Auxin and Cell Wall Invertase Related Signaling during Rice Grain Development
Abstract
:1. Introduction
2. Results
2.1. OsYUC12 and Other Endosperm YUCCA Genes Are Conserved in Cereal Species
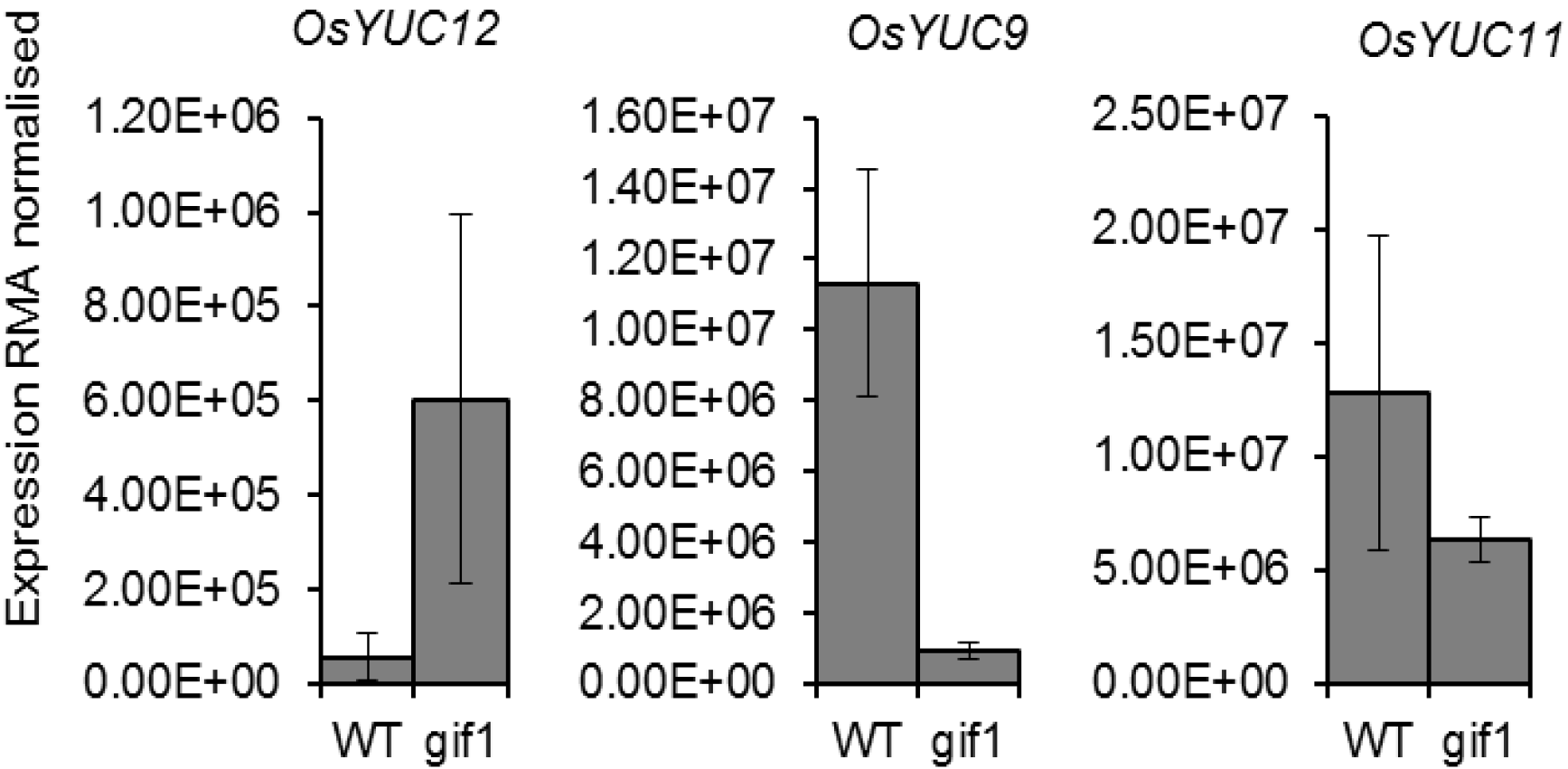
2.2. Expression of OsYUCCA Genes in the gif1 CWIN Mutant
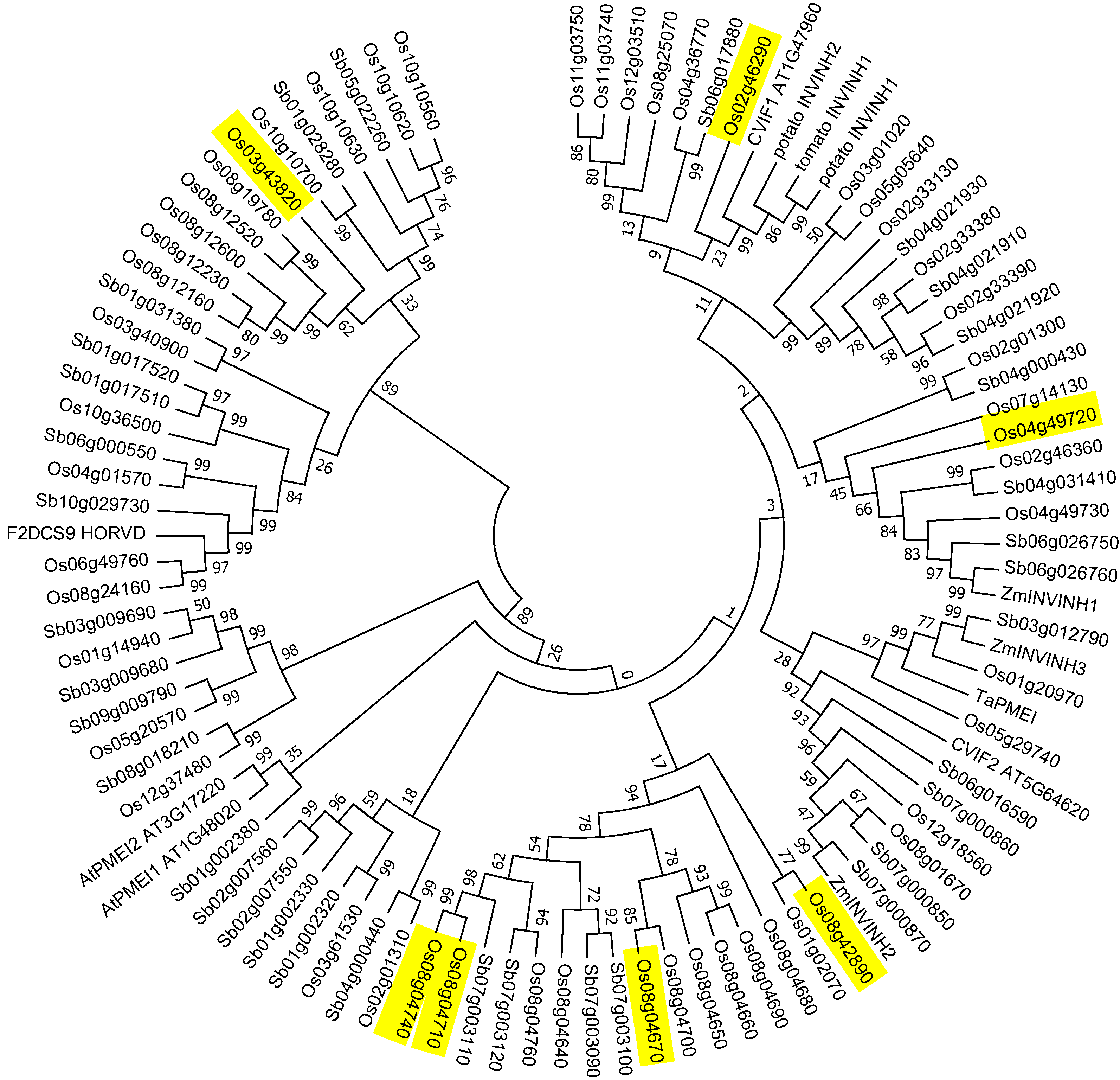
2.3. Invertase Inhibitor Phylogeny
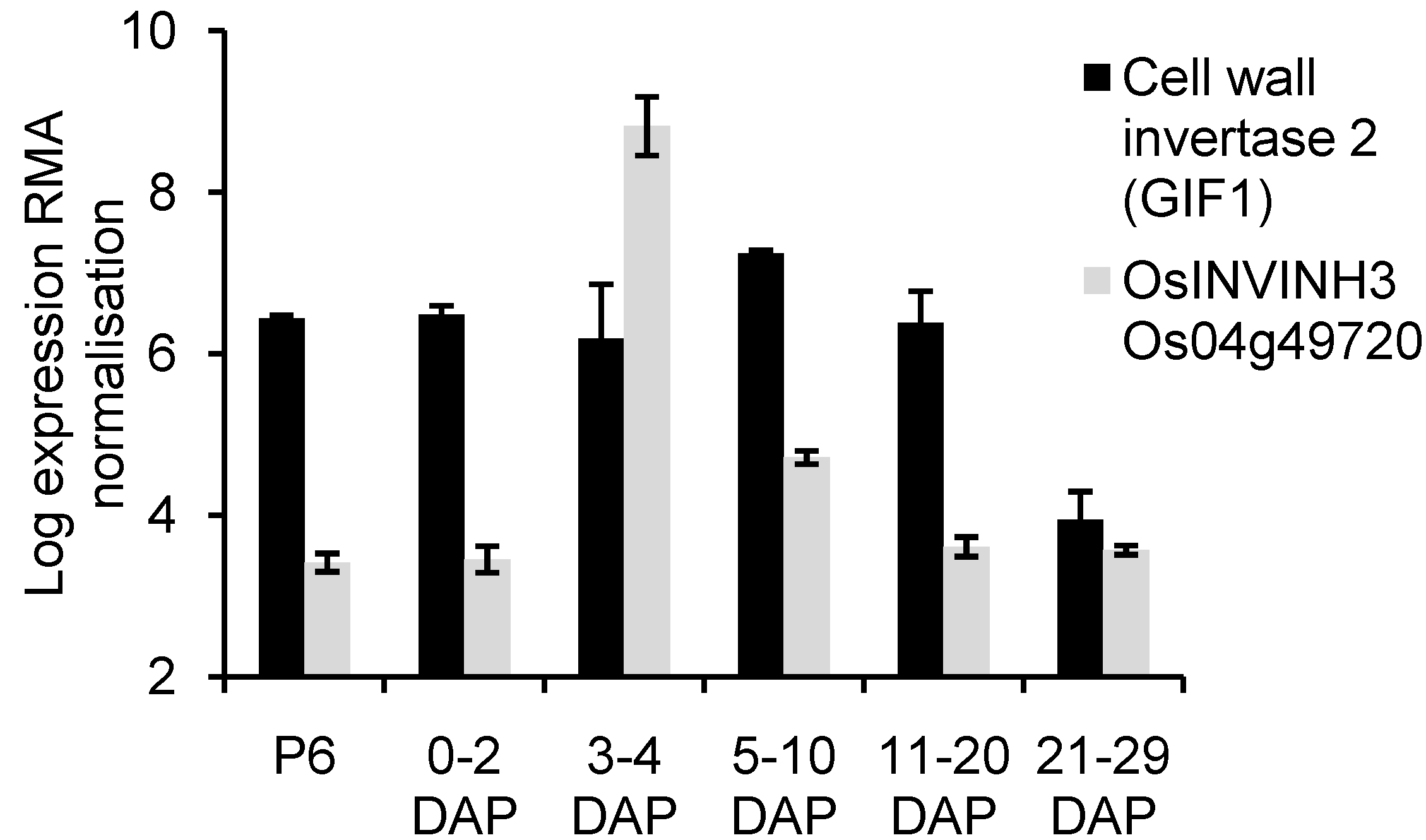
2.4. Analysis of OsCIN2/GIF1 and Putative INVINH Expression in Developing Caryopses Using Published Microarray Data
| Locus ID | Expression |
|---|---|
| Os02g46290.1 | 2 to 4 DAP, endosperm |
| Os03g43820.1 | 3-4 DAP, 5-10 DAP, endosperm |
| Os04g49720.1 | 3-4 DAP, 5-10 DAP, endosperm |
| Os08g04670.1 | 3-4 DAP, endosperm |
| Os08g04710.1 | 3-4 DAP, endosperm |
| Os08g04740.1 | 3-4 DAP, 5-10 DAP, endosperm |
| Os08g42890.1 | 3-4 DAP, 5-10 DAP, endosperm |
2.5. Coexpression Meta-Analysis Using Bait Genes OsIAA29, OsYUC12 and OsINVINH3
| Transcript ID | Description |
|---|---|
| Os12g13960.1 | Lipid transfer protein-like LTPL33 [37] |
| Os02g07628.1 | Defensin DEF5 [37] |
| Os11g45360.1 | Defensin-like DEFL15 [37] |
| Os01g70680.1 | Defensin DEF1 [37] |
| Os02g07624.1 | Defensin DEF4 [37] |
| Os11g18140.1 | homologous to ZmEBE 1 (embryo sac/basal endosperm transfer layer/embryo surrounding region [38] |
| Os06g17480.1 | Nuclear factor YC OsHAP3D /OsLEC1A [34] |
| Os05g07850.1 | Leucine-rich repeat receptor-like kinase [39] |
| Os11g14880.1 | Lipid transfer protein-like LTPL32 [37] |
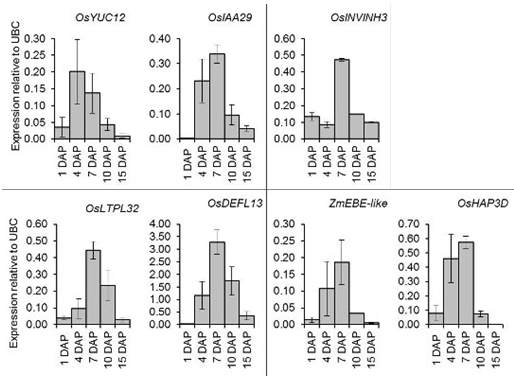
2.6. Quantitative Expression Analysis of Genes Coexpressed with OsIAA29, OsYUC12, and OsINVINH3
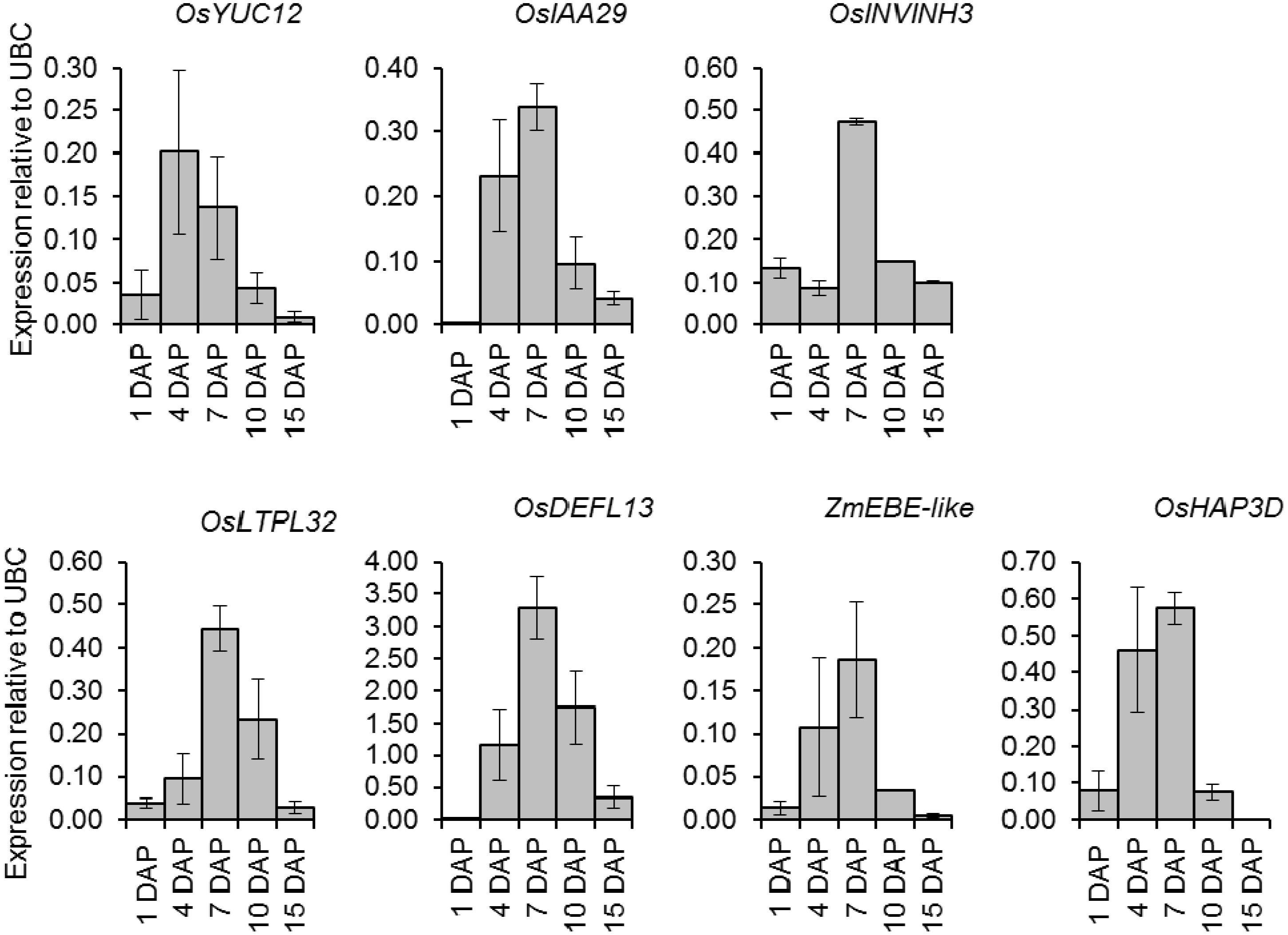
