Characterization of the Genetic Diversity of Acid Lime (Citrus aurantifolia (Christm.) Swingle) Cultivars of Eastern Nepal Using Inter-Simple Sequence Repeat Markers
Abstract
1. Introduction
2. Material and Methods
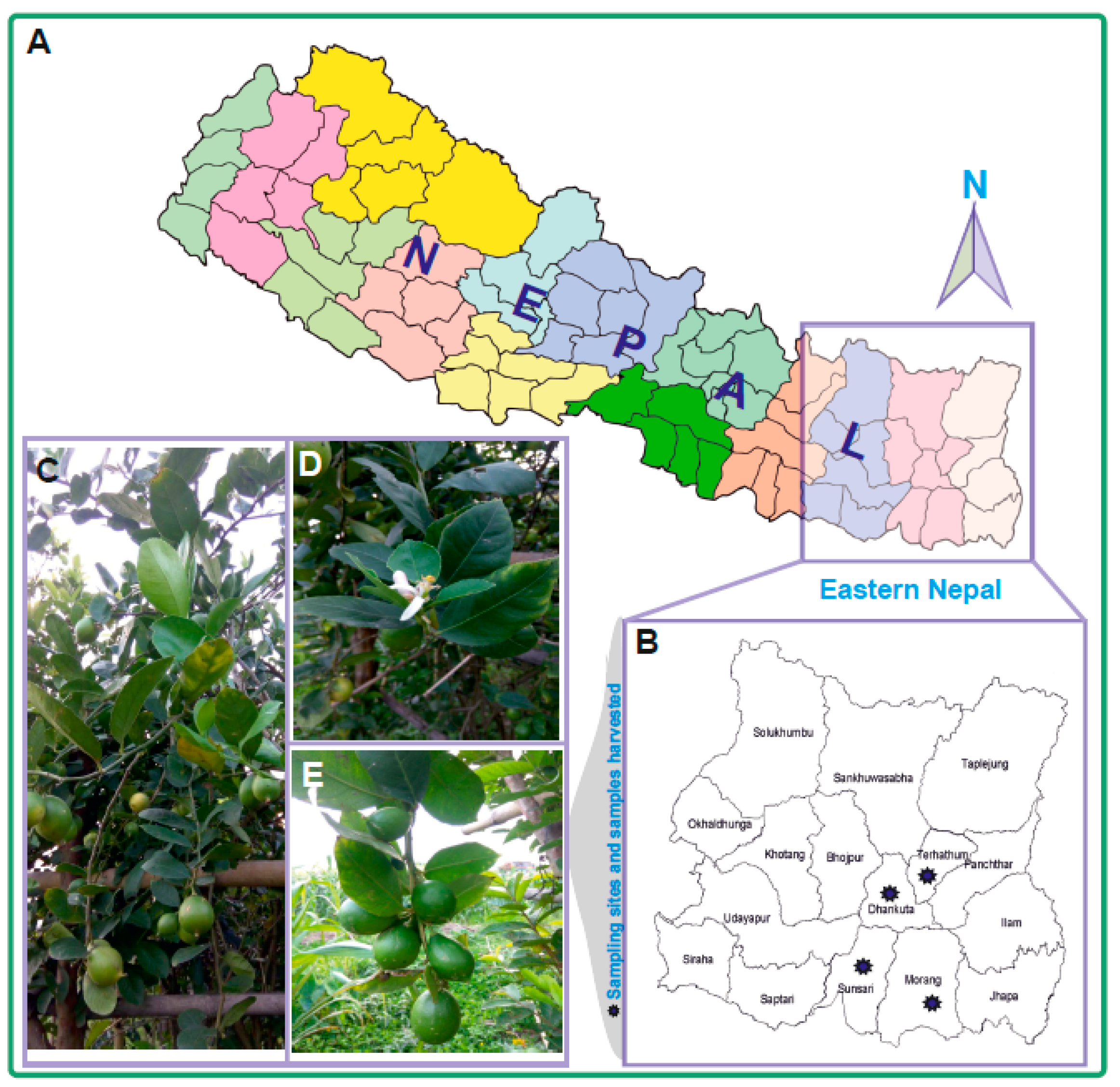
2.1. Plant Materials
2.2. DNA Extraction and PCR Amplification
2.3. Inter-Simple Sequence Repeat Profiling and Scoring of the Data
2.4. Data Analysis
3. Results
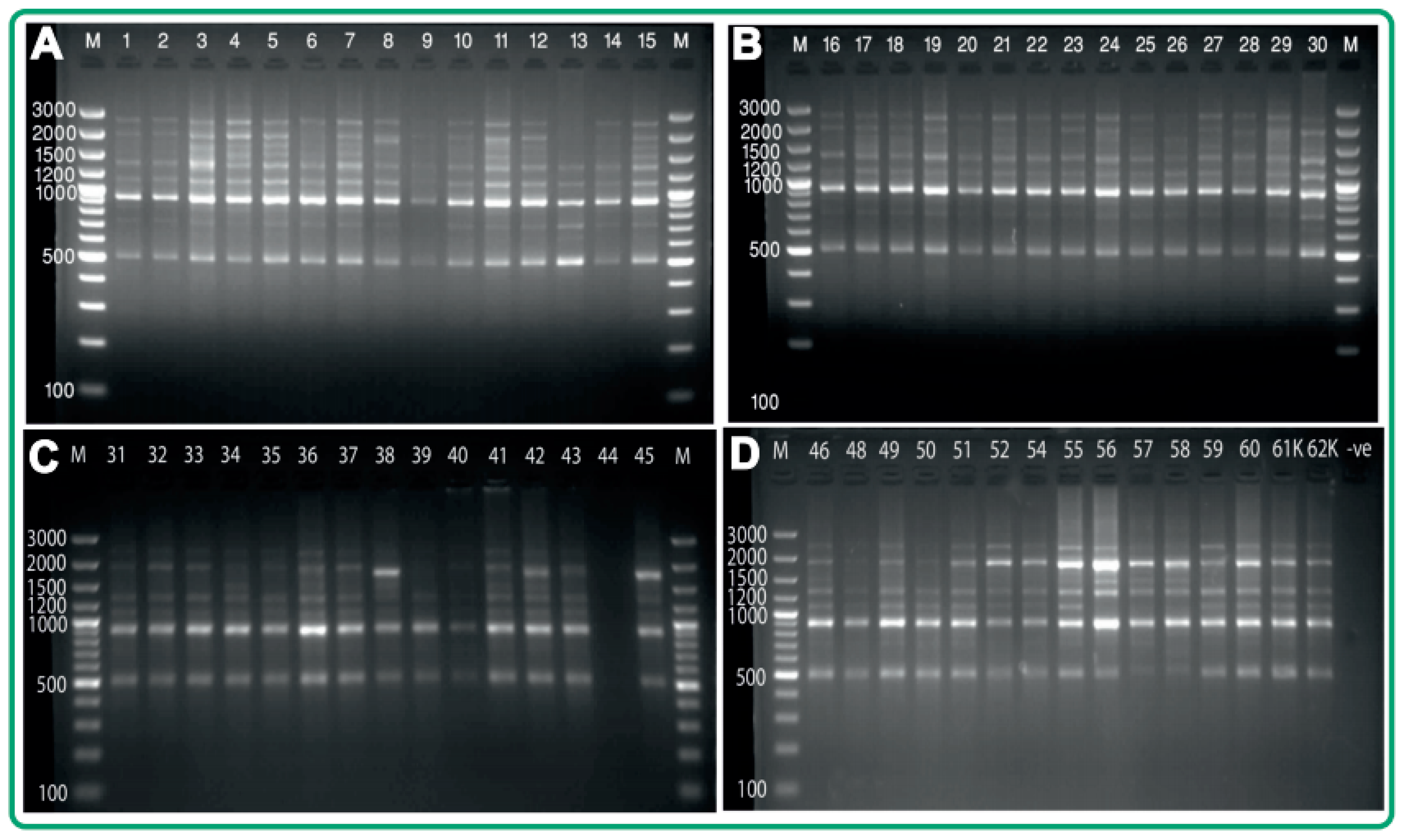
3.1. Inter-Simple Sequence Repeat Polymorphism in Acid Lime Cultivars
3.2. Genetic Diversity in Acid Lime Cultivars
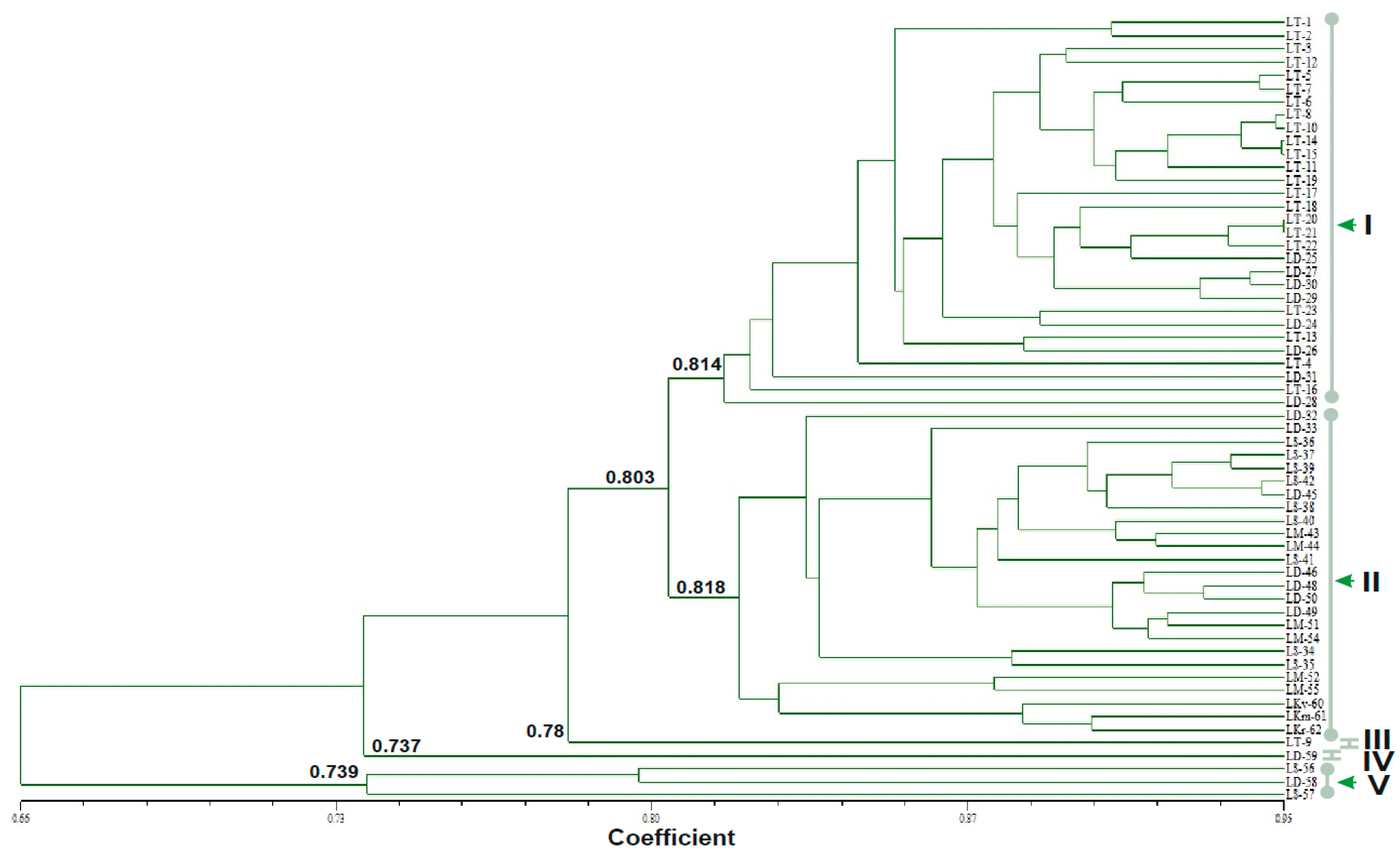
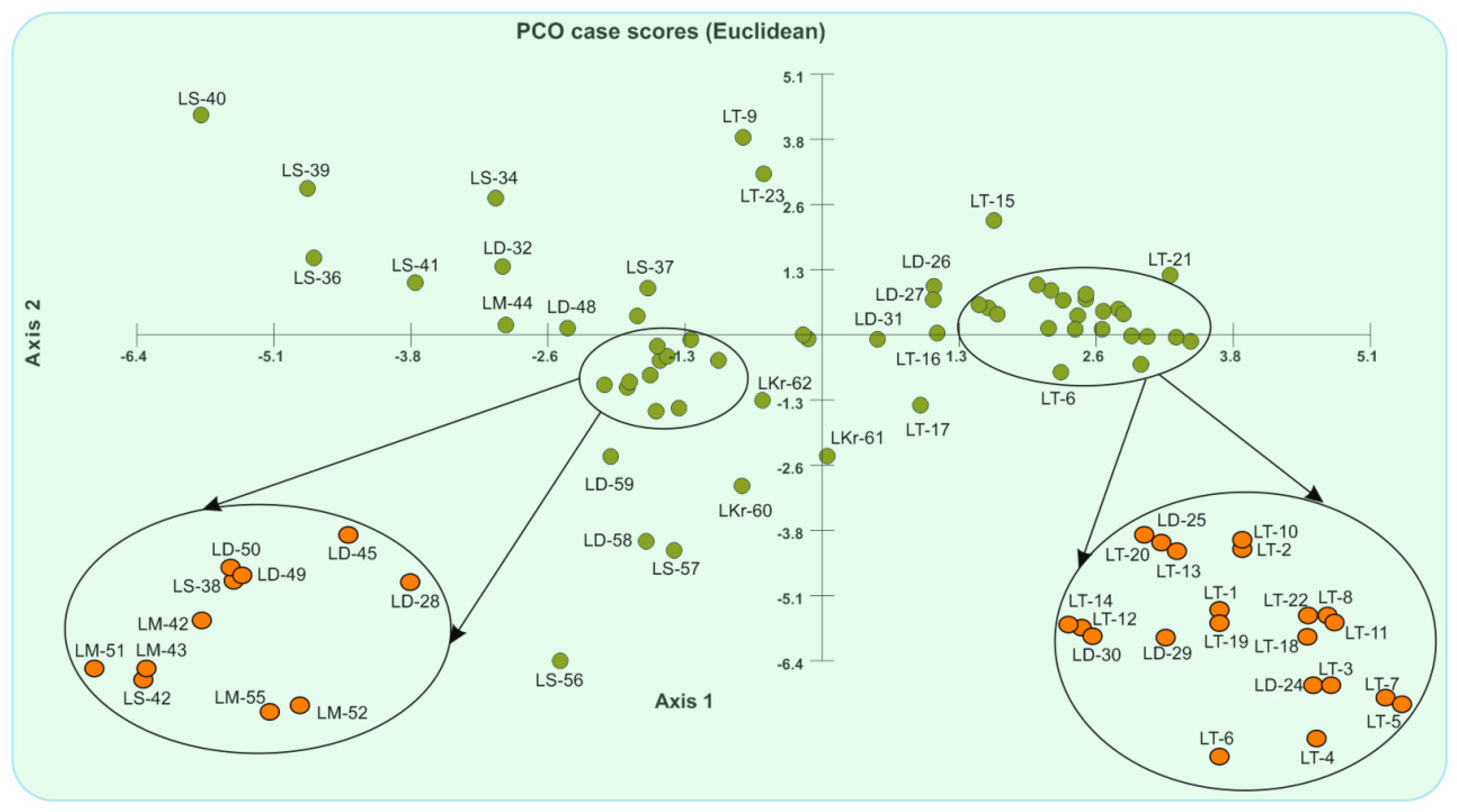
3.3. The Genetic Relationship Based on Unweighted Pair Group Method of Arithmetic Averages Cluster Analysis and Principal Coordinate Analysis
4. Discussion
4.1. Inter-Simple Sequence Repeat Polymorphism and Genetic Diversity Estimation in Nepalese Acid Lime Cultivars
4.2. Use of Inter-Simple Sequence Repeat-Based Genetic Diversity Estimates in an Acid Lime Breeding Program in Nepal
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Velasco, R.; Concetta, L. A genealogy of the Citrus family. Nat. Biotechnol. 2014, 32, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Hvarleva, T.T.; Kapari-Isaia, L.; Papayiannis, A.; Atanassov, A.; Hadjinicoli, A.; Kyriakou, A. Characterization of Citrus cultivars and clones in Cyprus through Microsatellite and RAPD analysis. Biotechnol. Biotechnol. Equip. 2008, 22, 787–794. [Google Scholar] [CrossRef]
- Ranjit, M.; Gokarna, G.C. Citrus Research and Development Action Plan; Chemonics International Inc.: Washington, DC, USA, 1997. [Google Scholar]
- Nicolosi, E.; Deng, Z.N.; Gentile, A.; La Malfa, S.; Continella, G.; Tribulato, E. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theor. Appl. Genet. 2000, 100, 1155–1166. [Google Scholar] [CrossRef]
- Barett, H.C.; Rhodes, A.M. A numerical taxonomic study of affinity relationship in cultivated Citrus and its clone relatives. Syst. Bot. 1976, 1, 105–136. [Google Scholar] [CrossRef]
