Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth
Abstract
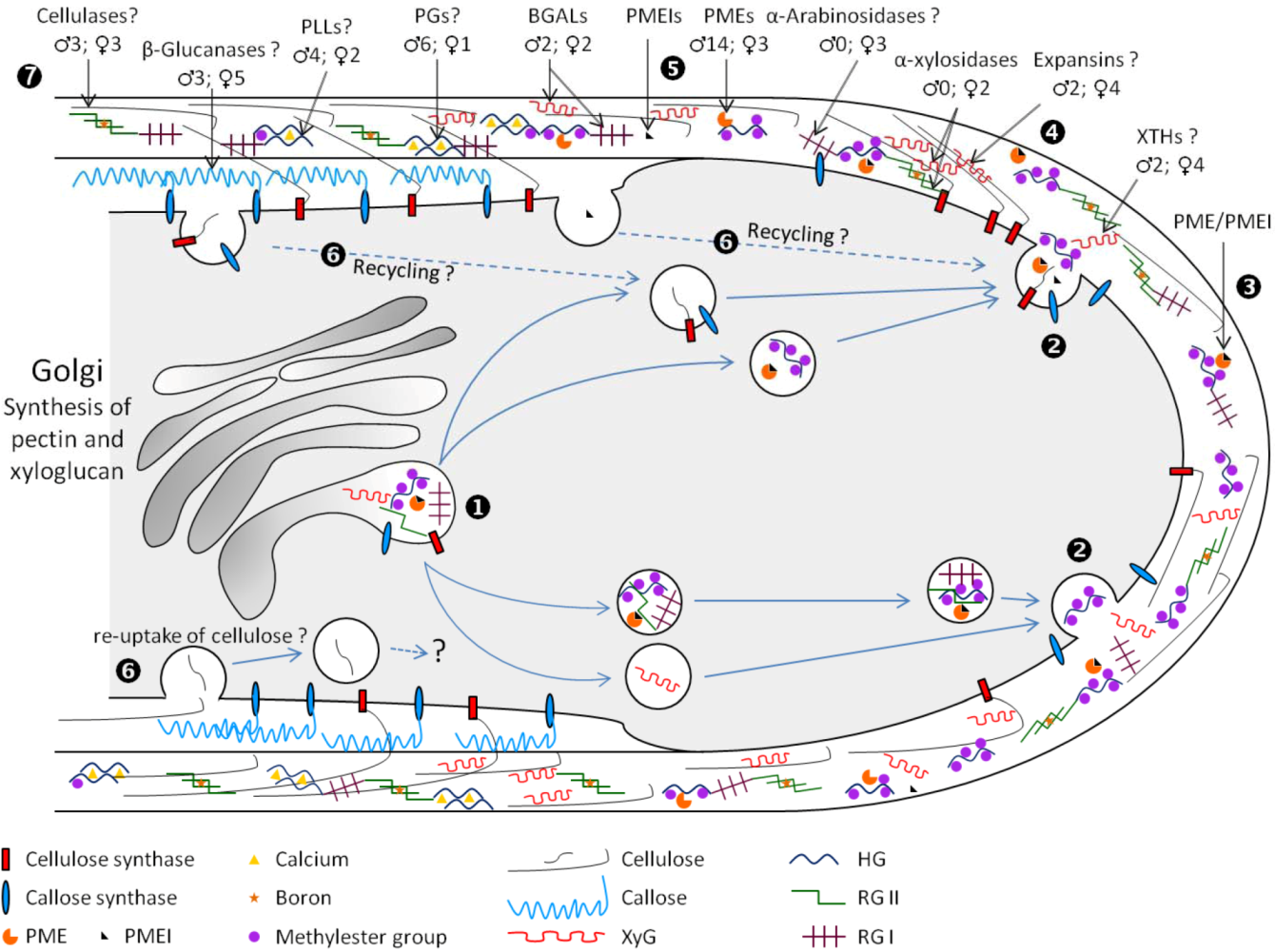
:1. Introduction
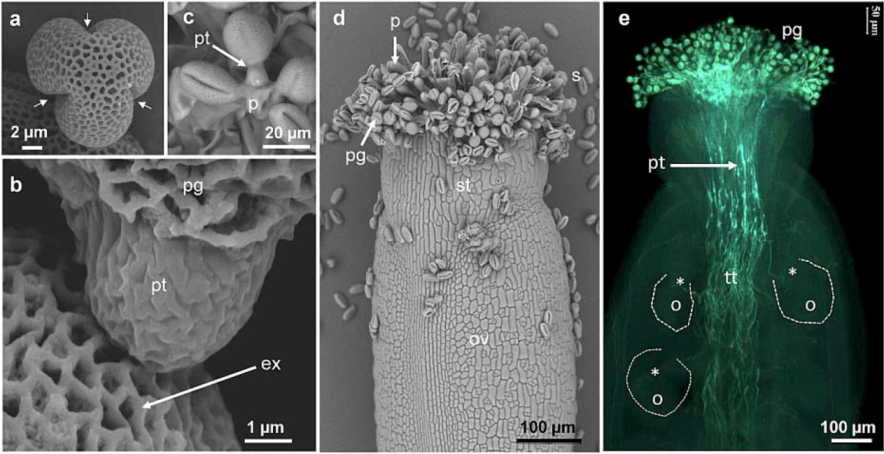
2. Cell Wall Polymers in Pollen Tubes
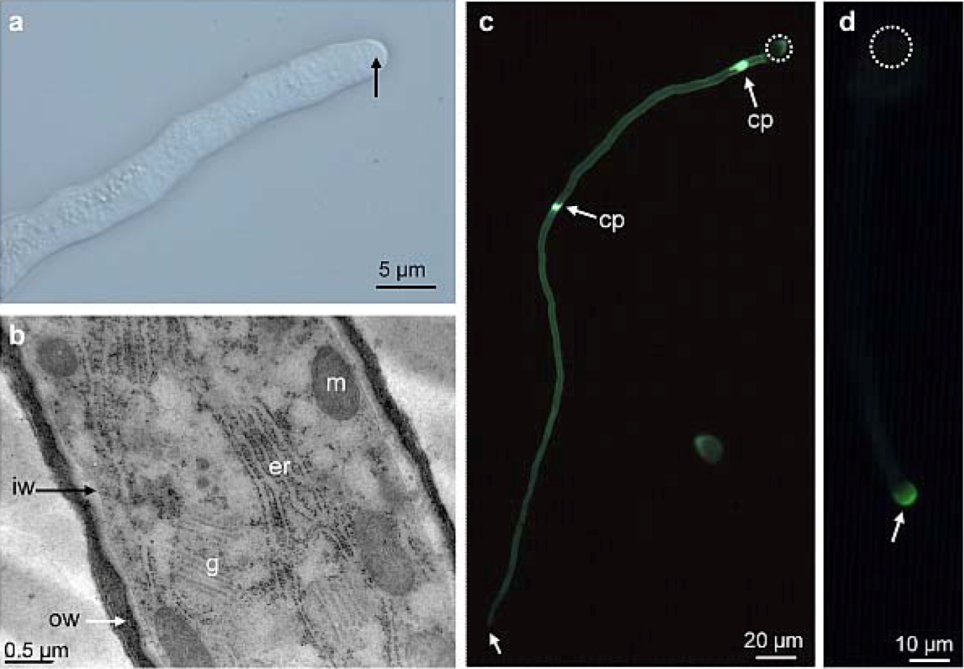
2.1. Distribution of Carbohydrate Epitopes in the Pollen Tube Cell Wall
| Probe a | Polysaccharide b | Epitope recognized c | Refs. |
|---|---|---|---|
| Antibody | |||
| CCRC-M1 | XyG | α-Fuc-(1→2)-β-Gal | [34] |
| LM15 | XyG | XXXG, XXLG, XLXG, XXGG | [35] |
| JIM5 | weakly methylesterified HG | α-MeGalA-(1→4)-α-GalA(4)-(1→4)-α-MeGalA | [36] |
| LM19 | weakly methylesterified HG | α-GalA-(1→4)(4) | [37] |
| JIM7 | partially methylesterified HG | α-GalA-(1→4)-α-MeGalA(4)-(1→4)-α-GalA | [36] |
| LM20 | partially methylesterified HG | α-MeGalA-(1→4)(4) | [37] |
| LM8 | xylogalacturonan | unknown | [38] |
| LM5 | (1→4)-β-d-galactan (RG-I) | β-Gal-(1→4)(3) | [39] |
| LM6 | (1→5)-α-l-arabinan (RG-I) | Branched α-Ara-(1→5)(5) | [40] |
| LM13 | (1→5)-α-l-arabinan (RG-I) | Linear α-Ara-(1→5)(5) | [41] |
| LAMP | callose | β-Glc-(1→3)(5) | [42] |
| Anti-RG-II | Monomeric and dimeric RG-II | unknown | [43] |
| Cytochemical reagent | |||
| Calcofluor white | β-Glucan | na | |
| Aniline blue | callose | na | [44] |
| PI | HG | na | [45] |
| Protein | |||
| CBH-I | cellulose | na | [22] |
| CBM3a | Crystalline cellulose | na | [46] |
2.2. Chemical Composition of Pollen Tube Cell Wall
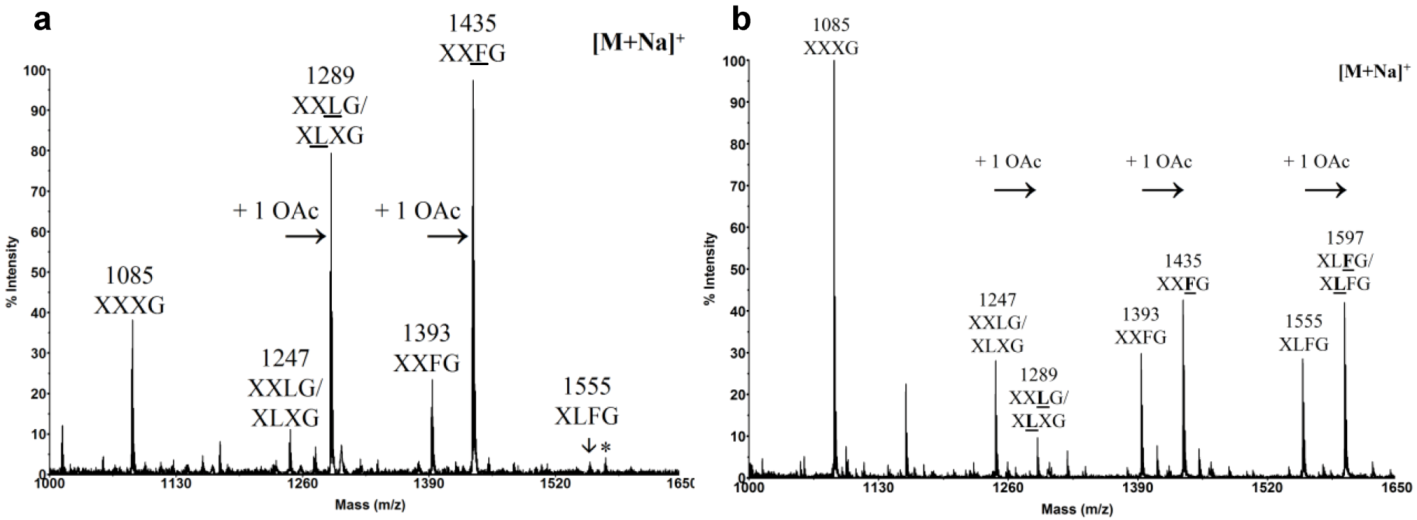
3. Cell Wall Polymer Biosynthesis in Pollen Tubes
3.1. Pectin Biosynthesis
3.1.1. Homogalacturonan
3.1.2. Xylogalacturonan
3.1.3. Rhamnogalacturonan-I
3.1.4. Rhamnogalacturonan-II
3.1.5. Pectin Methyltransferase and Pectin/XyG Acetyltransferase
3.2. Xyloglucan Biosynthesis
3.3. Cellulose Biosynthesis
3.4. Callose Biosynthesis
4. Cell Wall Remodeling during Pollen Tube Growth
4.1. Cell Expansion: Xyloglucan-Cellulose Reassembly
4.1.1. Expansins
| Cell wall metabolism | Locus | Protein name | Expression level | |||||
|---|---|---|---|---|---|---|---|---|
| Pollen grain | Pollen tube | Pistil | ||||||
| Dry | Imbibed | 4 h in vitro | Semi in vivo | stigma | ovary | |||
| Cell expansion | ||||||||
| CBMs | At1g20190 | Expansin A11 (EXPA11) | <50 | <50 | <50 | <50 | 1,614.7 | 421.6 |
| At1g26770 | Expansin A10 (EXPA10) | <50 | <50 | <50 | <50 | 1,893.9 | 641.8 | |
| At2g28950 | Expansin A6 (EXPA6) | <50 | <50 | <50 | <50 | 1,919.8 | 1,614.2 | |