2.7. Invertase Activity in Developing Rice Grains
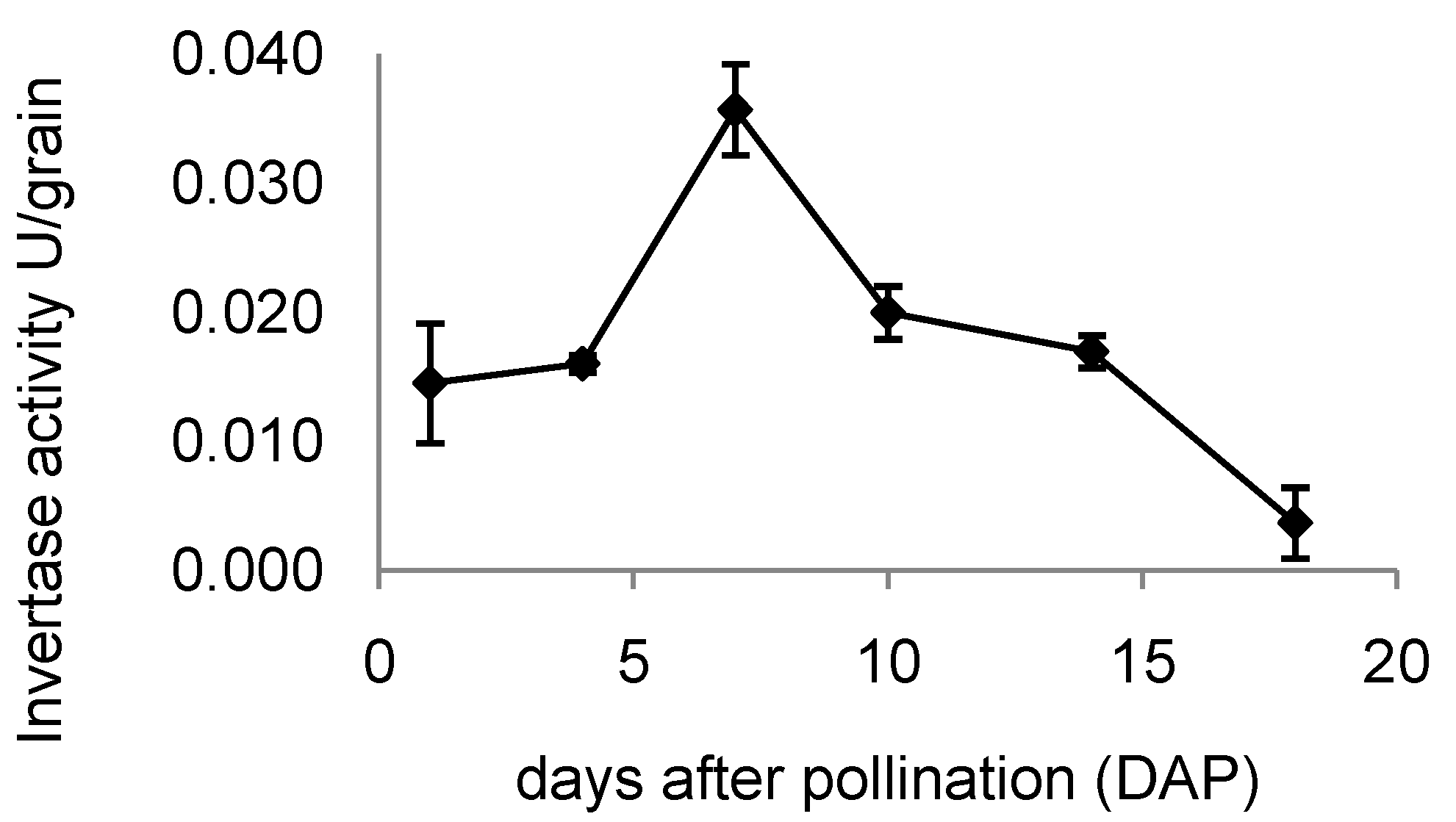
3. Discussion
3.1. Expression of IAA Related Genes, OsYUC12 and OsIAA29 Coincides with Endosperm Cellularisation
3.2. Expression of OsINVINH3 Coincides with Maximum Extracellular Invertase Activity
3.3. Rice Has 54 INVINH Homologs Several of Which Are Expressed Specifically in Developing Endosperm
3.4. Putative Signaling Proteins Coexpressed with OsYUC12, OsIAA29 and OsINVINH3
4. Experimental
4.1. Bioinformatic Analysis
4.2. Plant Material
4.3. Quantitative Reverse Transcriptase PCR
4.4. Invertase Assay
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Bernardi, J.; Lanubile, A.; Li, Q.-B.; Kumar, D.; Kladnik, A.; Cook, S.D.; Ross, J.J.; Marocco, A.; Chourey, P.S. Impaired auxin biosynthesis in the defective endosperm18 mutant is due to mutational loss of expression in the ZmYUC1 gene encoding endosperm-specific YUCCA1 protein in maize. Plant Physiol. 2012, 160, 1318–1328. [Google Scholar] [CrossRef]
- Kang, B.-H.; Xiong, Y.; Williams, D.S.; Pozueta-Romero, D.; Chourey, P.S. Miniature1-encoded cell wall invertase is essential for assembly and function of wall-in-growth in the maize endosperm transfer cell. Plant Physiol. 2009, 151, 1366–1376. [Google Scholar] [CrossRef]
- LeClere, S.; Schmelz, E.A.; Chourey, P.S. Sugar levels regulate tryptophan-dependent auxin biosynthesis in developing maize kernels. Plant Physiol. 2010, 153, 306–318. [Google Scholar] [CrossRef]