- Singh, A.K. Probable agricultural biodiversity heritage sites in India: V. The Garo, Khasi, and Jaintia Hills Region. Asian Agri-Hist. 2010, 14, 133–156. [Google Scholar]
- Dhillon, B.S.; Randhawa, J.S. Fruit Growth and Development in Citrus: Advance in Horticulture; Malhotra Publishing House: New Dehli, India, 1993; Volume 3. [Google Scholar]
- Penniston, K.L.; Nakada, S.Y.; Holmes, R.P.; Assimos, D.G. Quantitative assessment of citric acid in lemon juice, lime juice, and commercially-available fruit juice products. J. Endourol. 2008, 22, 567–570. [Google Scholar] [CrossRef] [PubMed]
- Aras, B.; Kalfazade, N.; Tugcu, V.; Kemahh, E.; Ozbay, B.; Polat, H.; Tasci, A.I. Can lemon juice be an alternative to potassium citrate in the treatment of urinary calcium stones in patients with hypocitraturia? A prospective randomized study. Urol. Res. 2008, 36, 313–317. [Google Scholar] [CrossRef] [PubMed]
- NCRP. Annual Report 2067/68 (2010/11); NCRP: Paripatle, Nepal, 2012. [Google Scholar]
- Paudyal, K.P.; Shrestha, R.L. Diversity Study and Selection of Lime (Citrus aurantifolia) Genotypes for Expansion of Production Period in Nepal. In Proceedings of the 4th National Horticulture Workshop, Kathmandu, Nepal, 2–4 March 2004; Nepal Agriculture Research Council: Kathmandu, Nepal, 2004. [Google Scholar]
- Dhakal, D.D.; Gotame, T.P.; Bhattarai, S.; Bhandari, H.N. Assessment of Lime and Lemon production in Nepal. J. Inst. Agric. Anim. Sci. 2002, 23, 49–58. [Google Scholar]
- MoAC. Statistical Information of Nepalese Agriculture. Agri. Business Promotion and Statistical Division, Ministry of Agriculture and Cooperatives; Singh Durbar: Kathmandu, Nepal, 2011.
- FAO. Food and Agriculture Organization of the United Nations Developments in International Citrus Trade in 2004–2005; FAO: Rome, Italy, 2006. [Google Scholar]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Rauf, S.; Iqbal, Z.; Shahzad, M. Genetic improvement of Citrus for disease resistance. Arch. Phytopathol. Plant Prot. 2013, 46, 2051–2061. [Google Scholar] [CrossRef]