| At2g39700 | Expansin A4 (EXPA4) | 4,069.6 | 3,962.1 | 3,950.8 | 2,106.8 | 321.3 | 429.6 | |
| At3g45970 | Expansin-like A1 (EXLA1) | <50 | <50 | <50 | <50 | 1,462.7 | 322.1 | |
| At3g60570 | Expansin B5 (EXPB5) | 1,020.2 | 1,139.5 | 1,347.1 | 3,002.9 | 53.9 | 52.7 | |
| Hemicellulose reassembly | ||||||||
| GH16 | At2g06850 | XTH4 | <50 | <50 | <50 | <50 | 129.8 | 1,174.4 |
| At4g03210 | XTH9 | <50 | <50 | <50 | <50 | 785.1 | 2,518.8 | |
| At4g30270 | XTH24 | <50 | <50 | <50 | <50 | 146.5 | 1,975.7 | |
| At5g65730 | XTH6 | <50 | <50 | <50 | <50 | 752.1 | 1,013.4 | |
| At1g32170 | XTH30 | 1,286.7 | 346.2 | 354.2 | 378.6 | 167.2 | 86.2 | |
| At4g18990 | XTH29 | 1,02.2 | 95.8 | 91.7 | <50 | <50 | <50 | |

4.1.2. Xyloglucan endo-Transglucosylase Hydrolase
4.2. Pectin Methylesterase and Pectin/Xyloglucan Acetylesterase
| Cell wall metabolism | Locus | Protein name | Expression level | |||||
|---|---|---|---|---|---|---|---|---|
| Pollen grain | Pollen tube | Pistil | ||||||
| Dry | Imbibed | 4 h in vitro | Semi in vivo | stigma | ovary | |||
| Esterase | ||||||||
| CE8 | At1g69940 | PPME1 | 7,600.7 | 5,098.6 | 4,702.65 | 3,209.4 | <50 | <50 |
| At2g26450 | PME13 | 4,168.8 | 4,391.9 | 4,361.9 | 4,954.5 | <50 | <50 | |
| At2g47030 | PME4/VGDH1 | 9,592.1 | 7,074.2 | 6,539.6 | 7,813.1 | 164.1 | <50 | |
| At2g47040 | PME5/VGD1 | 9,592.2 | 7,074.3 | 6,539.6 | 7,813.1 | 164.1 | <50 | |
| At3g05610 | PME21 | 7,557.3 | 4,778.3 | 4,618.9 | 5,347.8 | <50 | <50 | |
| At3g06830 | PME23 | 4,485.1 | 1,952.0 | 1,593.5 | 73.6 | <50 | <50 | |
| At3g14310 | PME3 | <50 | <50 | <50 | <50 | 284.2 | 1,047.5 | |
| At3g17060 | PME67 | 4,796.4 | 4,707.4 | 4,826.6 | 4,838.3 | <50 | <50 | |
| At3g49220 | PME34 | <50 | <50 | <50 | <50 | 1,370.3 | 1,408.2 | |
| At3g62170 | PME37 | 6,829.9 | 5,434.6 | 5,566.4 | 868.0 | <50 | <50 | |
| At4g15980 | PME43 | 1,930.8 | 905.9 | 592.5 | <50 | <50 | 51.5 | |
| At4g33230 | PME45 | 759.8 | 904.1 | 917.7 | 92.9 | <50 | <50 | |
| At5g07410 | PME48 | 7,600.8 | 5,098.7 | 4,702.65 | 3,209.4 | <50 | <50 | |
| At5g07420 | PME49 | 3,519.5 | 3,605.9 | 3,144.6 | 132.4 | <50 | <50 | |
| At5g07430 | PME50 | 8,606.5 | 7,181.6 | 7,258.6 | 7,198.7 | 90.3 | <50 | |
| At5g27870 | PME28 | 2,859.6 | 2,817.7 | 2,537.8 | 66.6 | <50 | <50 | |
| At5g47500 | PME68 | <50 | <50 | <50 | <50 | 687.9 | 1,308.9 | |