- Forestan, C.; Meda, S.; Varotto, S. ZmPIN1-mediated auxin transport is related to cellular differentiation during maize embryogenesis and endosperm development. Plant Physiol. 2010, 152, 1373–1390. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.D.; Hao, W.; Wang, L.Y.; Li, Q.; Zhang, L.X.; He, W.; Lu, B.R.; Lin, H.X.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Abu-Zaitoon, Y.M.; Bennett, K.; Normanly, J.; Nonhebel, H.M. A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific yucca. Physiol. Plant. 2012, 146, 487–499. [Google Scholar] [CrossRef]
- Jin, Y.; Ni, D.-A.; Ruan, Y.-L. Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell Online 2009, 21, 2072–2089. [Google Scholar] [CrossRef]
- Bate, N.J.; Niu, X.P.; Wang, Y.W.; Reimann, K.S.; Helentjaris, T.G. An invertase inhibitor from maize localizes to the embryo surrounding region during early kernel development. Plant Physiol. 2004, 134, 246–254. [Google Scholar] [CrossRef]
- Cho, J.I.; Lee, S.K.; Ko, S.H.; Kim, H.K.; Jun, S.H.; Lee, Y.H.; Bhoo, S.H.; Lee, K.W.; An, G.H.; Hahn, T.R.; et al. Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.). Plant Cell Rep. 2005, 24, 225–236. [Google Scholar] [CrossRef]
- Ishimaru, T.; Hirose, T.; Matsuda, T.; Goto, A.; Takahashi, K.; Sasaki, H.; Terao, T.; Ishii, R.; Ohsugi, R.; Yamagishi, T. Expression patterns of genes encoding carbohydrate-metabolizing enzymes and their relationship to grain filling in rice (Oryza sativa L.): Comparison of caryopses located at different positions in a panicle. Plant Cell Physiol. 2005, 46, 620–628. [Google Scholar] [CrossRef]
- Hothorn, M.; van den Ende, W.; Lammens, W.; Rybin, V.; Scheffzek, K. Structural insights into the pH-controlled targeting of plant cell-wall invertase by a specific inhibitor protein. Proc. Natl. Acad. Sci. USA 2010, 107, 17427–17432. [Google Scholar] [CrossRef]
