Physiological and Transcriptomic Response of Grey Poplar (Populus ×canescens Aiton Sm.) to Cadmium Stress
Abstract
1. Introduction
2. Results and Discussion
2.1. Microsatellite Analysis
2.2. Effect of Cd on Plant Growth and Chlorophyll Content
2.3. Effect of Cd on K, Ca, Mg and Zn Uptake and Translocation
2.4. Cadmium Accumulation, Translocation and TF
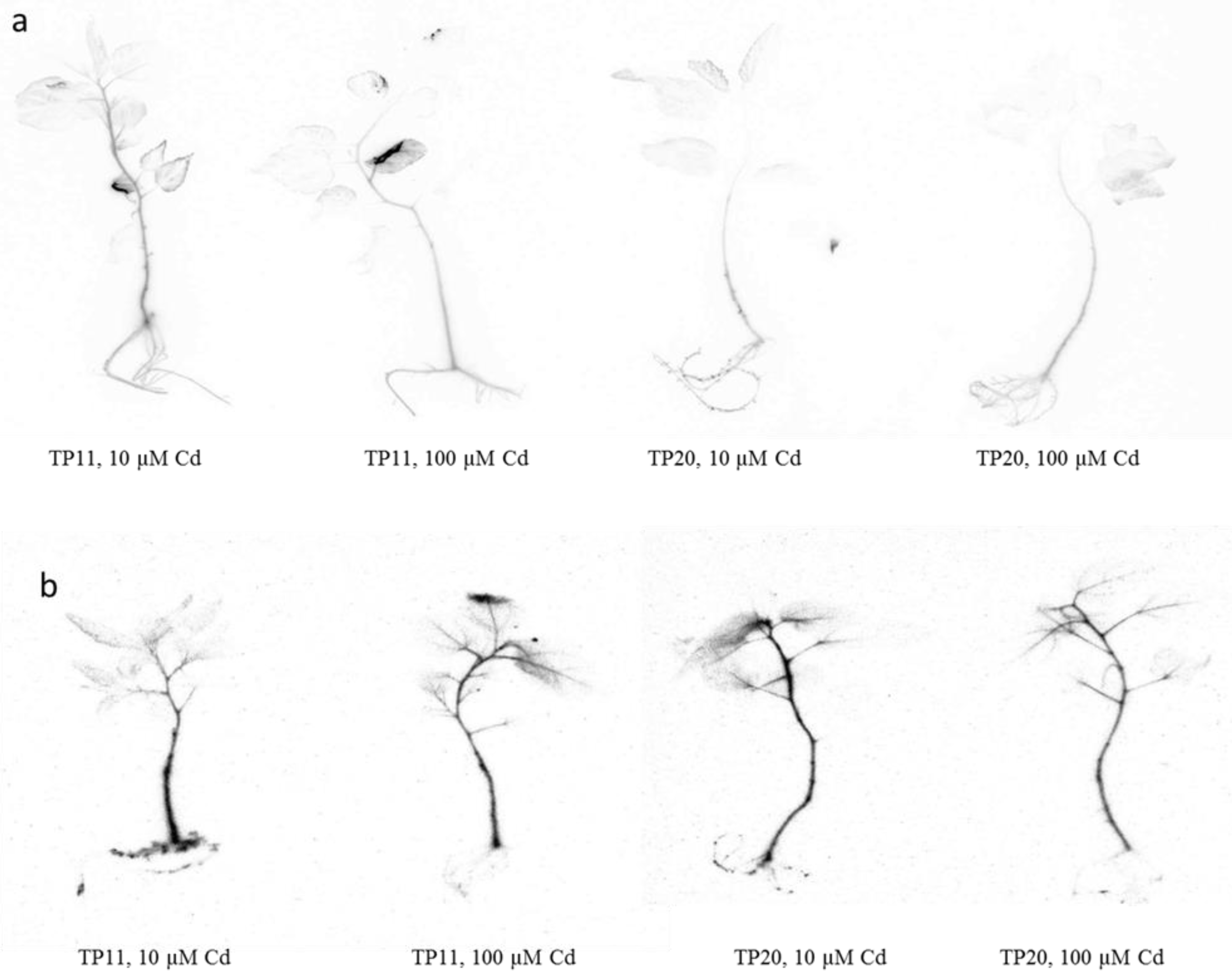
2.5. Cadmium Localization in Grey Poplar Plants
2.6. Analysis of Gene Expression Patterns
3. Materials and Methods
3.1. Plant Material and In Vitro Culture Establishment
3.2. Root Induction and Acclimatization
3.3. DNA Extraction, PCR and Genotyping
3.4. RNA Extraction and cDNA Synthesis
3.5. Gene Expression Analysis
3.6. Measurement of Cd and Selected Element Contents
3.7. Chlorophyll Measurement
3.8. Autoradiography Method
3.9. Data Analysis
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, D.; Yamaji, N.; Yamane, M.; Kashino-Fujii, M.; Sato, K.; Ma, J.F. The HvNramp5 Transporter Mediates Uptake of Cadmium and Manganese, But Not Iron. Plant Physiol. 2016, 172, 1899–1910. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhao, H.; Wu, L.; Liu, A.; Zhao, F.-J.; Xu, W. Heavy metal ATPase 3 (HMA3) confers cadmium hypertolerance on the cadmium/zinc hyperaccumulatorSedum plumbizincicola. New Phytol. 2017, 215, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Martinoia, E.; Lee, Y. Vacuolar Transporters for Cadmium and Arsenic in Plants and their Applications in Phytoremediation and Crop Development. Plant Cell Physiol. 2018, 59, 1317–1325. [Google Scholar] [CrossRef]
- Garmash, E.; Golovko, T.K. Effect of cadmium on growth and respiration of barley plants grown under two temperature regimes. Russ. J. Plant Physiol. 2009, 56, 343–347. [Google Scholar] [CrossRef]
- Li, S.; Yang, W.; Yang, T.; Chen, Y.; Ni, W. Effects of Cadmium Stress on Leaf Chlorophyll Fluorescence and Photosynthesis ofElsholtzia argyi—A Cadmium Accumulating Plant. Int. J. Phytoremediat. 2014, 17, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhou, W.; Dai, H.; Cao, F.; Zhang, G.; Wu, F. Selenium reduces cadmium uptake and mitigates cadmium toxicity in rice. J. Hazard. Mater. 2012, 236, 343–351. [Google Scholar] [CrossRef]
- Nazar, R.; Iqbal, N.; Masood, A.; Khan, M.I.R.; Syeed, S.; Khan, N.A. Cadmium Toxicity in Plants and Role of Mineral Nutrients in Its Alleviation. Am. J. Plant Sci. 2012, 3, 1476–1489. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 1–26. [Google Scholar] [CrossRef]
- Cui, W.; Wang, H.; Song, J.; Cao, X.; Rogers, H.J.; Francis, D.; Jia, C.; Sun, L.; Hou, M.; Yang, Y.; et al. Cell cycle arrest mediated by Cd-induced DNA damage in Arabidopsis root tips. Ecotoxicol. Environ. Saf. 2017, 145, 569–574. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Maistri, S.; Furini, A. How Plants Cope with Cadmium: Staking All on Metabolism and Gene Expression. J. Integr. Plant Biol. 2008, 50, 1268–1280. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, H.; Fukuoka, H.; Arao, T.; Ohyama, A.; Nunome, T.; Miyatake, K.; Negoro, S. Gene expression analysis in cadmium-stressed roots of a low cadmium-accumulating solanaceous plant, Solanum torvum. J. Exp. Bot. 2009, 61, 423–437. [Google Scholar] [CrossRef]
- Garbisu, C.; Hernández-Allica, J.; Barrutia, O.; Alkorta, I.; Becerril, J.M. Phytoremediation: A Technology Using Green Plants to Remove Contaminants from Polluted Areas. Rev. Environ. Health 2002, 17, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.D.; Van Der Ent, A.; Baker, A.J.M. Global Distribution and Ecology of Hyperaccumulator Plants. In Agromining: Farming for Metals; Springer: Cham, Switzerland, 2017; pp. 75–92. [Google Scholar]
- Brown, S.L.; Chaney, R.L.; Angle, J.S.; Baker, A.J.M. Zinc and Cadmium Uptake by Hyperaccumulator Thlaspi caerulescens and Metal Tolerant Silene vulgaris Grown on Sludge-Amended Soils. Environ. Sci. Technol. 1995, 29, 1581–1585. [Google Scholar] [CrossRef]
- Hernández-Allica, J.; Becerril, J.M.; Zárate, O.; Garbisu, C. Assessment of the Efficiency of a Metal Phytoextraction Process with Biological Indicators of Soil Health. Plant Soil 2006, 281, 147–158. [Google Scholar] [CrossRef]
- Epelde, L.; Becerril, J.M.; Kowalchuk, G.A.; Deng, Y.; Zhou, J.; Garbisu, C. Impact of Metal Pollution and Thlaspi caerulescens Growth on Soil Microbial Communities. Appl. Environ. Microbiol. 2010, 76, 7843–7853. [Google Scholar] [CrossRef]
