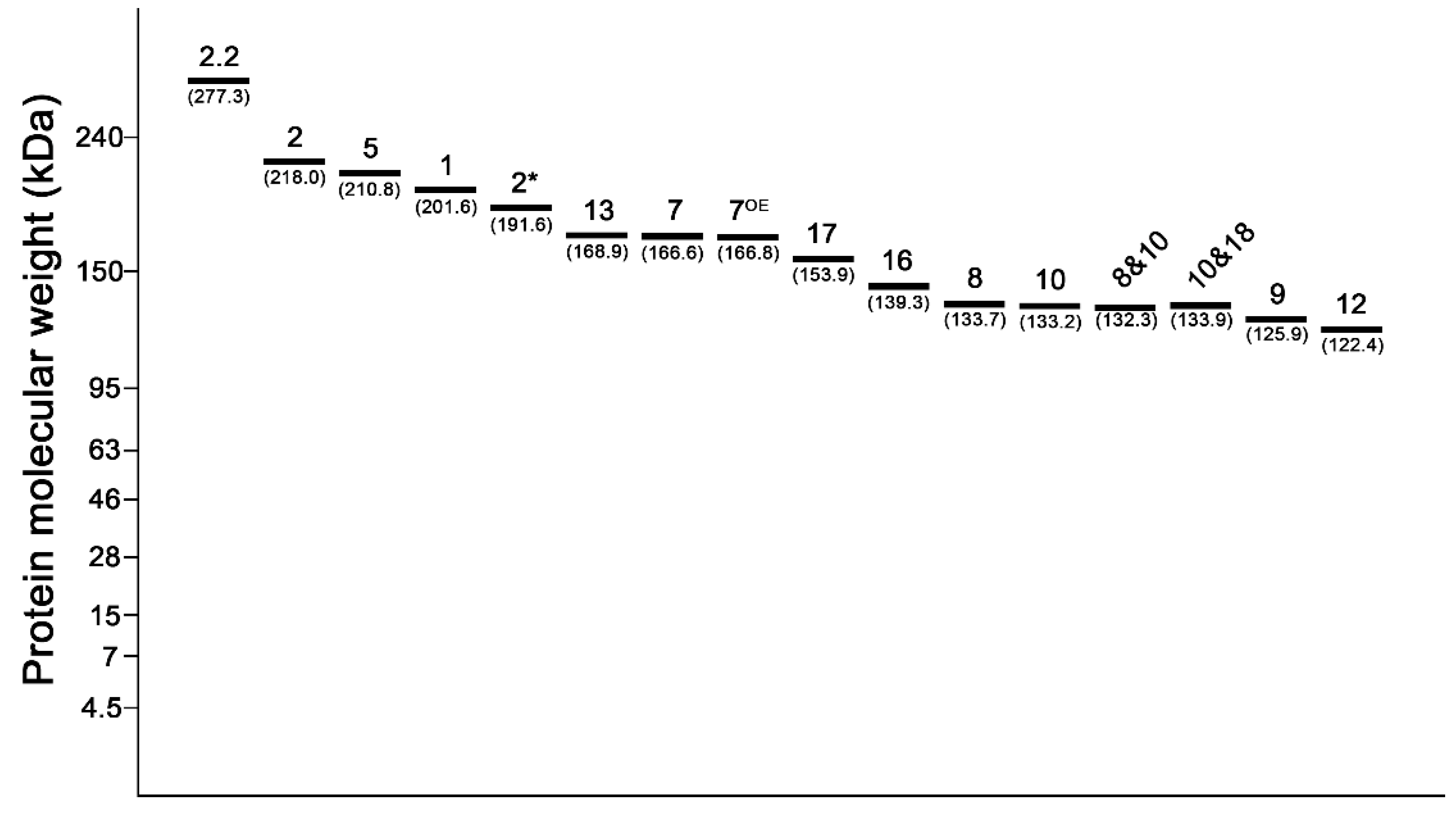
2.1. Identification of 1Ax1 and 1Ax2* at the Glu-A1 Locus by Lab-on-a-Chip
2.2. Electrophoresis Patterns of HMW-GS Encoded by the Glu-B1 Locus
2.3. Identification of the 1Dx5 + 1Dy10 Subunit from Other Subunits Encoded from the Glu-D1 Locus
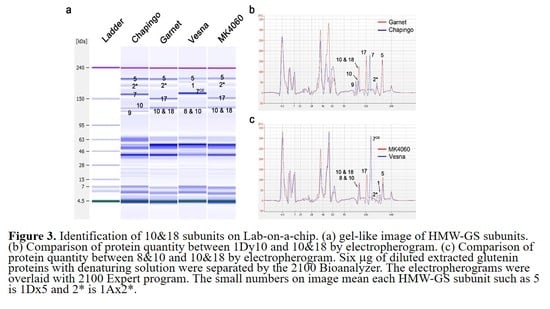
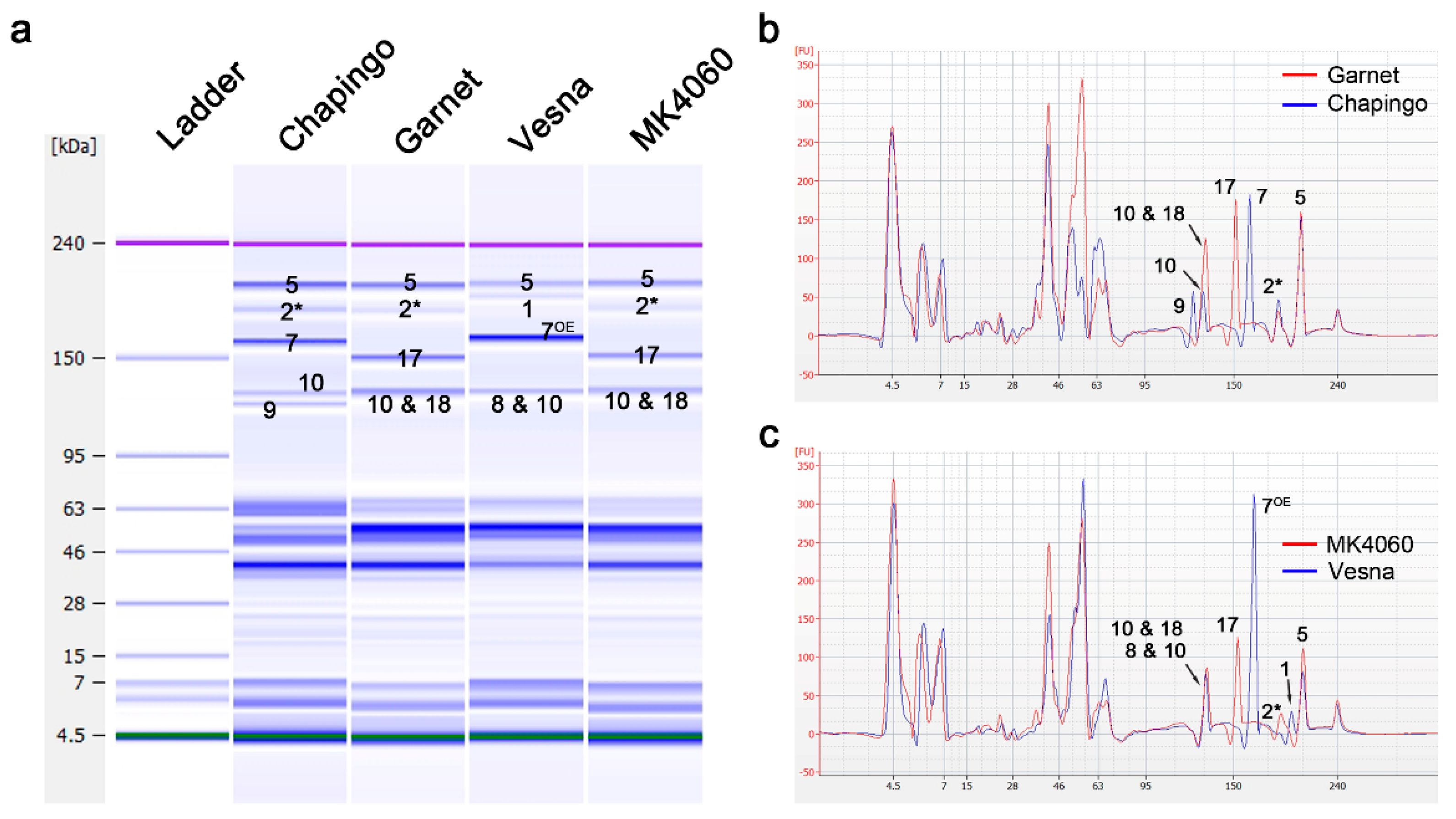
2.4. Discrimination of 8 & 10, 10 & 18, and 7OE Subunit by Analyzing the Electropherogram
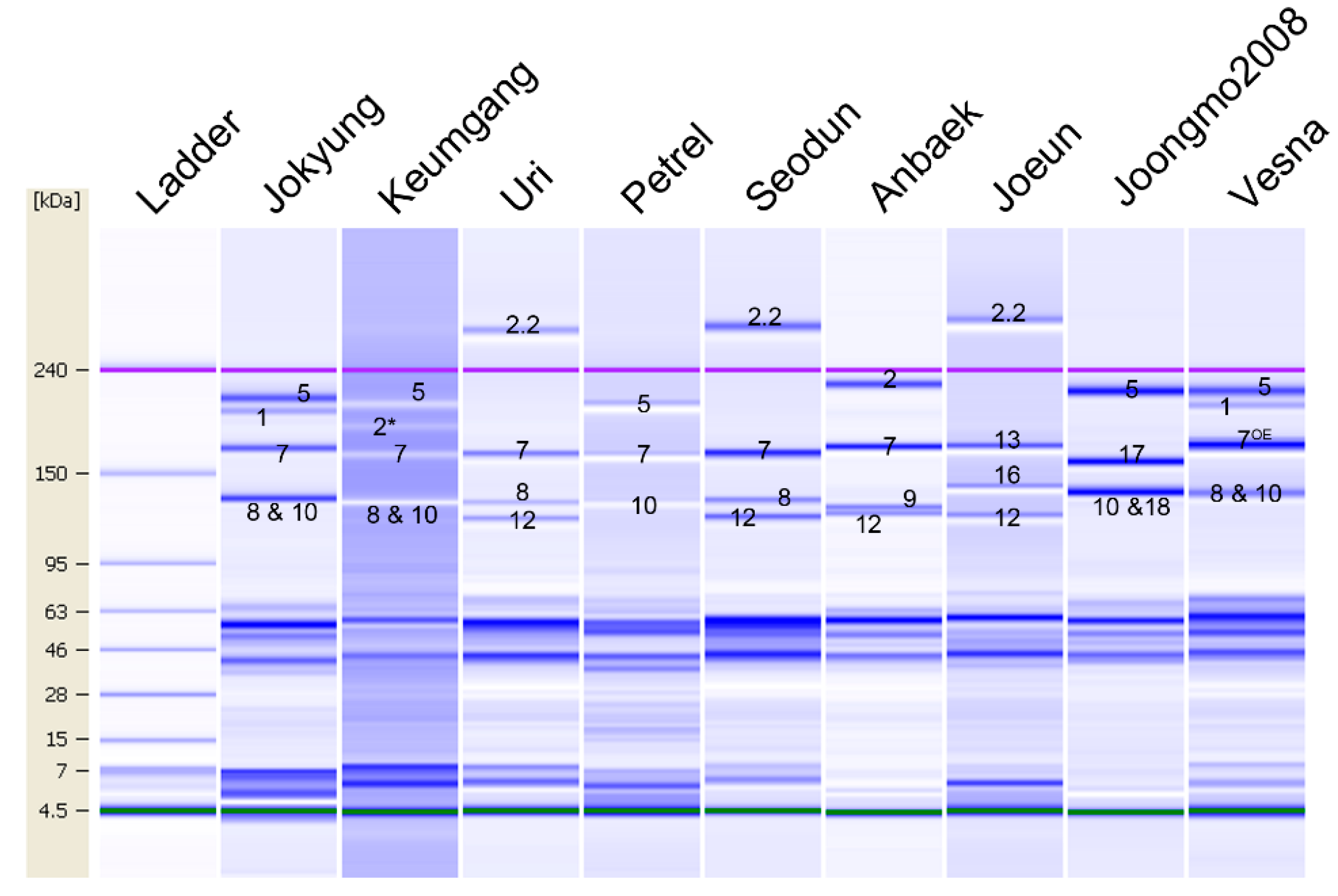
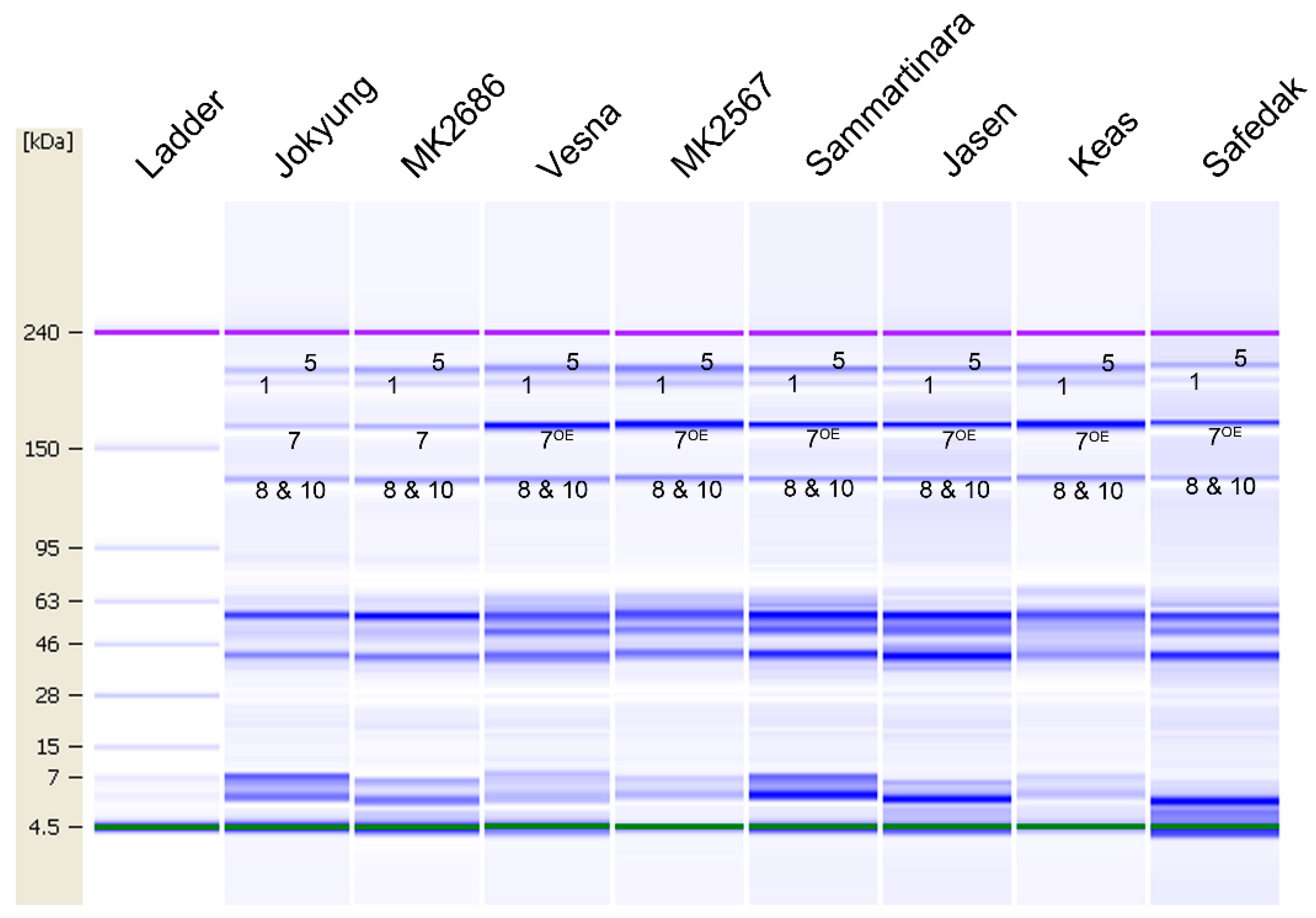
2.5. HMW-GS Composition Identification of Genetic Resources by Lab-on-a-Chip
3. Discussion
4. Materials and Methods
4.1. HMW-GS Composition Identification of Genetic Resources by Lab-on-a-Chip
4.2. Glutenin Proteins Extraction and HMW-GS Composition Identification of Genetic Resources by SDS-PAGE
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhang, Y.; Hu, M.; Liu, Q.; Sun, L.; Chen, X.; Lv, L.; Liu, Y.; Jia, X.; Li, H. Deletion of high-molecular-weight glutenin subunits in wheat significantly reduced dough strength and bread-baking quality. BMC Plant Biol. 2018, 18, 319. [Google Scholar] [CrossRef] [PubMed]
- Payne, P.I. Genetics of wheat storage proteins and the effect of allelic variation on bread-making quality. Annu. Rev. Plant Physiol. 1987, 38, 141–153. [Google Scholar] [CrossRef]
- Gao, L.; Ma, W.; Chen, J.; Wang, K.; Li, J.; Wang, S.; Bekes, F.; Appels, R.; Yan, Y. Characterization and comparative analysis of wheat high molecular weight glutenin subunits by SDS-PAGE, RP-HPLC, HPCE, and MALDI-TOF-MS. J. Agric. Food Chem. 2010, 58, 2777–2786. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, K.; Xiao, L.; Wang, S.; Du, L.; Cao, X.; Zhang, X.; Zhou, Y.; Yan, Y.; Ye, X. Comprehensive Identification and Bread-Making Quality Evaluation of Common Wheat Somatic Variation Line AS208 on Glutenin Composition. PLoS ONE 2016, 11, e0146933. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jang, Y.R.; Beom, H.R.; Altenbach, S.B.; Lee, M.K.; Lim, S.H.; Lee, J.Y. Improved Method for Reliable HMW-GS Identification by RP-HPLC and SDS-PAGE in Common Wheat Cultivars. Molecules 2017, 22, 1055. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Huang, L.; Gong, F.; Liu, J.; Wang, Y.; Jin, Y.; He, Y.; He, J.; Jiang, Q.; Zheng, Y.; et al. Enriching LMW-GS alleles and strengthening gluten properties of common wheat through wide hybridization with wild emmer. 3 Biotech 2019, 9, 355. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Song, S.; Zhao, L.; Shen, L.; Song, Y.; Wang, X.; Yu, K.; Liu, Z.; Li, Y.; Yang, W. Mechanisms, origin and heredity of Glu-1Ay silencing in wheat evolution and domestication. Theor. Appl. Genet. 2018, 131, 1561–1575. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Inazu, A.; Hirano, H. Allelic variation in high-molecular-weight glutenin subunit loci of Glu-1 in Japanese common wheats. Euphtica 1999, 106, 131–138. [Google Scholar] [CrossRef]
- Geisslitz, S.; America, A.H.; Scherf, K.A. Mass spectrometry of in-gel digests reveals differences in amino acid sequences of high-molecular-weight glutenin subunits in spelt and emmer compared to common wheat. Anal. Bioanal. Chem. 2020, 412, 1277–1289. [Google Scholar] [CrossRef]
- Jackson, E.A.; Holt, L.M.; Payne, P.I. Characterisation of high molecular weight gliadin and low-molecular-weight glutenin subunits of wheat endosperm by two-dimensional electrophoresis and the chromosomal localisation of their controlling genes. Theor. Appl. Genet. 1983, 66, 29–37. [Google Scholar] [CrossRef]
- Payne, P.I.; Holt, L.M.; Lawrence, G.J.; Law, C.N. The genetics of gliadin and glutenin, the major storage proteins of the wheat endosperm. Plant Foods Hum. Nutr. 1982, 31, 229–241. [Google Scholar] [CrossRef]