- Fu, F.-F.; Xue, H.-W. Coexpression analysis identifies rice starch regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 2010, 154, 927–938. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Lin, H.N.; Childs, K.L.; Hansey, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. Genome-wide atlas of transcription during maize development. Plant J. 2011, 66, 553–563. [Google Scholar] [CrossRef]
- Dash, S.; van Hemert, J.; Hong, L.; Wise, R.P.; Dickerson, J.A. PLEXdb: Gene expression resources for plants and plant pathogens. Nucleic Acids Res. 2011, 40, D1194–D1201. [Google Scholar]
- Hong, M.J.; Kim, D.Y.; Lee, T.G.; Jeon, W.B.; Seo, Y.W. Functional characterization of pectin methylesterase inhibitor (PMEI) in wheat. Genes Genet. Syst. 2010, 85, 97–106. [Google Scholar] [CrossRef]
- Rausch, T.; Greiner, S. Plant protein inhibitors of invertases. Biochim. Biophys. Acta 1696, 1696, 253–261. [Google Scholar] [CrossRef]
- Liu, X.; Song, B.T.; Zhang, H.L.; Li, X.Q.; Xie, C.H.; Liu, J. Cloning and molecular characterization of putative invertase inhibitor genes and their possible contributions to cold-induced sweetening of potato tubers. Mol. Genet. Genomics 2010, 284, 147–159. [Google Scholar] [CrossRef]
- Mangelsen, E.; Kilian, J.; Harter, K.; Jansson, C.; Wanke, D.; Sundberg, E. Transcriptome analysis of high-temperature stress in developing barley caryopses: Early stress responses and effects on storage compound biosynthesis. Mol. Plant 2011, 4, 97–115. [Google Scholar] [CrossRef]
- Scognamiglio, M.A.; Ciardiello, M.A.; Tamburrini, M.; Carratore, V.; Rausch, T.; Camardella, L. The plant invertase inhibitor shares structural properties and disulfide bridges arrangement with the pectin methylesterase inhibitor. J. Protein Chem. 2003, 22, 363–369. [Google Scholar] [CrossRef]