- Dominquez, A.; de Mendoza, A.; Guerri, J.; Cambra, M.; Navarro, L.; Moreno, P.; Pena, L. Pathogen-derived resistance to Citrus Tristeza Virus (CTV) in transgenic Mexican lime (Citrus aurantifolia (Christ.) Swing.) plants expressing its p25 coat protein gene. Mol. Breed. 2002, 10, 1–10. [Google Scholar] [CrossRef]
- Viloria, Z.; Grosser, J.W. Acid Citrus fruit improvement via interploid hybridization using allotetraploid somatic hybrid and autotetraploid breeding parents. J. Am. Soc. Hortic. Sci. 2005, 130, 392–402. [Google Scholar]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2006, 10, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Nei, J.; Colowit, P.M.; Mackill, D.J. Evaluation of genetic diversity in rice sub species using microsatellite markers. Crop Sci. 2002, 42, 601–607. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter-Simple Sequence Repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- Shrestha, S. Molecular Systematic of Weedy Sporobolus Species of Australia; The University of Queensland: Brisbane, Australia, 2001. [Google Scholar]
- Shahsavar, A.R.; Izadpanah, K.; Tafazoli, E.; Tabatabaei, B.E. Characterization of Citrus germplasm including unknown variants by Inter-Simple Sequence Repeat (ISSR) markers. Sci. Hortic. 2007, 112, 310–314. [Google Scholar] [CrossRef]
- Fang, D.Q.; Roose, M.L. Identification of closely related Citrus cultivars with Inter-Simple Sequence Repeat markers. Theor. Appl. Genet. 1997, 95, 408–417. [Google Scholar] [CrossRef]
- Kijas, J.M.H.; Thomas, M.R.; Fowler, J.C.S.; Roose, M.L. Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor. Appl. Genet. 1997, 94, 701–706. [Google Scholar] [CrossRef]
- Sanker, A.A.; Moore, G.A. Evaluation of Inter–Simple Sequence Repeat analysis for mapping in Citrus and extension of the genetic linkage map. Theor. Appl. Genet. 2001, 102, 206–214. [Google Scholar] [CrossRef]
- Fang, D.Q.; Roose, M.L.; Krueger, R.R.; Federici, C.T. Fingerprinting trifoliate orange germplasm accessions with isozymes, RFLPS and Inter-Simple Sequence Repeat markers. Theor. Appl. Genet. 1997, 95, 211–219. [Google Scholar] [CrossRef]