- Yasmeen, F.; Khurshid, H.; Ghafoor, A. Genetic divergence for high-molecular weight glutenin subunits (HMW-GS) in indigenous landraces and commercial cultivars of bread wheat of Pakistan. Genet. Mol. Res. 2015, 14, 4829–4839. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Sun, G.; Liu, W.; Sun, D.; Peng, Y.; Ren, X. High-molecular-weight glutenin subunit compositions in current Chinese commercial wheat cultivars and the implication on Chinese wheat breeding for quality. Cereal Chem. 2020, 97, 762–771. [Google Scholar] [CrossRef]
- Fu, X.; Jia, D.; Li, M.; He, M.; Shi, Z.; Cai, X.; Guo, J. Composition analysis of HMW-GS in Australian wheat cultivars. Hunan Agric. Sci. Technol. 2011, 12, 1010–1012. [Google Scholar]
- Zheng, W.; Li, S.; Liu, Z.; Zhou, Q.; Feng, Y.; Chai, S. Molecular marker assisted gene stacking for disease resistance and quality genes in the dwarf mutant of an elite common wheat cultivar Xiaoyan22. BMC Genet. 2020, 21, 45. [Google Scholar] [CrossRef]
- Vázquez, D.; Berger, A.G.; Cuniberti, M.; Bainotti, C.; de Miranda, M.Z.; Scheeren, P.L.; Jobet, C.; Zúñiga, J.; Cabrera, G.; Verges, R. Influence of cultivar and environment on quality of Latin American wheats. J. Cereal Sci. 2012, 56, 196–203. [Google Scholar] [CrossRef]
- Tabiki, T.; Ikeguchi, S.; Ikeda, T.M. Effects of high-molecular-weight and low-molecular-weight glutenin subunit alleles on common wheat flour quality. Breed. Sci. 2006, 56, 131–136. [Google Scholar] [CrossRef][Green Version]
- Guo, H.; Wu, J.; Lu, Y.; Yan, Y. High-Molecular-Weight Glutenin 1Bx17 and 1By18 Subunits Encoded by Glu-B1i Enhance Rheological Properties and Breadmaking Quality of Wheat Dough. J. Food Qual. 2019, 2019, 1958747. [Google Scholar] [CrossRef]
- Ragupathy, R.; Naeem, H.A.; Reimer, E.; Lukow, O.M.; Sapirstein, H.D.; Cloutier, S. Evolutionary origin of the segmental duplication encompassing the wheat GLU-B1 locus encoding the overexpressed Bx7 (Bx7OE) high molecular weight glutenin subunit. Theor. Appl. Genet. 2008, 116, 283–296. [Google Scholar] [CrossRef]