| At5g49180 | PME58 | <50 | <50 | <50 | 1,361.1 | 446.2 | 1,175.3 | |
| CE12 | At4g19410 | Putative PAE | <50 | <50 | <50 | <50 | 438.9 | 1,095.7 |

4.3. Pectin lyase
| Cell wall metabolism | Locus | Protein name | Expression level | |||||
|---|---|---|---|---|---|---|---|---|
| Pollen grain | Pollen tube | Pistil | ||||||
| Dry | Imbibed | 4 h in vitro | Semi in vivo | stigma | ovary | |||
| Lyase | ||||||||
| PL1 | At1g04680 | Probable pectate lyase 1 | <50 | <50 | <50 | <50 | 681.0 | 1,457.2 |
| At1g14420 | Probable pectate lyase 3 | 3,922.2 | 3,514.5 | 3,591.2 | 1,171.3 | <50 | <50 | |
| At2g02720 | Probable pectate lyase 6 | 3,608.0 | 3,599.7 | 3,533.1 | 4,007.9 | <50 | <50 | |
| At3g01270 | Probable pectate lyase 7 | 6,694.0 | 6,572.3 | 6,546.1 | 7,161.1 | 166.1 | 112.7 | |
| At4g24780 | Probable pectate lyase 18 | <50 | <50 | <50 | <50 | 1,969.1 | 1,173.3 | |
| At5g09280 | Major pollen allergen like protein | <50 | <50 | <50 | 3,192.1 | <50 | <50 | |
| At5g15110 | Probable pectate lyase 18 | 3,865.4 | 3,782.2 | 3,390.4 | 625.1 | <50 | <50 | |

4.4. Glycoside Hydrolases
| Family | Known activities | Possible function during pollen tube growth |
|---|---|---|
| GH3 | β-d-xylosidase (EC 3.2.1.37) α-l-arabinofuranosidase (EC 3.2.1.55) β-glucosidase (EC 3.2.1.21) | Degradation of xylan/arabinoxylan Degradation of RG-I, arabinogalactan proteins. |
| GH9 | Endo-(1→4)-β-glucanase (EC 3.2.1.4) Cellobiohydrolase (EC 3.2.1.91) β-glucosidase (EC 3.2.1.21) | Degradation of cellulose and XyG backbone May control the diameter of the pollen tube. May help to digest the cell wall of the stigma to facilitate the pollen tube penetration in the female tissues. |
| GH10 | Endo-(1→4)-β-xylanase (EC 3.2.1.8) | Degradation of xylan |
| GH17 | Endo-Glucan (1→3)-β-glucosidase (EC 3.2.1.39) Glucan (1→3)-β-glucosidase (EC 3.2.1.58) Endo-(1→3-1→4)-β-glucanase (EC 3.2.1.73) β-(1→3)-glucanosyltransglycosylase (EC 2.4.1.) | Degradation of callose or β-mixed-(1→3, 1→4) glucans. May promote pollen germination and control the mechanical properties of the pollen tube cell wall during elongation. |
| GH28 | Polygalacturonase (EC 3.2.1.15) Exo-polygalacturonase (EC 3.2.1.67) Exo-polygalacturonosidase (EC 3.2.1.82) Rhamnogalacturonase (EC 3.2.1.171) Rhamnogalacturonan α-l-rhamnopyranohydrolase (EC 3.2.1.40) Endo-xylogalacturonan hydrolase (EC 3.2.1.-) | Degradation of weakly esterified HG: cell wall loosening. May control the stiffness of the tube during elongation and/or help to digest the cell wall of the stigma thus facilitating the penetration of the tube. Degradation of RG-I backbone Degradation of xylogalacturonan |