- Broadhurst, C.L.; Chaney, R.L. Growth and Metal Accumulation of an Alyssum murale Nickel Hyperaccumulator Ecotype Co-cropped with Alyssum montanum and Perennial Ryegrass in Serpentine Soil. Front. Plant Sci. 2016, 7, 451. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Miyauchi, K.; Inoue, C.; Endo, G. Development of suitable hydroponics system for phytoremediation of arsenic-contaminated water using an arsenic hyperaccumulator plant Pteris vittata. Biosci. Biotechnol. Biochem. 2015, 80, 1–5. [Google Scholar] [CrossRef]
- Tlustoš, P.; Břendová, K.; Száková, J.; Najmanová, J.; Koubová, K. The long-term variation of Cd and Zn hyperaccumulation by Noccaea spp and Arabidopsis halleri plants in both pot and field conditions. Int. J. Phytoremediat. 2016, 18, 110–115. [Google Scholar] [CrossRef]
- He, J.; Ma, C.; Ma, Y.; Li, H.; Kang, J.; Liu, T.; Polle, A.; Peng, C.; Luo, Z.-B. Cadmium tolerance in six poplar species. Environ. Sci. Pollut. Res. 2013, 20, 163–174. [Google Scholar] [CrossRef]
- Martinsen, G.D.; Whitham, T.G.; Turek, R.J.; Keim, P. Hybrid populations selectively filter gene introgression between species. Evol. Int. J. Org. Evol. 2001, 55, 1325–1335. [Google Scholar] [CrossRef]
- Lexer, C.; Fay, M.F.; Joseph, J.A.; Nica, M.-S.; Heinze, B. Barrier to gene flow between two ecologically divergent Populus species, P. alba (white poplar) and P. tremula (European aspen): The role of ecology and life history in gene introgression. Mol. Ecol. 2005, 14, 1045–1057. [Google Scholar] [CrossRef]
- Suvanto, L.I.; Latva-Karjanmaa, T.B. Clone identification and clonal structure of the European aspen (Populus tremula). Mol. Ecol. 2005, 14, 2851–2860. [Google Scholar] [CrossRef]
- Puschenreiter, M.; Türktaş, M.; Sommer, P.; Wieshammer, G.; Laaha, G.; Wenzel, W.W.; Hauser, M.-T. Differentiation of metallicolous and non-metallicolous Salix caprea populations based on phenotypic characteristics and nuclear microsatellite (SSR) markers. Plant Cell Environ. 2010, 33, 1641–1655. [Google Scholar] [CrossRef] [PubMed]
- Anwar, G.M.; Helmey, R.K.; Mostafa, Y.M. Assesment of genetic diversityin garlic clones using SSR and ISSR markers. Egypt. J. Genet. Cytol. 2017, 45, 333–345. Available online: http://journal.esg.net.eg/index.php/EJGC/article/view/233 (accessed on 22 July 2019). [CrossRef]
- Dimsoski, P.; Toth, G.P. Development of DNA-based microsatellite marker technology for studies of genetic diversity in stressor impacted populations. Ecotoxicology 2001, 10, 229–232. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.D.F. Microsatellite markers: What they mean and why they are so useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Landhäusser, S.M.; Lieffers, V.J.; Zwiazek, J.J. Signals controlling root suckering and adventitious shoot formation in aspen (Populus tremuloides). Tree Physiol. 2006, 26, 681–687. [Google Scholar] [CrossRef]
- Baryla, A.; Carrier, P.; Franck, F.; Coulomb, C.; Sahut, C.; Havaux, M. Leaf chlorosis in oilseed rape plants (Brassica napus) grown on cadmium-polluted soil: Causes and consequences for photosynthesis and growth. Planta 2001, 212, 696–709. [Google Scholar] [CrossRef] [PubMed]
- Paunov, M.; Koleva, L.; Vassilev, A.; Vangronsveld, J.; Goltsev, V. Effects of Different Metals on Photosynthesis: Cadmium and Zinc Affect Chlorophyll Fluorescence in Durum Wheat. Int. J. Mol. Sci. 2018, 19, 787. [Google Scholar] [CrossRef]
- Ondrasek, G.; Rengel, Z.; Romic, D. Humic acids decrease uptake and distribution of trace metals, but not the growth of radish exposed to cadmium toxicity. Ecotoxicol. Environ. Saf. 2018, 151, 55–61. [Google Scholar] [CrossRef]
- Zhan, Y.-H.; Zhang, C.; Zheng, Q.-X.; Huang, Z.-A.; Yu, C.-L. Cadmium stress inhibits the growth of primary roots by interfering auxin homeostasis in Sorghum bicolor seedlings. J. Plant Biol. 2017, 60, 593–603. [Google Scholar] [CrossRef]
- Schützendübel, A.; Nikolova, P.; Rudolf, C.; Polle, A. Cadmium and H2O2-induced oxidative stress in Populus × canescens roots. Plant Physiol. Biochem. 2002, 40, 577–584. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, J.; Zhang, J. Effects of external phosphorus on the cell ultrastructure and the chlorophyll content of maize under cadmium and zinc stress. Environ. Pollut. 2007, 147, 750–756. [Google Scholar] [CrossRef]