- Geng, Y.; Pang, B.; Hao, C.; Tang, S.; Zhang, X.; Li, T. Expression of wheat high molecular weight glutenin subunit 1Bx is affected by large insertions and deletions located in the upstream flanking sequences. PLoS ONE 2014, 9, e105363. [Google Scholar] [CrossRef]
- Cooper, J.K.; Stromberger, J.A.; Morris, C.F.; Bai, G.; Haley, S.D. End-Use Quality and Agronomic Characteristics Associated with the Glu-B1al High-Molecular-Weight Glutenin Allele in US Hard Winter Wheat. Crop Sci. 2016, 56, 2348–2353. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, C.; Song, W.; Zhang, Y.; Gao, D.; Zhang, X.; Zhao, L.; Yang, X.; Liu, D.; Song, Q. Quality differences between NILs of wheat variety Longmai 20 possessing HMW-GS 7 OE+ 8* and 17+ 18. Cereal Res. Commun. 2020, 1–6. [Google Scholar] [CrossRef]
- Wang, G.; Snape, J.; Hu, H.; Rogers, W. The high-molecular-weight glutenin subunit compositions of Chinese bread wheat varieties and their relationship with bread-making quality. Euphytica 1993, 68, 205–212. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, Y.; Zhang, B.; Florides, C.G.; Gao, Z.; Wang, Z.; Zhang, X.; Wei, Y. Comparison of quality properties between high-molecular-weight glutenin subunits 5+ 10 and 2+ 12 near-isogenic lines under three common wheat genetic backgrounds. Cereal Chem. 2018, 95, 575–583. [Google Scholar] [CrossRef]
- Hernández-Estrada, Z.J.; Rayas-Duarte, P.; Cárdenas, J.D.D.F. Creep recovery of wet gluten and high-molecular-weight glutenin subunit composition: Relationship with viscoelasticity of dough and breadmaking quality of hard red winter wheat. Cereal Chem. 2017, 94, 223–229. [Google Scholar] [CrossRef]
- Payne, P.I.; Lawrence, G.J. Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1, and Glu-D1 which code for high-molecular-weight subunits of glutenin in hexaploid wheat. Cereal Res. Commun. 1983, 11, 29–35. [Google Scholar]