- Filho, H.D.C.; Machado, M.A.; Targon, M.L.P.N.; Moreira, M.C.P.Q.D.G.; Pompeu, J. Analysis of the genetic diversity among mandarins (Citrus spp.) using RAPD markers. Euphytica 1998, 102, 133–139. [Google Scholar] [CrossRef]
- Novelli, V.M.; Machado, M.A.; Lopes, C.R. Iso-enzymatic polymorphism in Citrus spp. and P. trifoliata (L.) Raf. (Rutaceae). Genet. Mol. Biol. 2000, 23, 163–168. [Google Scholar] [CrossRef]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by Simple Sequence Repeat (SSR)—Anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Buczkowska, K.; Rabska, M.; Gonera, P.; Pawlaczyk, E.M.; Wawrzyniak, P.; Czolpinska, M.; Baczkiewicz, A. Effectiveness of ISSR markers for determination of the Aneura pinguis cryptic species and Aneura maxima. Biochem. Syst. Ecol. 2016, 68, 27–35. [Google Scholar] [CrossRef]
- Celinski, K.; Zbrankova, V.; Wojnicka-Poltorak, A.; Chudzinska, E. Biogeography and evolutionary factors determine genetic differentiation of Pinus mugo (Turra) in the Tatra Mountains (Central Europe). J. Mt. Sci. 2015, 12, 549–557. [Google Scholar] [CrossRef]
- Bornet, B.; Branchard, M. Non-anchored Simple Sequence Repeat markers: Reproducible and specific tools for genome fingerprinting. Plant Mol. Biol. Rep. 2001, 19, 209–215. [Google Scholar] [CrossRef]
- Tautz, D.; Renz, M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984, 12, 4127–4138. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.P.; Gupta, V.S.; Aggarwal, R.K.; Ranjekar, P.K.; Brar, D.S. Genetic diversity and phylogenetic relationship as revealed by Inter-Simple Sequence Repeat (ISSR) polymorphism in the genus Oryza. Theor. Appl. Genet. 2000, 100, 1311–1320. [Google Scholar] [CrossRef]
- Meloni, M.; Perini, D.; Filigheddu, R.; Binelli, G. Genetic variation in five mediterranean populations of Juniperus phoenicea as revealed by Inter-Simple Sequence Repeat (ISSR) markers. Ann. Bot. 2006, 97, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.; Chyi, Y.S.; Romero-Severson, J.; Owen, J.L. Amplification of DNA markers from evolutionarily diverse genomes using single primers of Simple-Sequence Repeats. Theor. Appl. Genet. 