| GH31 | α-xylosidase (EC 3.2.1.177) | Degradation of XyG or RG-II side chains |
| GH35 | β-galactosidase (EC 3.2.1.23) Exo-β-(1→4)-galactanase (EC 3.2.1.-) | Degradation of RG-I, XyG and/or arabinogalactan proteins. Turn over of pollen tube cell wall. Pollen tube guidance |
| GH43 | β-d-xylosidase (EC 3.2.1.37) α-l-arabinofuranosidase (EC 3.2.1.55) | Degradation of xylan/arabinoxylan Degradation of RG-I, arabinogalactan proteins, arabinoxylan. |
| GH51 | α-l-arabinofuranosidase (EC 3.2.1.55) | Degradation of RG-I, arabinogalactan proteins, arabinoxylan. |
| Cell wall metabolism | Locus | Protein name | Expression level | |||||
|---|---|---|---|---|---|---|---|---|
| Pollen grain | Pollen tube | Pistil | ||||||
| Dry | Imbibed | 4 h in vitro | Semi in vivo | stigma | ovary | |||
| Glycoside Hydrolases | ||||||||
| GH3 | At1g02640 | β-d-xylosidase | <50 | <50 | <50 | <50 | 66.1 | 227.3 |
| At1g78060 | β-d-xylosidase | <50 | <50 | <50 | <50 | 149.2 | 401.8 | |
| At3g19620 | β-d-xylosidase | <50 | <50 | <50 | <50 | 388.6 | 76.9 | |
| At3g47000 | β-d-xylosidase | <50 | <50 | <50 | <50 | 190.2 | 330.6 | |
| At3g62710 | β-d-xylosidase | 4,780.4 | 4,342.3 | 4,813.6 | 6,223.6 | 73 | 51.7 | |
| At5g09730 | β-d-xylosidase 3/α-l-Arabinofuranosidase | <50 | <50 | <50 | <50 | 803.5 | 2,807.8 | |
| At5g10560 | β-d-xylosidase | <50 | <50 | <50 | <50 | 191.5 | 508.8 | |
| At5g20940 | β-d-xylosidase | <50 | <50 | <50 | <50 | <50 | 62.9 | |
| At5g20950 | β-d-xylosidase | <50 | <50 | <50 | <50 | 741.4 | 1,468.4 | |
| At5g49360 | β-d-xylosidase 1/α-l-Arabinofuranosidase | <50 | <50 | <50 | <50 | 193.9 | 921.4 | |
| At5g64570 | β-d-xylosidase | <50 | <50 | <50 | <50 | 324.3 | 1,586.8 | |
| At1g70710 | Endo-(1→4)-β-glucanase (CEL1) | <50 | <50 | <50 | <50 | 732.8 | 1,921.9 | |
| At1g71380 | Endo-(1→4)-β-glucanase (CEL3) | <50 | 71.0 | 665.4 | 2,823.2 | <50 | 57.8 | |
| At2g44560 | Endo-(1→4)-β-glucanase | 2,345.4 | 2,152.9 | 2,492.6 | 1,224.1 | <50 | <50 | |
| At3g43860 | Endo-(1→4)-β-glucanase | 5,932.2 | 5,925.4 | 5,738.7 | 3,735.4 | <50 | <50 | |
| GH9 | At4g02290 | Endo-(1→4)-β-glucanase (CEL4) | <50 | <50 | <50 | <50 | <50 | 1,223.5 |