- Courcoux, P.; Serot, T.; Larre, C.; Popineau, Y. Characterization and identification of wheat cultivars by multi-dimensional analysis of reversed-phase high-performance liquid chromatograms. J. Chromatogr. A 1992, 596, 225–232. [Google Scholar] [CrossRef]
- Wang, S.; Yu, Z.; Cao, M.; Shen, X.; Li, N.; Li, X.; Ma, W.; Weissgerber, H.; Zeller, F.; Hsam, S.; et al. Molecular mechanisms of HMW glutenin subunits from 1S(l) genome of Aegilops longissima positively affecting wheat breadmaking quality. PLoS ONE 2013, 8, e58947. [Google Scholar]
- Peng, Y.; Yu, K.; Zhang, Y.; Islam, S.; Sun, D.; Ma, W. Two Novel Y-Type High Molecular Weight Glutenin Genes in Chinese Wheat Landraces of the Yangtze-River Region. PLoS ONE 2015, 10, e0142348. [Google Scholar] [CrossRef]
- Yan, Y.; Hsam, S.; Yu, J.; Jiang, Y.; Zeller, F. Allelic variation of the HMW glutenin subunits in Aegilops tauschii accessions detected by sodium dodecyl sulphate, acid polyacrylamide gel and capillary electrophoresis. Euphytica 2003, 130, 377–385. [Google Scholar] [CrossRef]
- Živančev, D.; Horvat, D.; Torbica, A.; Belović, M.; Šimić, G.; Magdić, D.; Đukić, N. Benefits and limitations of lab-on-a-chip method over reversed-phase high-performance liquid chromatography method in gluten proteins evaluation. J. Chem. 2015, 2015, 430328. [Google Scholar] [CrossRef]
- Rasheed, A.; Wen, W.; Gao, F.; Zhai, S.; Jin, H.; Liu, J.; Guo, Q.; Zhang, Y.; Dreisigacker, S.; Xia, X.; et al. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor. Appl. Genet. 2016, 129, 1843–1860. [Google Scholar] [CrossRef]
- Ravel, C.; Faye, A.; Ben-Sadoun, S.; Ranoux, M.; Dardevet, M.; Dupuits, C.; Exbrayat, F.; Poncet, C.; Sourdille, P.; Branlard, G. SNP markers for early identification of high molecular weight glutenin subunits (HMW-GSs) in bread wheat. Theor. Appl. Genet. 2020, 133, 751–770. [Google Scholar] [CrossRef]