1994, 89, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Tsumura, Y.; Ohba, K.; Strauss, S.H. Diversity and inheritance of Inter-Simple Sequence Repeat polymorphisms in Douglasfir (Pseudotsuga menziesii) and Sugi (Cryptomeria japonica). Theor. Appl. Genet. 1996, 92, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Ratnaparkhe, M.B.; Tekeoglu, M.; Muehlbauer, F.J. Inter-Simple Sequence Repeat (ISSR) polymorphisms are useful for finding markers associated with disease resistance gene clusters. Theor. Appl. Genet. 1998, 97, 515–519. [Google Scholar] [CrossRef]
- Biswas, M.K.; Chai, L.; Amar, M.H.; Zhang, X.; Deng, X. Comparative analysis of genetic diversity in Citrus germplasm collection using AFLP, SSAP, SAMPL and SSR markers. Sci. Hortic. 2011, 129, 798–803. [Google Scholar] [CrossRef]
- Shrestha, R.L.; Dhakal, D.; Gautam, D.; Paudyal, K.P.; Shrestha, S. Genetic diversity assessment of Acid lime (Citrus aurantifolia) landraces in Nepal, using SSR markers. Am. J. Plant Sci. 2012, 3, 1674–1681. [Google Scholar] [CrossRef]
- Kumar, M.; Parthiban, S.; SaralaDevi, D.; Ponnuswami, V. Genetic diversity analysis of Acid lime (Citrus aurantifolia Swingle) cultivars. Bioscan 2013, 8, 481–484. [Google Scholar]
- Munankarmi, N.M.; Shrestha, R.L.; Rana, N.; Shrestha, J.K.C.; Shrestha, S.; Koirala, R.; Shrestha, S. Genetic diversity assessment of Acid lime (Citrus aurantifolia, Swingle) landraces of Nepal using RAPD markers. Int. J. Appl. Sci. Biotechnol. 2014, 2, 315–327. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Pub Cold Spring Harbour Laboratory Press: New York, NY, USA, 2001; Volume III. [Google Scholar]
- Marak, C.K.; Laskar, M.A. Analysis of phenetic relationship between Citrus indica Tanaka and a few commercially important Citrus species by ISSR markers. Sci. Hortic. 2010, 124, 345–348. [Google Scholar] [CrossRef]