- Jain, M.; Nijhawan, A.; Arora, R.; Agarwal, P.; Ray, S.; Sharma, P.; Kapoor, S.; Tyagi, A.K.; Khurana, J.P. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 2007, 143, 1467–1483. [Google Scholar] [CrossRef]
- Xue, H.W. Institute of Plant Physiology and Ecology, Shanghai Institutes of Biological Sciences: Shanghai, China, Unpublished data. 2009.
- Li, M.N.; Xu, W.Y.; Yang, W.Q.; Kong, Z.S.; Xue, Y.B. Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol. 2007, 144, 1797–1812. [Google Scholar] [CrossRef]
- Fujita, M.; Horiuchi, Y.; Ueda, Y.; Mizuta, Y.; Kubo, T.; Yano, K.; Yamaki, S.; Tsuda, K.; Nagata, T.; Niihama, M.; et al. Rice expression atlas in reproductive development. Plant Cell Physiol. 2010, 51, 2060–2081. [Google Scholar] [CrossRef]
- Gao, L.-L.; Xue, H.-W. Global analysis of expression profiles of rice receptor-like kinase genes. Mol. Plant 2012, 5, 143–153. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method—A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- Mutwil, M.; Klie, S.; Tohge, T.; Giorgi, F.M.; Wilkins, O.; Campbell, M.M.; Fernie, A.R.; Usadel, B.; Nikoloski, Z.; Persson, S. PlaNet: Combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell 2011, 23, 895–910. [Google Scholar] [CrossRef]
- Mutwil, M.; Obro, J.; Willats, W.G.T.; Persson, S. GeneCAT—Novel webtools that combine blast and co-expression analyses. Nucleic Acids Res. 2008, 36, W320–W326. [Google Scholar] [CrossRef]