- Liu, L.; Wang, A.; Appels, R.; Ma, J.; Xia, X.; Lan, P.; He, Z.; Bekes, F.; Yan, Y.; Ma, W. A MALDI-TOF based analysis of high molecular weight glutenin subunits for wheat breeding. J. Cereal Sci. 2009, 50, 295–301. [Google Scholar] [CrossRef]
- Shin, D.; Cha, J.K.; Lee, S.M.; Ko, J.M.; Lee, J.H. Validation and selection of functional allele-specific molecular markers to analyze High-Molecular-Weight glutenin subunit composition in wheat. Korean J. Breed. Sci. 2020, 52, 235–243. [Google Scholar] [CrossRef]
- Živančev, D.R.; Torbica, A.M.; Tomić, J.M.; Janić Hajnal, E.P.; Belović, M.M.; Mastilović, J.S.; Kevrešan, Ž.S. Effect of Climate Change on Wheat Quality and HMW Glutenin Subunit Composition in the Pannonian Plain. Cereal Chem. 2016, 93, 90–99. [Google Scholar] [CrossRef]
- Rhazi, L.; Bodard, A.L.; Fathollahi, B.; Aussenac, T. High throughput microchip-based separation and quantitation of high-molecular-weight glutenin subunits. J. Cereal Sci. 2009, 49, 272–277. [Google Scholar] [CrossRef]
- Uthayakumaran, S.; Batey, I.L.; Wrigley, C.W. On-the-spot identification of grain variety and wheat-quality type by Lab-on-a-chip capillary electrophoresis. J. Cereal Sci. 2005, 41, 371–374. [Google Scholar] [CrossRef]
- Uthayakumaran, S.; Listiohadi, Y.; Baratta, M.; Batey, I.L.; Wrigley, C.W. Rapid identification and quantitation of high-molecular-weight glutenin subunits. J. Cereal Sci. 2006, 44, 34–39. [Google Scholar] [CrossRef]
- Balázs, G.; Tömösközi, S.; Harasztos, A.; Németh, V.; Tamás, Á.; Morgounov, A.; Belan, I.; Ma, W.; Békés, F. Advantages and Limitation of Lab-on-a-chip Technique in the Analysis of Wheat Proteins. Cereal Res. Commun. 2012, 40, 562–572. [Google Scholar] [CrossRef]
- Bradová, J.; Matějová, E. Comparison of the results of SDS PAGE and chip electrophoresis of wheat storage proteins. Chromatographia 2008, 67, 83–88. [Google Scholar] [CrossRef]