- Fang, D.Q.; Krueger, R.R.; Roose, M.L. Phylogenetic relationships among selected Citrus germplasm accessions revealed by Inter-Simple Sequence Repeat (ISSR) markers. J. Am. Soc. Hortic. Sci. 1998, 123, 612–617. [Google Scholar]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Jaccard, P. Nouvelles recherche sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Transue, D.K.; Fairbanks, D.J.; Robison, L.R.; Anderson, W.R. Species identification by RAPD analysis of Grain Amaranth genetic resources. Crop Sci. 1994, 34, 1385–1389. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of genetic map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Sokal, R.R. Ecological Parameters Inferred from Spatial Correlograms. In Contemporary Quantitative Ecology and Related Ecometrics; Patil, G.P., Rosenzweig, M.L., Eds.; International Cooperative Publishing House: Fairland, MD, USA, 1979; pp. 167–196. [Google Scholar]
- Sneath, P.H.A.; Sokal, R.R. Taxonomy, the Principle and Practice of Numerical Classification; Pub W.H. Freeman and Company: San Francisco, CA, USA, 1973. [Google Scholar]
- Sokal, R.R.; Michener, C.D. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 1958, 38, 1409–1438. [Google Scholar]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 685–686. [Google Scholar] [CrossRef]
- Mantel, N.A. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Kumar, S.; Jena, S.N.; Nair, N.K. ISSR polymorphism in Indian wild orange (Citrus indica Tanaka, Rutaceae) and related wild species in North-East India. Sci. Hortic. 2010, 123, 350–359. [Google Scholar] [CrossRef]
- Teklewood, A.; Becker, H.C. Geographic pattern of genetic diversity among 43 Ethiopian Mustard (Brassica carinata A. Braun) accession as revealed by RAPD analysis. Genet. Resour. Crop Evol. 2006, 53, 1173–1185. [Google Scholar] [CrossRef]
- Vemireddy, L.R.; Archak, S.; Nagaraju, J. Capillary electrophoresis is essential for microsatellite marker based detection and quantification of adulteration of Basmati Rice (Oryza sativa). J. Agric. Food Chem. 2007, 55, 8112–8117. [Google Scholar] [CrossRef] [PubMed]
- Jeya Prakash, S.P.; Biji, K.R.; Mimchael, G.S.; Ganesha, M.K.; Chandra, B.R. Genetic diversity analysis of Sorghum (Sorghum bicolor L. Moench) accessions using RAPD markers. Indian J. Crop Sci. 2006, 1, 109–112. [Google Scholar]