| At5g49720 | Endo-(1→4)-β-glucanase (RSW2/KOR1) | <50 | <50 | <50 | <50 | 822.2 | 1,027.5 | |
| GH10 | At4g33850 | GH 10 protein | 1,571.9 | 1,369.8 | 1,495.2 | 205.7 | <50 | <50 |
| At4g33860 | Endo-(1→4)-β xylanase, putative | 1,571.9 | 1,369.8 | 1,495.2 | 205.7 | <50 | <50 | |
| At2g05790 | Putative β-(1→3)-glucanase | <50 | <50 | <50 | <50 | 1,054.1 | 2,723.2 | |
| At3g07320 | Putative β-(1→3)-glucanase | <50 | <50 | <50 | <50 | 482.9 | 1,241.9 | |
| At3g55430 | Putative β-(1→3)-glucanase | 3,815.6 | 3,499.7 | 3,184.8 | 1,146.9 | 504.2 | 824.6 | |
| GH17 | At4g26830 | Putative β-(1→3)-glucanase | <50 | <50 | 221.1 | 3,284.7 | <50 | <50 |
| At5g20390 | Putative β-(1→3)-glucanase | 3,190.6 | 3,251.1 | 3,427.8 | 365 | <50 | <50 | |
| At5g42100 | Glucan endo-(1→3)-β-glucosidase 10 | <50 | <50 | <50 | <50 | 726.5 | 1,402.3 | |
| At5g55180 | β-(1→3)-glucanase like | <50 | <50 | <50 | <50 | 258.2 | 1,247.9 | |
| At5g64790 | β-(1→3)-glucanase | 2,007.8 | 1,991.2 | 1,972.5 | 1,439.2 | <50 | <50 | |
| At1g02790 | Exopolygalacturonase (PGA4) | 6,048.4 | 5,842.1 | 5,885.1 | 5,376.4 | 137.3 | 82.5 | |
| At2g23900 | Putative polygalacturonase | 3,341.6 | 3,090.6 | 3,228.7 | 2,308.4 | <50 | <50 | |
| At3g07820 | Polygalacturonase (PGA3) | 6,791.1 | 7,077.4 | 6,983.5 | 7,731.2 | 137.5 | 63.1 | |
| At3g07850 | Exopolygalacturonase | 7,694.0 | 7,415.6 | 7,187.2 | 3,814.6 | <50 | <50 | |
| At3g14040 | Exopolygalacturonase | 7,694.0 | 7,415.6 | 7,187.2 | 3,814.6 | <50 | <50 | |
| At4g23820 | Putative polygalacturonase | <50 | <50 | <50 | <50 | 726.1 | 1,214.5 | |
| At5g48140 | Putative polygalacturonase | 5,423.3 | 5,327.8 | 5,417.7 | 5,266.6 | <50 | <50 | |
| At1g68560 | α-xylosidase (XYL1) | <50 | <50 | <50 | <50 | 500.4 | 765.4 | |
| At3g45940 | α-xylosidase (XYL2) | <50 | <50 | <50 | <50 | 101.3 | 171.9 | |
| At2g16730 | β-galactosidase (BGAL13) | 3,753.9 | 3,752.6 | 3,219.2 | 550.5 | <50 | <50 | |
| At2g28470 | β-galactosidase (BGAL8) | <50 | <50 | <50 | <50 | 2,437.4 | 2,075.1 | |
| At4g35010 | β-galactosidase (BGAL11) | 4,248.3 | 4,056.9 | 4,264.4 | 5,783.1 | 77.7 | <50 | |
| At4g36360 | β-galactosidase (BGAL3) | <50 | <50 | <50 | <50 | 598.5 | 1,462.5 | |
| At3g49880 | β-xylosidase | <50 | <50 | <50 | <50 | 77.8 | 134.7 | |
| At3g10740 | α-l-Arabinofuranosidase | <50 | <50 | <50 | <50 | 184.9 | 402.8 | |

5. Mechanical Properties of the Cell Wall Network during Pollen Tube Growth