- Kapoor, D.; Kaur, S.; Bhardwaj, R. Physiological and Biochemical Changes inBrassica junceaPlants under Cd-Induced Stress. BioMed Res. Int. 2014, 2014, 1–13. [Google Scholar] [CrossRef]
- Dotaniya, M.L.; Meena, V.D.; Basak, B.B.; Meena, R.S. Potassium Uptake by Crops as Well as Microorganisms. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Springer: New Delhi, India, 2016; pp. 267–280. [Google Scholar]
- Sadeghipour, O. Enhancing Cadmium Tolerance in Common Bean Plants by Potassium Application. Philipp. Agric. Sci. 2018, 101, 167–175. [Google Scholar]
- Liu, S.; Yang, R.; Tripathi, D.K.; Li, X.; Jiang, M.; Lv, B.; Ma, M.; Qibing, C. RETRACTED: Signalling cross-talk between nitric oxide and active oxygen in Trifolium repens L. plants responses to cadmium stress. Environ. Pollut. 2018, 239, 53–68. [Google Scholar] [CrossRef]
- Zhang, G.; Fukami, M.; Sekimoto, H. Influence of cadmium on mineral concentrations and yield components in wheat genotypes differing in Cd tolerance at seedling stage. Field Crop. Res. 2002, 77, 93–98. [Google Scholar] [CrossRef]
- Rivetta, A.; Negrini, N.; Cocucci, M. Involvement of Ca2+-calmodulin in Cd2+ toxicity during the early phases of radish (Raphanus sativus L.) seed germination. Plant Cell Environ. 1997, 20, 600–608. [Google Scholar] [CrossRef]
- Tkalec, M.; Štefanić, P.P.; Cvjetko, P.; Šikić, S.; Pavlica, M.; Balen, B. The Effects of Cadmium-Zinc Interactions on Biochemical Responses in Tobacco Seedlings and Adult Plants. PLoS ONE 2014, 9, e87582. [Google Scholar] [CrossRef]
- Courbot, M.; Willems, G.; Motte, P.; Arvidsson, S.; Roosens, N.; Saumitou-Laprade, P.; Verbruggen, N. A Major Quantitative Trait Locus for Cadmium Tolerance in Arabidopsis halleri Colocalizes with HMA4, a Gene Encoding a Heavy Metal ATPase. Plant Physiol. 2007, 144, 1052–1065. [Google Scholar] [CrossRef]
- Morel, M.; Crouzet, J.; Gravot, A.; Auroy, P.; Leonhardt, N.; Vavasseur, A.; Richaud, P. AtHMA3, a P1B-ATPase Allowing Cd/Zn/Co/Pb Vacuolar Storage in Arabidopsis. Plant Physiol. 2008, 149, 894–904. [Google Scholar] [CrossRef]
- Di Lonardo, S.; Capuana, M.; Arnetoli, M.; Gabbrielli, R.; Gonnelli, C. Exploring the metal phytoremediation potential of three Populus alba L. clones using an in vitro screening. Environ. Sci. Pollut. Res. 2010, 18, 82–90. [Google Scholar] [CrossRef]
- Nikolić, N.; Zoric, L.; Cvetković, I.; Pajević, S.; Borišev, M.; Orlović, S. Assessment of cadmium tolerance and phytoextraction ability in young Populus deltoides L. and Populus × euramericana plants through morpho-anatomical and physiological responses to growth in cadmium enriched soil. IForest-Biogeosci. For. 2017, 10, 635–644. [Google Scholar] [CrossRef]
- Redovniković, I.R.; De Marco, A.; Proietti, C.; Hanousek, K.; Sedak, M.; Bilandžić, N.; JakovljeviĆ, T. Poplar response to cadmium and lead soil contamination. Ecotoxicol. Environ. Saf. 2017, 144, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Pietrini, F.; Zacchini, M.; Iori, V.; Pietrosanti, L.; Ferretti, M.; Massacci, A. Spatial distribution of cadmium in leaves and its impact on photosynthesis: Examples of different strategies in willow and poplar clones. Plant Biol. 2009, 12, 355–363. [Google Scholar] [CrossRef]
- Vollenweider, P.; Cosio, C.; Günthardt-Goerg, M.S.; Keller, C. Localization and effects of cadmium in leaves of a cadmium-tolerant willow (Salix viminalis L.). Environ. Exp. Bot. 2006, 58, 25–40. [Google Scholar] [CrossRef]
- Cosio, C.; DeSantis, L.; Frey, B.; Diallo, S.; Keller, C. Distribution of cadmium in leaves of Thlaspi caerulescens. J. Exp. Bot. 2005, 56, 765–775. [Google Scholar] [CrossRef]
- Kumar, M.; Brar, A.; Yadav, M.; Chawade, A.; Vivekanand, V.; Pareek, N. Chitinases—Potential Candidates for Enhanced Plant Resistance towards Fungal Pathogens. Agriculture 2018, 8, 88. [Google Scholar] [CrossRef]
- Cao, J.; Tan, X. Comprehensive Analysis of the Chitinase Family Genes in Tomato (Solanum lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Békésiová, B.; Hraška, Š.; Libantova, J.; Moravčíková, J.; Matušíková, I. Heavy-metal stress induced accumulation of chitinase isoforms in plants. Mol. Biol. Rep. 2007, 35, 579–588. [Google Scholar] [CrossRef]