- Rasheed, A.; Safdar, T.; Gul-Kazi, A.; Mahmood, T.; Akram, Z.; Mujeeb-Kazi, A. Characterization of HMW-GS and evaluation of their diversity in morphologically elite synthetic hexaploid wheats. Breed. Sci. 2012, 62, 365–370. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Van den Broeck, H.C.; America, A.H.; Smulders, M.J.; Bosch, D.; Hamer, R.J.; Gilissen, L.J.; van der Meer, I.M. A modified extraction protocol enables detection and quantification of celiac disease-related gluten proteins from wheat. J. Chromatogr. B 2009, 877, 975–982. [Google Scholar] [CrossRef]
- Hlozáková, T.K.; Gregová, E.; Šliková, S.; Gálová, Z.; Chňapek, M.; Drábeková, J. Determination of HMW–GS in wheat using SDS–PAGE and Lab-on-chip methods. Potravin. Slovak J. Food Sci. 2019, 13, 477–481. [Google Scholar] [CrossRef]
- Nakamura, H.; Sasaki, H.; Hirano, H.; Yamashita, A. A high molecular weight subunit of wheat glutenin seed storage protein correlates with its flour quality. Jpn. J. Breed. 1990, 40, 485–494. [Google Scholar] [CrossRef]
- He, Z.H.; Liu, L.; Xia, X.C.; Liu, J.J.; Peña, R.J. Composition of HMW and LMW glutenin subunits and their effects on dough properties, pan bread, and noodle quality of Chinese bread wheats. Cereal Chem. 2005, 82, 345–350. [Google Scholar] [CrossRef]

| Variety Name | Glu-1 Locus Allele | SDS-PAGE Allele | Reference | ||||
|---|---|---|---|---|---|---|---|
| Glu-A1 | Glu-B1 | Glu-D1 | Glu-A1 | Glu-B1 | Glu-D1 | ||
| Jokyung | a | b | d | 1 | 7 + 8 | 5 + 10 | Jang et al. [5] |
| Keumgang | b | b | d | 2* | 7 + 8 | 5 + 10 | Jang et al. [5] |
| Uri | c | b | f | null | 7 + 8 | 2.2 + 12 | Jang et al. [5] |
| Petrel | c | a | d | null | 7 | 5 + 10 | Liu et al. [34] |
| Seodun | c | b | f | null | 7 + 8 | 2.2 + 12 | Jang et al. [5] |
| Anbaek | c | c | a | null | 7 + 9 | 2 + 12 | Jang et al. [5] |
| Joeun | c | f | f | null | 13 + 16 | 2.2 + 12 | Jang et al. [5] |
| Joongmo2008 | c | i | d | null | 17 + 18 | 5 + 10 | Jang et al. [5] |
| Vesna | a | al | d | 1 | 7OE + 8 | 5 + 10 | Shin et al. [35] |
| HMW-GS | Saekeumgang | Chapingo | Hanbaek |