6. Conclusions
Acknowledgments
References
- Lord, E.M.; Russell, S.D. The mechanisms of pollination and fertilization in plants. Annu. Rev. Cell Dev. Biol. 2002, 18, 81–105. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Nasrallah, J.B.; Nasrallah, M.E. Pollen-pistil interactions and developmental regulation of pollen tube growth in Arabidopsis. Development 1994, 120, 3405–3418. [Google Scholar]
- Lennon, K.A.; Roy, S.; Hepler, P.K.; Lord, E.M. The structure of the transmitting tissue of Arabidopsis thaliana (L.) and the path of pollen tube growth. Sex. Plant Reprod. 1998, 11, 49–59. [Google Scholar]
- Palanivelu, R.; Preuss, D. Pollen tube targeting and axon guidance: Parallels in tip growth mechanisms. Trends Cell Biol. 2000, 10, 517–524. [Google Scholar] [CrossRef]
- Kim, S.; Mollet, J.C.; Dong, J.; Zhang, K.; Park, S.Y.; Lord, E.M. Chemocyanin, a small basic protein from the lily stigma, induces pollen tube chemotropism. Proc. Natl. Acad. Sci. USA 2003, 100, 16125–16130. [Google Scholar]
- McCormick, S.; Yang, H. Is there more than one way to attract a pollen tube? Trends Plant Sci. 2005, 10, 260–263. [Google Scholar]
- Boavida, L.C.; Vieira, A.M.; Becker, J.D.; Feijò, J.A. Gametophyte interaction and sexual reproduction: How plants make a zygote. Int. J. Dev. Biol. 2005, 49, 615–632. [Google Scholar]
- Johnson, M.A.; Lord, E.M. Extracellular guidance cues and intracellular signaling pathways that direct pollen tube growth. In The Pollen Tube: A Cellular and Molecular Perspective; Malho, R., Ed.; Springer: Berlin, Germany, 2006; Volume 3, pp. 223–242. [Google Scholar]
- Mollet, J.C.; Faugeron, C.; Morvan, H. Cell adhesion, separation and guidance in compatible plant reproduction. Annu. Plant Rev. 2007, 25, 69–90. [Google Scholar]
- Wang, H.J.; Huang, J.C.; Jauh, G.Y. Pollen germination and tube growth. Adv. Bot. Res. 2010, 54, 1–52. [Google Scholar]
- Boisson-Dernier, A.; Kessler, S.A.; Grossniklaus, U. The walls have ears: The role of plant CrRLK1Ls in sensing and transducing extracellular signals. J. Exp. Bot. 2011, 62, 1581–1591. [Google Scholar]
- Geitmann, A.; Steer, M. The Architecture and properties of the pollen tube cell wall. In The Pollen Tube: A Cellular and Molecular Perspective; Malhó, R., Ed.; Springer: Berlin, Germany, 2006; Volume 3, pp. 177–200. [Google Scholar]