- Gálusová, T.; Rybanský, Ľ.; Mészáros, P.; Spieß, N.; Piršelová, B.; Kuna, R.; Libantova, J.; Moravčíková, J.; Hauptvogel, P.; Matušíková, I. Variable responses of soybean chitinases to arsenic and cadmium stress at the whole plant level. Plant Growth Regul. 2014, 76, 147–155. [Google Scholar] [CrossRef]
- Breithaupt, C.; Kurzbauer, R.; Schaller, F.; Stintzi, A.; Schaller, A.; Huber, R.; Macheroux, P.; Clausen, T. Structural Basis of Substrate Specificity of Plant 12-Oxophytodienoate Reductases. J. Mol. Biol. 2009, 392, 1266–1277. [Google Scholar] [CrossRef]
- Xiang, C.-B.; Oliver, D.J. Glutathione Metabolic Genes Coordinately Respond to Heavy Metals and Jasmonic Acid in Arabidopsis. Plant Cell 1998, 10, 1539. [Google Scholar] [CrossRef]
- Maksymiec, W.; Krupa, Z. The effects of short-term exposition to Cd, excess Cu ions and jasmonate on oxidative stress appearing in Arabidopsis thaliana. Environ. Exp. Bot. 2006, 57, 187–194. [Google Scholar] [CrossRef]
- Liu, T.; Liu, S.; Guan, H.; Ma, L.; Chen, Z.; Gu, H.; Qu, L. Transcriptional profiling of Arabidopsis seedlings in response to heavy metal lead (Pb). Environ. Exp. Bot. 2009, 67, 377–386. [Google Scholar] [CrossRef]
- Velazhahan, R.; Datta, S.K.; Muthukrishnan, S. The PR-5 family: Thaumatin-like proteins. In Pathogenesis-Related Proteins in Plants; CRC Press: Boca Raton, FL, USA, 1999; pp. 107–129. [Google Scholar] [CrossRef]
- Sarowar, S.; Kim, Y.J.; Kim, E.N.; Kim, K.D.; Hwang, B.K.; Islam, R.; Shin, J.S. Overexpression of a pepper basic pathogenesis-related protein 1 gene in tobacco plants enhances resistance to heavy metal and pathogen stresses. Plant Cell Rep. 2005, 24, 216–224. [Google Scholar] [CrossRef]
- Fecht-Christoffers, M.M.; Braun, H.-P.; Lemaitre-Guillier, C.; Vandorsselaer, A.; Horst, W.J. Effect of Manganese Toxicity on the Proteome of the Leaf Apoplast in Cowpea. Plant Physiol. 2003, 133, 1935–1946. [Google Scholar] [CrossRef]
- Júnior, W.V.A.; Neto, C.F.D.O.; Filho, B.G.D.S.; Amarante, C.B.D.; Cruz, E.D.; Okumura, R.S.; Barbosa, A.V.C.; De Sousa, D.J.P.; Teixeira, J.; Botelho, A.D.S. Effect of cadmium on young plants of Virola surinamensis. AoB Plants 2019, 11, plz022. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Donghwan, S.; Won-Yong, S.; InHwan, H.; Youngsook, L. Arabidopsis metallothioneins 2a and 3 enhance resistance to cadmium when expressed in Vicia faba guard cells. Plant Mol. Biol. 2004, 54, 805–815. [Google Scholar] [CrossRef]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-Transferase Enzymes in Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef]
- Khan, N.; Hu, C.-M.; Khan, W.A.; Hou, X. Genome-Wide Identification, Classification, and Expression Divergence of Glutathione-Transferase Family in Brassica rapa under Multiple Hormone Treatments. BioMed Res. Int. 2018, 2018, 1–19. [Google Scholar] [CrossRef]
- Srivastava, D.; Verma, G.; Chauhan, A.S.; Pande, V.; Chakrabarty, D. Rice (Oryza sativa L.) tau class glutathione S-transferase (OsGSTU30) overexpression in Arabidopsis thaliana modulates a regulatory network leading to heavy metal and drought stress tolerance. Metallomics 2019, 11, 375–389. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, Y.-J.; Zeng, Q. Overexpression of three orthologous glutathione S-transferases from Populus increased salt and drought resistance in Arabidopsis. Biochem. Syst. Ecol. 2019, 83, 57–61. [Google Scholar] [CrossRef]
- Li, L.; Hou, M.; Cao, L.; Xia, Y.; Shen, Z.; Hu, Z. Glutathione S-transferases modulate Cu tolerance in Oryza sativa. Environ. Exp. Bot. 2018, 155, 313–320. [Google Scholar] [CrossRef]
- Du, J.; Ren, J.; Ye, X.; Hou, A.; Fu, W.; Mei, F. Genome-wide identification and expression analysis of the glutathione S-transferase (GST) family under different developmental tissues and abiotic stresses in Chinese cabbage (Brassica rapa ssp. pekinensis). PeerJ 2018. [Google Scholar] [CrossRef]
- Lan, T.; Yang, Z.-L.; Yang, X.; Liu, Y.-J.; Wang, X.-R.; Zeng, Q. Extensive Functional Diversification of the Populus Glutathione S-Transferase Supergene Family. Plant Cell 2009, 21, 3749–3766. [Google Scholar] [CrossRef]
- Kieffer, P.; Dommes, J.; Hoffmann, L.; Hausman, J.-F.; Renaut, J. Quantitative changes in protein expression of cadmium-exposed poplar plants. Proteomics 2008, 8, 2514–2530. [Google Scholar] [CrossRef] [PubMed]