- Cao, P.; Jung, K.-H.; Choi, D.; Hwang, D.; Zhu, J.; Ronald, P. The rice oligonucleotide array database: An atlas of rice gene expression. Rice 2012, 2. [Google Scholar] [CrossRef]
- Obayashi, T.; Hayashi, S.; Saeki, M.; Ohta, H.; Kinoshita, K. ATTED-II provides coexpressed gene networks for arabidopsis. Nucleic Acids Res. 2009, 37, D987–D991. [Google Scholar] [CrossRef]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of hap family genes in rice. Mol. Genet. Genomics 2008, 279, 279–289. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.-H.; Schaller, G.E.; Kieber, J.J. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef]
- Saiga, S.; Möller, B.; Watanabe-Taneda, A.; Abe, M.; Weijers, D.; Komeda, Y. Control of embryonic meristem initiation in arabidopsis by phd-finger protein complexes. Development 2012, 139, 1391–1398. [Google Scholar] [CrossRef]
- Silverstein, K.A.T.; Moskal, W.A.; Wu, H.C.; Underwood, B.A.; Graham, M.A.; Town, C.D.; VandenBosch, K.A. Small cysteine-rich peptides resembling antimicrobial peptides have been under-predicted in plants. Plant J. 2007, 51, 262–280. [Google Scholar] [CrossRef]
- Magnard, J.L.; Lehouque, G.; Massonneau, A.E.; Frangne, N.; Heckel, T.; Gutierrez-Marcos, J.F.; Perez, P.; Dumas, C.; Rogowsky, P.M. ZmEBE genes show a novel, continuous expression pattern in the central cell before fertilization and in specific domains of the resulting endosperm after fertilization. Plant Mol. Biol. 2003, 53, 821–836. [Google Scholar] [CrossRef]
- Hwang, S.G.; Kim, D.S.; Jang, C.S. Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and arabidopsis. Genetica 2011, 139, 1023–1032. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive AUX/IAA gene family in rice (Oryza sativa). Funct. Integr. Genomics 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef]