- Ortiz, R.; Madsen, S.; Vuylsteke, D. Classification of African plantain landraces and Banana cultivars using a phenotypic distance index of quantitative descriptors. Theor. Appl. Genet. 1998, 96, 904–911. [Google Scholar] [CrossRef]
- Smith, J.S.C.; Smith, O.S. The description and assessment of distance between inbred lines of maize, 11. The utility of morphological, biochemical and genetic descriptors and a scheme for testing of distinctiveness between inbred lines. Maydica 1998, 34, 151–161. [Google Scholar]
- Shrestha, R.L.; Dhakal, D.; Gautam, D.; Paudyal, K.P.; Shrestha, S. Study of fruits diversity and selection of elite Acid lime (Citrus aurantifolia Swingle) genotypes in Nepal. Am. J. Plant Sci. 2012, 3, 1098–1104. [Google Scholar] [CrossRef]
- Pangali, P.L.; Hossain, M.; Gerpasio, R.V. Asian Rice Blows: The Returning Crises? International Rice Research Institute (IRRI): Los Banus, Philippines, 1997. [Google Scholar]
| Above 1200 m asL | 600–1200 m asL | Less Than 600 m asL | ||||||
|---|---|---|---|---|---|---|---|---|
| Cult. No. | Altitude (m asL) | VDC-Ward No. | Cult. No. | Altitude, (m asL) | VDC-Ward No. | Cult. No. | Altitude, (m asL) | VDC-Ward No. |
| LD-24 | 1290 | Balehara-8 | LD-25 | 1180 | Balara-1 | LD-45 | 135 | Sunpur-2 |
| LD-46 | 1278 | Bodhe-2 | LD-26 | 1175 | Balara-1 | LD-58 | 135 | Sunpur-2 |
| LD-50 | 1638 | Rajarani-9 | LD-27 | 1175 | Balara-1 | LM-43 | 135 | Sunpur-2 |
| LT-1 | 1605 | Okhre-8 | LD-28 | 1175 | Balara-1 | LM-44 | 135 | Sunpur-2 |
| LT-2 | 1285 | Okhre-1 | LD-29 | 1175 | Balara-1 | LM-51 | 125 | Pathari-2 |
| LT-3 | 1305 | Okhre-8 | LD-30 | 1175 | Balara-1 | LM-52 | 125 | Pathari-2 |
| LT-8 | 1505 | Okhre-8 | LD-31 | 1150 | Dhnk-3 | LM-54 | 125 | Pathari-2 |
| LT-9 | 1500 | Okhre-5 | LD-32 | 1130 | Balhra-3 | LM-55 | 125 | Pathari-2 |
| LT-12 | 1310 | Fachamara-7 | LD-33 | 1130 | Balhra-1 | LS-34 | 128 | Narsing-2 |
| LT-13 | 1315 | Fachamara-7 | LD-48 | 1181 | Bodhe-1 | LS-35 | 128 | Narsing-4 |
| LT-14 | 1308 | Fachamara-7 | LD-49 | 1185 | Bodhe-1 | LS-36 | 128 | Narsing-4 |
| LT-15 | 1655 | Fachmara-9 | LD-59 | 1175 | Balara-1 | LS-37 | 128 | Narsing-4 |