- Tognetti, V.B.; Van Aken, O.; Morreel, K.; Vandenbroucke, K.; Van De Cotte, B.; De Clercq, I.; Chiwocha, S.; Fenske, R.; Prinsen, E.; Boerjan, W.; et al. Perturbation of Indole-3-Butyric Acid Homeostasis by the UDP-Glucosyltransferase UGT74E2 Modulates Arabidopsis Architecture and Water Stress Tolerance. Plant Cell 2010, 22, 2660–2679. [Google Scholar] [CrossRef]
- Hu, Y.F.; Zhou, G.; Na, X.F.; Yang, L.; Bin Nan, W.; Liu, X.; Zhang, Y.Q.; Li, J.L.; Bi, Y. Cadmium interferes with maintenance of auxin homeostasis in Arabidopsis seedlings. J. Plant Physiol. 2013, 170, 965–975. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Van Der Schoot, J.; Pospíšková, M.; Vosman, B.; Smulders, M.J.M. Development and characterization of microsatellite markers in black poplar (Populus nigra L.). Theor. Appl. Genet. 2000, 101, 317–322. [Google Scholar] [CrossRef]
- Smulders, M.J.M.; Van Der Schoot, J.; Arens, P.; Vosman, B. Trinucleotide repeat microsatellite markers for black poplar (Populus nigra L.). Mol. Ecol. Notes 2001, 1, 188–190. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Gunter, L.E.; Yang, Z.K.; Yin, T.; Sewell, M.M.; DiFazio, S.P. Characterization of microsatellites revealed by genomic sequencing of Populus trichocarpa. Can. J. For. Res. 2004, 34, 85–93. [Google Scholar] [CrossRef]
- Loo, M.V.; Joseph, J.A.; Heinze, B.; Fay, M.F.; Lexer, C. Clonality and spatial genetic structure in Populus × canescens and its sympatric backcross parent P. alba in a Central European hybrid zone. New Phytol. 2008, 177, 506–516. [Google Scholar]
- Politov, D.V.; Belokon, M.M.; Belokon, Y.S.; Polyakova, T.A.; Shatokhina, A.V.; Mudrik, E.A.; Azarova, A.B.; Filippov, M.V.; Shestibratov, K.A. Application of Microsatellite Loci for Molecular Identification of Elite Genotypes, Analysis of Clonality, and Genetic Diversity in Aspen Populus tremula L. (Salicaceae). Int. J. Plant Genom. 2015, 2015, 1–11. [Google Scholar] [CrossRef]
- Pokorna, E.; Cizkova, L.; Machova, P.; Cvrckova, H.; Burianek, V.; Komarkova, M. Characterization of genetic diversity of local population of grey poplar (Populus × canescens Aiton Sm.) using SSR marers and phenotypic evaluation. Rep. For. Res. 2018, 63, 281–289. [Google Scholar]
- Guichoux, E.; Lagache, L.; Wagner, S.; Chaumeil, P.; Léger, P.; Lepais, O.; Lepoittevin, C.; Malausa, T.; Revardel, E.; Salin, F.; et al. Current trends in microsatellite genotyping. Mol. Ecol. Resour. 2011, 11, 591–611. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3. [Google Scholar] [CrossRef]
- Brentner, L.B.; Mukherji, S.T.; Merchie, K.M.; Yoon, J.M.; Schnoor, J.L.; Van Aken, B. Expression of glutathione S-transferases in poplar trees (Populus trichocarpa) exposed to 2,4,6-trinitrotoluene (TNT). Chemosphere 2008, 73, 657–662. [Google Scholar] [CrossRef]
- Basa, B.; Solti, Á.; Sárvári, É.; Tamás, L. Housekeeping gene selection in poplar plants under Cd-stress: Comparative study for real-time PCR normalisation. Funct. Plant Biol. 2009, 36, 1079–1087. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, P.; Schröder, P.; Dommes, J.; Hoffmann, L.; Renaut, J.; Hausman, J.-F. Proteomic and enzymatic response of poplar to cadmium stress. J. Proteom. 2009, 72, 379–396. [Google Scholar] [CrossRef]
- He, J.; Li, H.; Luo, J.; Ma, C.; Li, S.; Qu, L.; Gai, Y.; Jiang, X.; Janz, D.; Polle, A.; et al. A Transcriptomic Network Underlies Microstructural and Physiological Responses to Cadmium in Populus × canescens. Plant Physiol. 2013, 162, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Blaudez, D.; Chalot, M.; Martin, F. Cloning and expression of multiple metallothioneins from hybrid poplar. New Phytol. 2004, 164, 83–93. [Google Scholar] [CrossRef]
- Gaudet, M.; Pietrini, F.; Beritognolo, I.; Iori, V.; Zacchini, M.; Massacci, A.; Mugnozza, G.S.; Sabatti, M. Intraspecific variation of physiological and molecular response to cadmium stress in Populus nigra L. Tree Physiol. 2011, 31, 1309–1318. [Google Scholar] [CrossRef]
- Sumanta, N.; Haque, C.; Nishika, J.; Suprakash, R. Spectrophotometric Analysis of Chlorophylls and Carotenoids from Commonly Grown Fern Species by Using Various Extracting Solvents. Res. J. Chem. Sci. 2014, 4, 63–69. [Google Scholar]
- Hoagland, D.R.; Arnon, D.I. Growing Plants without Soil by the Water-Culture Method. Grow Plants Soil Water-Cult Method. 1938. Available online: https://www.cabdirect.org/cabdirect/abstract/19381900944 (accessed on 15 September 2020).