- LeClere, S.; Schmelz, E.A.; Chourey, P.S. Cell wall invertase-deficient miniature1 kernels have altered phytohormone levels. Phytochemistry 2008, 69, 692–699. [Google Scholar] [CrossRef]
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef]
- Amien, S.; Kliwer, I.; Marton, M.L.; Debener, T.; Geiger, D.; Becker, D.; Dresselhaus, T. Defensin-like ZmES4 mediates pollen tube burst in maize via opening of the potassium channel kzm1. PLoS Biol. 2010, 8. [Google Scholar] [CrossRef]
- Costa, L.M.; Yuan, J.; Rouster, J.; Paul, W.; Dickinson, H.; Gutierrez-Marcos, J.F. Maternal control of nutrient allocation in plant seeds by genomic imprinting. Curr. Biol. 2012, 22, 160–165. [Google Scholar]
- Li, M.; Singh, R.; Bazanova, N.; Milligan, A.S.; Shirley, N.; Langridge, P.; Lopato, S. Spatial and temporal expression of endosperm transfer cell-specific promoters in transgenic rice and barley. Plant Biotechnol. J. 2008, 6, 465–476. [Google Scholar] [CrossRef]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. Leafy cotyledon1-like defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef]
- Junker, A.; Mönke, G.; Rutten, T.; Keilwagen, J.; Seifert, M.; Thi, T.M.N.; Renou, J.-P.; Balzergue, S.; Viehöver, P.; Hähnel, U.; et al. Elongation-related functions of leafy cotyledon1 during the development of Arabidopsis thaliana. Plant J. 2012, 71, 427–442. [Google Scholar]
- Boisson-Dernier, A.; Kessler, S.A.; Grossniklaus, U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Botany 2011, 62, 1581–1591. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mrna using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Li, Q.F.; Sun, S.S.M.; Yuan, D.Y.; Yu, H.X.; Gu, M.H.; Liu, Q.Q. Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol. Biol. Rep. 2010, 28, 49–57. [Google Scholar]