|---|---|---|---|
| 1Ax2* | 11.5 ± 2.68 | 13.7 ± 1.26 | 11.0 ± 0.98 |
| 1Dx5 | - | 33.7 ± 3.29 | 34.3 ± 5.11 |
| 1Dx2.2 | 23.7 ± 5.08 | - | - |
| 1Bx7 | 31.3 ± 5.56 | 27.5 ± 1.85 | 25.8 ± 1.63 |
| 1By8 | 13.1 ± 3.29 | - | - |
| 1By9 | - | 9.9 ± 1.98 | - |
| 8 & 10 | - | - | 29.0 ± 5.28 |
| 1Dy10 | - | 15.2 ± 3.04 | - |
| 1Dy12 | 20.4 ± 5.49 | - | - |
| HMW-GS | Chapingo | Garnet | Vesna | MK4060 |
|---|---|---|---|---|
| 1Ax1 | - | - | 9.5 ± 1.12 | - |
| 1Ax2* | 13.7 ± 1.26 | 6.4 ± 0.34 | - | 9.8 ± 1.12 |
| 1Bx7 | 27.5 ± 1.85 | - | - | - |
| 1Bx17 | - | 30.2 ± 0.94 | - | 30.2 ± 2.62 |
| 1Bx7OE | - | - | 45.3 ± 2.71 | - |
| 1By9 | 9.9 ± 1.98 | - | - | - |
| 8&10 | - | - | 20.4 ± 1.28 | |
| 10&18 | - | 29.2 ± 4.25 | - | 28.9 ± 2.15 |
| 1Dx5 | 33.7 ± 3.29 | 34.3 ± 3.11 | 24.8 ± 1.06 | 31.1 ± 0.96 |
| 1Dy10 | 15.2 ± 3.04 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, D.; Cha, J.-K.; Lee, S.-M.; Kabange, N.R.; Lee, J.-H. Rapid and Easy High-Molecular-Weight Glutenin Subunit Identification System by Lab-on-a-Chip in Wheat (Triticum aestivum L.). Plants 2020, 9, 1517. https://doi.org/10.3390/plants9111517
Shin D, Cha J-K, Lee S-M, Kabange NR, Lee J-H. Rapid and Easy High-Molecular-Weight Glutenin Subunit Identification System by Lab-on-a-Chip in Wheat (Triticum aestivum L.). Plants. 2020; 9(11):1517. https://doi.org/10.3390/plants9111517
Chicago/Turabian StyleShin, Dongjin, Jin-Kyung Cha, So-Myeong Lee, Nkulu Rolly Kabange, and Jong-Hee Lee. 2020. "Rapid and Easy High-Molecular-Weight Glutenin Subunit Identification System by Lab-on-a-Chip in Wheat (Triticum aestivum L.)" Plants 9, no. 11: 1517. https://doi.org/10.3390/plants9111517
APA StyleShin, D., Cha, J.-K., Lee, S.-M., Kabange, N. R., & Lee, J.-H. (2020). Rapid and Easy High-Molecular-Weight Glutenin Subunit Identification System by Lab-on-a-Chip in Wheat (Triticum aestivum L.). Plants, 9(11), 1517. https://doi.org/10.3390/plants9111517