| SSR | Allele Size | |
|---|---|---|
| TP11 | TP20 | |
| WPMS5 | 298 | 276/310 |
| WPMS15 | 189/192 | 189/192 |
| WPMS16 | 165/171 | 165/171 |
| WPMS18 | 222/228 | 219/222 |
| WPMS19 | 195/204 | 186 |
| WPMS20 | 178 | 178 |
| ORPM14 | 141 | 141 |
| ORPM16 | 223 | 223 |
| ORPM20 | 180/196 | 197/201 |
| ORPM30 | 221/229 | 207/219 |
| ORPM60 | 200/206 | 206 |
| ORPM127 | 189 | 189 |
| ORPM193 | 175/190 | 175/190 |
| ORPM220 | 217/229 | 217 |
| ORPM312 | 195 | 195 |
| Cd (µM) | Root Length | Root Weight | Shoot Weight | Chl a | Chl b | Chl a + b | ||
|---|---|---|---|---|---|---|---|---|
| 2 days | TP11 | 0 | 10.7 ± 2.0 d | 47.3 ± 6.6 e | 196.7 ± 59 bc | 2.49 ± 0.3 ab | 0.89 ± 0.1 ab | 3.38 ± 0.4 ab |
| 10 | 14.7 ± 0.8 abc | 75.0 ± 3.6 bcd | 255.7 ± 49 bc | 1.02 ± 0.2 ef | 0.24 ± 0.1 cd | 1.26 ± 0.2 de | ||
| 100 | 15.3 ± 1.3 abc | 85.3 ± 4.2 abc | 226.0 ± 56 bc | 0.62 ± 0.2 f | 0.21 ± 0.1 d | 0.83 ± 0.2 e | ||
| TP20 | 0 | 10.8 ± 2.3 d | 52.3 ± 5.8 e | 199.0 ± 63 bc | 2.27 ± 0.2 b | 0.65 ± 0.1 abcd | 2.92 ± 0.3 b | |
| 10 | 13.6 ± 0.5 bcd | 71.3 ± 5.5 cd | 157.7 ± 39 c | 2.10 ± 0.1 bc | 0.43 ± 0.2 bcd | 2.52 ± 0.1 bc | ||
| 100 | 15.1 ± 0.8 abc | 84.3 ± 5.5 abc | 151.0 ± 45 c | 1.47 ± 0.3 cde | 0.33 ± 0.1 bcd | 1.83 ± 0.3 cd | ||
| 10 days | TP11 | 0 | 12.2 ± 1.0 cd | 79.3 ± 5.1 abcd | 440.7 ± 39 a | 3.07 ± 0.5 a | 0.84 ± 0.1 ab | 3.91 ± 0.3 a |
| 10 | 15.5 ± 1.2 abc | 89.7 ± 6.7 ab | 448.7 ± 45 a | 2.17 ± 0.1 bc | 0.78 ± 0.1 abc | 2.95 ± 0.2 b | ||
| 100 | 15.8 ± 1.7 ab | 93.0 ± 7.9 a | 301.4 ± 40 abc | 1.08 ± 0.2 cde | 1.08 ± 0.1 a | 2.55 ± 0.2 bc | ||
| TP20 | 0 | 12.3 ± 0.5 cd | 62.3 ± 4.9 de | 328.3 ± 69 ab | 3.09 ± 0.3 a | 0.81 ± 0.2 abc | 3.90 ± 0.4 a | |
| 10 | 16.0 ± 0.4 ab | 77.1 ± 7.8 abcd | 291.8 ± 91 abc | 2.03 ± 0.3 bcd | 0.69 ± 0.3 abcd | 2.72 ± 0.6 bc | ||
| 100 | 17.1 ± 0.3 a | 81.3 ± 7.7 abc | 274.1 ± 43 bc | 1.40 ± 0.1 de | 0.52 ± 0.2 abcd | 1.91 ± 0.1 cd |
| K Content (g kg−1) | Ca Content (g kg−1) | Mg Content (g kg−1) | Zn Content (g kg−1) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Roots | Shoots | Roots | Shoots | Roots | Shoots | Roots | Shoots | |||
| 2 days | TP11 | Cont | 13.24 ± 1.93 hij | 30.19 ± 1.84 a | 5.05 ± 0.21 fgh | 6.60 ± 1.11 cde | 1.64 ± 0.15 bcd | 1.50 ± 0.11 cd | 0.22 ± 0.05 ab | 0.16 ± 0.06 abcdef |
| Cd10 | 10.40 ± 0.87 j | 23.57 ± 2.75 bcde | 4.04 ± 0.31 h | 5.93 ± 0.70 def | 1.42 ± 0.26 cd | 1.64 ± 0.18 bcd | 0.20 ± 0.04 abc | 0.17 ± 0.01 abcde | ||
| Cd100 | 12.50 ± 1.19 hij | 24.29 ± 2.58 bcde | 4.69 ± 0.36 fgh | 8.54 ± 0.44 ab | 2.02 ± 0.43 abcd | 2.41 ± 0.52 abc | 0.25 ± 0.04 a | 0.17 ± 0.03 abcd | ||
| TP20 | Cont | 12.22 ± 2.68 hij | 21.24 ± 1.60 cdef | 5.03 ± 0.91 gh | 7.07 ± 0.42 abcd | 1.53 ± 0.51 bcd | 1.85 ± 0.99 abcd | 0.14 ± 0.02 bcdef | 0.08 ± 0.00 f | |
| Cd10 | 16.03 ± 3.12 fgh | 22.81 ± 2.02 bcdef | 4.39 ± 0.58 gh | 6.86 ± 0.38 bcd | 2.22 ± 0.11 abcd | 1.95 ± 0.18 abcd | 0.17 ± 0.07 abcdef | 0.08 ± 0.03 ef | ||
| Cd100 | 16.25 ± 0.38 ghi | 25.60 ± 1.52 abcd | 4.75 ± 1.04 gh | 6.68 ± 0.84 abcd | 2.83 ± 0.20 a | 1.94 ± 0.01 abcd | 0.18 ± 0.02 abcd | 0.09 ± 0.03 def | ||
| 10 days | TP11 | Cont | 11.70 ± 0.98 ij | 27.43 ± 2.47 abc | 5.04 ± 1.03 gh | 6.64 ± 0.61 abcd | 1.67 ± 0.00 bcd | 1.73 ± 0.51 abcd | 0.21 ± 0.03 abc | 0.14 ± 0.01 bcdef |