- Geitmann, A. How to shape a cylinder: Pollen tube as a model system for the generation of complex cellular geometry. Sex. Plant Reprod. 2010, 23, 63–71. [Google Scholar]
- Nguema-Ona, E.; Coimbra, S.; Vicré-Gibouin, M.; Mollet, J.C.; Driouich, A. Arabinogalactan-proteins in root and pollen tube cells: Distribution and functional aspects. Ann. Bot. 2012, 110, 383–404. [Google Scholar]
- Cheung, A.Y.; Wu, H.M. Structural and signalling networks for the polar cell growth machinery in pollen tubes. Annu. Rev. Plant Biol. 2008, 59, 547–572. [Google Scholar]
- Moscatelli, A.; Ciampolini, F.; Rodigheiro, S.; Onelli, E.; Cresti, M.; Santo, N.; Idilli, A. Distinct endocytosis pathways identified in tobacco pollen tubes using charged nanogold. J. Cell Sci. 2007, 120, 3804–3819. [Google Scholar]
- Bove, J.; Vaillancourt, B.; Kroeger, J.; Hepler, P.K.; Wiseman, P.W.; Geitmann, A. Magnitude and direction of vesicle dynamics in growing pollen tubes using spatiotemporal image correlation spectroscopy and fluorescence recovery after photobleaching. Plant Physiol. 2008, 147, 1646–1658. [Google Scholar]
- Moscatelli, A.; Idilli, A.I. Pollen tube growth: A delicate equilibrium between secretory and endocytic pathways. J. Integr. Plant Biol. 2009, 51, 727–739. [Google Scholar]
- Zonia, L. Spatial and temporal integration of signalling networks regulating pollen tube growth. J. Exp. Bot. 2010, 61, 1939–1957. [Google Scholar]
- Roy, S.; Eckard, K.J.; Lancelle, S.; Hepler, P.K.; Lord, E.M. High-pressure freezing improves the ultrastructural preservation of in vivo grown lily pollen tubes. Protoplasma 1997, 20, 87–98. [Google Scholar]
- Li, Y.Q.; Faleri, C.; Geitmann, A.; Zhang, H.Q.; Cresti, M. Immunogold localization of arabinogalactan proteins, unesterified and esterified pectins in pollen grains and pollen tubes of Nicotiana tabacum L. Protoplasma 1995, 189, 26–36. [Google Scholar]
- Ferguson, C.; Teeri, T.T.; Siika-aho, M.; Read, S.M.; Bacic, A. Location of cellulose and callose in pollen tubes and grains of Nicotiana tabacum. Planta 1998, 206, 452–460. [Google Scholar]
- Lennon, K.A.; Lord, E.M. The in vivo pollen tube cell of Arabidopsis thaliana. I. Tube cell cytoplasm and wall. Protoplasma 2000, 214, 45–56. [Google Scholar]
- Derksen, J.; Knuiman, B.; Hoedemaekers, K.; Guyon, A.; Bonhomme, S.; Pierson, E.S. Growth and cellular organization of Arabidopsis pollen tubes in vitro. Sex. Plant Reprod. 2002, 15, 133–139. [Google Scholar]