| LT-16 | 1405 | Fachamara-7 | LKm-61 | 1285 | Balara-1 | LS-38 | 128 | Narsing-4 |
| LT-17 | 1750 | Fachmara-7 | LKr-62 | 1285 | Balara-1 | LS-39 | 128 | Narsing-4 |
| LT-18 | 1710 | Fachmara-9 | LKv-60 | 1285 | Balara-1 | LS-40 | 128 | Narsing-4 |
| LT-19 | 1350 | Fachamara-7 | LT-4 | 1155 | Okhre-1 | LS-41 | 128 | Narsing-4 |
| LT-20 | 1410 | Fachamara-8 | LT-5 | 1155 | Okhre-3 | LS-42 | 128 | Narsing-4 |
| LT-21 | 1485 | Fachamara-1 | LT-6 | 1150 | Okhre-3 | LS-56 | 128 | Narsing-4 |
| LT-22 | 1505 | Sudap-1 | LT-7 | 1145 | Okhre-2 | LS-57 | 128 | Narsing-4 |
| LT-23 | 1308 | Sudap-7 | LT-10 | 1135 | Okhre-3 | |||
| LT-11 | 1130 | Okhre-3 | ||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Munankarmi, N.N.; Rana, N.; Bhattarai, T.; Shrestha, R.L.; Joshi, B.K.; Baral, B.; Shrestha, S. Characterization of the Genetic Diversity of Acid Lime (Citrus aurantifolia (Christm.) Swingle) Cultivars of Eastern Nepal Using Inter-Simple Sequence Repeat Markers. Plants 2018, 7, 46. https://doi.org/10.3390/plants7020046
Munankarmi NN, Rana N, Bhattarai T, Shrestha RL, Joshi BK, Baral B, Shrestha S. Characterization of the Genetic Diversity of Acid Lime (Citrus aurantifolia (Christm.) Swingle) Cultivars of Eastern Nepal Using Inter-Simple Sequence Repeat Markers. Plants. 2018; 7(2):46. https://doi.org/10.3390/plants7020046
Chicago/Turabian StyleMunankarmi, Nabin Narayan, Neesha Rana, Tribikram Bhattarai, Ram Lal Shrestha, Bal Krishna Joshi, Bikash Baral, and Sangita Shrestha. 2018. "Characterization of the Genetic Diversity of Acid Lime (Citrus aurantifolia (Christm.) Swingle) Cultivars of Eastern Nepal Using Inter-Simple Sequence Repeat Markers" Plants 7, no. 2: 46. https://doi.org/10.3390/plants7020046
APA StyleMunankarmi, N. N., Rana, N., Bhattarai, T., Shrestha, R. L., Joshi, B. K., Baral, B., & Shrestha, S. (2018). Characterization of the Genetic Diversity of Acid Lime (Citrus aurantifolia (Christm.) Swingle) Cultivars of Eastern Nepal Using Inter-Simple Sequence Repeat Markers. Plants, 7(2), 46. https://doi.org/10.3390/plants7020046