| Cd10 | 14.76 ± 2.31 hij | 27.36 ± 1.73 abc | 3.78 ± 0.31 h | 5.49 ± 0.96 defg | 1.13 ± 0.22 d | 1.83 ± 0.45 abcd | 0.14 ± 0.01 bcdef | 0.21 ± 0.04 abc | ||
| Cd100 | 20.35 ± 1.33 defg | 26.88 ± 2.84 ab | 4.41 ± 0.72 gh | 8.13 ± 0.81 a | 2.02 ± 0.04 abcd | 2.64 ± 0.77 ab | 0.16 ± 0.03 bcdef | 0.22 ± 0.03 ab | ||
| TP20 | Cont | 9.96 ± 1.54 j | 19.40 ± 0.59 defg | 4.66 ± 0.63 fgh | 6.45 ± 1.06 cd | 2.14 ± 0.12 abcd | 1.64 ± 0.25 bcd | 0.18 ± 0.04 abcd | 0.08 ± 0.00 f | |
| Cd10 | 15.26 ± 2.72 hij | 26.46 ± 2.00 abcd | 3.70 ± 5.60 h | 6.30 ± 1.04 cd | 1.13 ± 0.12 d | 1.73 ± 0.31 abcd | 0.20 ± 0.02 abc | 0.13 ± 0.03 bcdef | ||
| Cd100 | 14.00 ± 3.49 hij | 26.81 ± 2.10 ab | 5.00 ± 0.51 fgh | 7.98 ± 0.51 abc | 1.93 ± 0.13 abcd | 2.10 ± 0.18 abcd | 0.13 ± 0.03 cdef | 0.15 ± 0.03 bcdef | ||
| Translocation Factor (TF) | ||
|---|---|---|
| 2 days | TP11 | TP20 |
| Cd 10 | 0.34 | 0.54 |
| Cd 100 | 0.27 | 0.42 |
| 10 days | ||
| Cd 10 | 0.61 | 1.06 |
| Cd 100 | 0.57 | 0.63 |
| 2 Days | 10 Days | |||
|---|---|---|---|---|
| Root | Shoot | Root | Shoot | |
| Endochitinase 2 | ||||
| TP11 | ||||
| Control | 1.15 ± 0.7 cde | 0.97 ± 0.5 cde | 1.05 ± 0.2 cde | 0.90 ± 0.2 cde |
| 10 Cd | 6.85 ± 0.8 a | 1.18 ± 0.2 cde | 1.83 ± 0.2 c | 0.47 ± 0.1 de |
| 100 Cd | 3.36 ± 1.1 b | 1.99 ± 0.3 c | 1.55 ± 0.3 cd | 0.49 ± 0.2 de |
| TP20 | ||||
| Control | 1.03 ± 0.1 cde | 0.91 ± 0.2 cde | 1.03 ± 0.1 cde | 0.90 ± 0.1 cde |
| 10 Cd | 7.20 ± 0.9 a | 1.60 ± 0.3 cd | 1.40 ± 0.3 cde | 0.72 ± 0.2 cde |
| 100 Cd | 4.63 ± 0.5 b | 0.21 ± 0.1 e | 1.77 ± 0.4 cd | 0.93 ± 0.2 cde |
| 12-oxophytodienoate reductase 1 | ||||
| TP11 | ||||
| Control | 0.55 ± 0.1 g | 1.71 ± 0.9 fg | 0.92 ± 0.4 g | 1.56 ± 0.3 fg |
| 10 Cd | 1.95 ± 0.1 fg | 6.09 ± 1.8 defg | 35.36 ± 8.4 a | 13.89 ± 1.7 c |
| 100 Cd | 4.83 ± 1.8 defg | 2.10 ± 0.1 fg | 24.26 ± 3.9 b | 8.00 ± 1.1 cdef |
| TP20 | ||||
| Control | 0.99 ± 0.4 g | 1.60 ± 0.6 fg | 0.87 ± 0.1 g | 1.59 ± 0.5 fg |
| 10 Cd | 3.00 ± 0.7 efg | 0.93 ± 0.1 g | 6.57 ± 1.5 defg | 26.03 ± 0.9 b |
| 100 Cd | 9.34 ± 0.9 cde | 1.97 ± 0.3 fg | 9.87 ± 1.9 cd | 10.24 ± 0.9 cd |
| thaumatin-like protein | ||||
| TP11 | ||||
| Control | 0.95 ± 0.4 b | 1.79 ± 0.2 b | 0.97 ± 0.5 b | 2.25 ± 1.5 b |
| 10 Cd | 0.25 ± 0.1 b | 0.25 ± 0.2 b | 1.32 ± 0.4 b | 1.24 ± 1.0 b |
| 100 Cd | 1.35 ± 0.3 b | 1.75 ± 1.4 b | 0.95 ± 0.2 b | 1.36 ± 0.6 b |
| TP20 | ||||
| Control | 0.90 ± 0.1 b | 1.85 ± 0.6 b | 0.91 ± 0.2 b | 1.81 ± 0.3 b |
| 10 Cd | 0.47 ± 0.2 b | 2.24 ± 0.9 b | 0.92 ± 0.3 b | 1.56 ± 0.8 b |
| 100 Cd | 6.32 ± 1.2 a | 0.29 ± 0.1 b | 1.20 ± 0.6 b | 1.54 ± 0.6 b |
| photosystem II 10 kDa polypeptide | ||||
| TP11 | ||||
| Control | - | 1.42 ± 1.1 abc | - | 1.23 ± 0.5 abc |
| 10 Cd | - | 1.88 ± 0.3 ab | - | 2.25 ± 0.2 ab |
| 100 Cd | - | 0.73 ± 0.1 bc | - | 2.43 ± 1.1 a |
| TP20 | ||||
| Control | - | 1.25 ± 0.6 abc | - | 1.17 ± 0.1 abc |
| 10 Cd | - | 0.78 ± 0.1 bc | - | 2.34 ± 0.3 a |
| 100 Cd | - | 0.04 ± 0.0 c | - | 1.20 ± 0.1 abc |
| Metallothionein 2a | ||||
| TP11 | ||||
| Control | 2.86 ± 1.2 bcdef | 1.10 ± 0.4 defgh | 2.84 ± 1.0 bcdefg | 1.11 ± 0.5 defgh |
| 10 Cd | 0.74 ± 0.6 efgh | 1.23 ± 0.2 cdefgh | 9.68 ± 1.8 a | 1.42 ± 0.3 bcdefgh |
| 100 Cd | 3.50 ± 1.3 b | 0.77 ± 0.2 defgh | 2.98 ± 0.7 bcd | 1.05 ± 0.6 defgh |
| TP20 | ||||
| Control | 2.74 ± 0.5 bcdefg | 1.14 ± 0.6 defgh | 2.90 ± 1.3 bcde | 1.04 ± 0.1 defgh |
| 10 Cd | 0.84 ± 0.2 defgh | 0.63 ± 0.2 gh | 3.43 ± 0.5 bc | 1.23 ± 0.1 cdefgh |