- Tomlinson, K.L.; McHugh, S.; Labbe, H.; Grainger, J.L.; James, L.E.; Pomeroy, K.M.; Mullin, J.W.; Miller, S.S.; Dennis, D.T.; Miki, B.L.A. Evidence that the hexose-to-sucrose ratio does not control the switch to storage product accumulation in oilseeds: Analysis of tobacco seed development and effects of overexpressing apoplastic invertase. J. Exp. Botany 2004, 55, 2291–2303. [Google Scholar] [CrossRef]
- Fournier, E. Colorimetric quantification of carbohydrates. In Current Protocols in Food Analytical Chemistry; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
French, S.R.; Abu-Zaitoon, Y.; Uddin, M.M.; Bennett, K.; Nonhebel, H.M. Auxin and Cell Wall Invertase Related Signaling during Rice Grain Development. Plants 2014, 3, 95-112. https://doi.org/10.3390/plants3010095
French SR, Abu-Zaitoon Y, Uddin MM, Bennett K, Nonhebel HM. Auxin and Cell Wall Invertase Related Signaling during Rice Grain Development. Plants. 2014; 3(1):95-112. https://doi.org/10.3390/plants3010095
Chicago/Turabian StyleFrench, Sarah Russell, Yousef Abu-Zaitoon, Md. Myn Uddin, Karina Bennett, and Heather M. Nonhebel. 2014. "Auxin and Cell Wall Invertase Related Signaling during Rice Grain Development" Plants 3, no. 1: 95-112. https://doi.org/10.3390/plants3010095
APA StyleFrench, S. R., Abu-Zaitoon, Y., Uddin, M. M., Bennett, K., & Nonhebel, H. M. (2014). Auxin and Cell Wall Invertase Related Signaling during Rice Grain Development. Plants, 3(1), 95-112. https://doi.org/10.3390/plants3010095