| 100 Cd | 1.66 ± 0.1 bcdefgh | 0.07 ± 0.0 h | 2.10 ± 0.1 bcdefgh | 0.66 ± 0.0 fgh |
| phi class glutathione S-transferase | ||||
| TP11 | ||||
| Control | 1.90 ± 0.7 d | 1.04 ± 0.1 d | 1.91 ± 0.8 d | 1.24 ± 0.7 d |
| 10 Cd | 2.97 ± 0.4 d | 3.92 ± 0.1 d | 20.43 ± 0.9 a | 0.94 ± 0.2 d |
| 100 Cd | 1.33 ± 0.8 d | 2.75 ± 0.2 d | 15.65 ± 1.6 b | 0.05 ± 0.4 d |
| TP20 | ||||
| Control | 1.80 ± 0.2 d | 1.21 ± 0.9 d | 1.82 ± 0.4 d | 1.12 ± 0.5 d |
| 10 Cd | 2.89 ± 0.8 d | 2.97 ± 0.3 d | 16.03 ± 1.3 b | 2.59 ± 1.5 d |
| 100 Cd | 8.50 ± 4.9 c | 2.21 ± 0.0 d | 17.50 ± 1.3 ab | 1.99 ± 0.9 d |
| Uridine diphosphate glycosyltransferase 74E2 | ||||
| TP11 | ||||
| Control | 1.02 ± 0.1 defg | 1.72 ± 0.1 cdef | 1.04 ± 0.3 defg | 1.76 ± 0.5 cdef |
| 10 Cd | 1.13 ± 0.4 defg | 3.73 ± 0.4 ab | 4.98 ± 1.2 a | 0.81 ± 0.2 efg |
| 100 Cd | 1.55 ± 0.2 cdefg | 1.33 ± 0.3 cdefg | 2.32 ± 0.5 cd | 0.33 ± 0.1 g |
| TP20 | ||||
| Control | 1.01 ± 0.1 defg | 1.81 ± 0.7 cde | 1.01 ± 0.1 defg | 1.73 ± 0.2 cdef |
| 10 Cd | 1.05 ± 0.2 defg | 1.17 ± 0.3 defg | 2.56 ± 0.8 bc | 2.60 ± 0.4 bc |
| 100 Cd | 1.36 ± 0.5 cdefg | 0.45 ± 0.1 fg | 1.37 ± 0.2 cdefg | 1.39 ± 0.2 cdefg |
| Gene | Accession Number | Gene Description | Primer Sequence (5′–3′) | Reference |
|---|---|---|---|---|
| LOC112328551 | XR_002983567 | 18S Ribosomal RNA | F: AGAAACGGCTACCACATCCAA R: CCAGACTTGCCCTCCAATGG | [83] |
| LOC18109220 | EF147878.1 | Elongation factor 1-α | F: CCACACCTGTCACATTGCTG R: ACCAGCATCACCGTTCTTCAG | [84] |
| LOC7470435 | XM_002306184.3 | Endochitinase 2 | F: TACGGGCAATGTGGAAAAGC R: ATTGTGGCATGAGGGCTTTG | [85] |
| OPR1 | NM_106318.4 | 12-Oxophytodienoate reductase 1 | F: CGGACAAGCAGGAGACTCAAA R: CCACCGTCTTCATTCTTGGC | [83] |
| UGT74E2 | NM_100448.4 | Uridine diphosphate glycosyltransferase 74E2 | F: CACAAATCCGTGGGATGCTT R: TCTGTCCACTGTGGCATTGC | [83] |
| LOC105129022 | XM_011030918 | Thaumatin-like protein | F: ACCACACAAGCACGCATTTG R: TGAACCATAGCCTTGGCATG | [85] |
| LOC18095761 | 18095761 | Photosystem II 10 kDa polypeptide | F: ATGGTGCTAATGTGGATGGC R: AACAGCCCAGATTAGCAAGC | [86] |
| MT2a | AY594297.1 | Metallothionein 2a | F: ATCATCGCATCGACGGATTG R: CCAGAGCTGCAAATCCAAGAAG | [87] |
| GSTF4 | GQ377243.1 | Phi class glutathione S-transferase | F: CTTAGCCTCGTTTTCCTCCA R: CTTAGCCTCGTTTTCCTCCA | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komárková, M.; Chromý, J.; Pokorná, E.; Soudek, P.; Máchová, P. Physiological and Transcriptomic Response of Grey Poplar (Populus ×canescens Aiton Sm.) to Cadmium Stress. Plants 2020, 9, 1485. https://doi.org/10.3390/plants9111485
Komárková M, Chromý J, Pokorná E, Soudek P, Máchová P. Physiological and Transcriptomic Response of Grey Poplar (Populus ×canescens Aiton Sm.) to Cadmium Stress. Plants. 2020; 9(11):1485. https://doi.org/10.3390/plants9111485
Chicago/Turabian StyleKomárková, Martina, Jakub Chromý, Eva Pokorná, Petr Soudek, and Pavlína Máchová. 2020. "Physiological and Transcriptomic Response of Grey Poplar (Populus ×canescens Aiton Sm.) to Cadmium Stress" Plants 9, no. 11: 1485. https://doi.org/10.3390/plants9111485
APA StyleKomárková, M., Chromý, J., Pokorná, E., Soudek, P., & Máchová, P. (2020). Physiological and Transcriptomic Response of Grey Poplar (Populus ×canescens Aiton Sm.) to Cadmium Stress. Plants, 9(11), 1485. https://doi.org/10.3390/plants9111485