- Dardelle, F.; Lehner, A.; Ramdani, Y.; Bardor, M.; Lerouge, P.; Driouich, A.; Mollet, J.C. Biochemical and immunocytological characterizations of Arabidopsis thaliana pollen tube cell wall. Plant Physiol. 2010, 153, 1563–1576. [Google Scholar]
- Chebli, Y.; Kaneda, M.; Zerzour, R.; Geitmann, A. The cell wall of the Arabidopsis thaliana pollen tube—Spatial distribution, recycling and network formation of polysaccharides. Plant Physiol. 2012, 160, 1940–1955. [Google Scholar]
- Derksen, J.; Li, Y.Q.; Knuiman, B.; Geurts, H. The wall of Pinus sylvestris L. pollen tubes. Protoplasma 1999, 208, 26–36. [Google Scholar]
- Yatomi, R.; Nakamura, S.; Nakamura, N. Immunochemical and cytochemical detection of wall components of germinated pollen of Gymnosperms. Grana 2002, 41, 21–28. [Google Scholar]
- Qin, Y.; Chen, D.; Zhao, J. Localization of arabinogalactan proteins in anther, pollen, and pollen tube of Nicotiana tabacum L. Protoplasma 2007, 231, 43–53. [Google Scholar]
- Parre, E.; Geitmann, A. Pectin and the role of the physical properties of the cell wall in pollen tube growth of Solanum chacoense. Planta 2005, 220, 582–592. [Google Scholar]
- Hasegawa, Y.; Nakamura, S.; Kakizoe, S.; Sato, M.; Nakamura, N. Immunocytochemical and chemical analyses of Golgi vesicles isolated from the germinated pollen of Camellia japonica. J. Plant Res. 1998, 111, 421–429. [Google Scholar]
- Wu, J.Z.; Lin, Y.; Zhang, X.L.; Pang, D.W.; Zhao, J. IAA stimulates pollen tube growth and mediates the modification of its wall composition and structure in Torenia fournieri. J. Exp. Bot. 2008, 59, 2529–2543. [Google Scholar]
- Abreu, I.; Oliveira, M. Immunolocalisation of arabinogalactan proteins and pectins in Actinidia deliciosa pollen. Protoplasma 2004, 224, 123–128. [Google Scholar]
- Puhlmann, J.; Bucheli, E.; Swain, M.J.; Dunning, N.; Albersheim, P.; Darvill, A.G.; Hahn, M.G. Generation of monoclonal antibodies against plant cell wall polysaccharides. I. Characterization of a monoclonal antibody to a terminal alpha-(1,2)-linked fucosyl-containing epitope. Plant Physiol. 1994, 104, 699–710. [Google Scholar]
- Marcus, S.E.; Verhertbruggen, Y.; Herve, C.; Ordaz-Ortiz, J.J.; Farkas, V.; Pedersen, H.L.; Willats, W.G.T.; Knox, J.P. Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 2008, 8, 60–71. [Google Scholar]
- Clausen, M.H.; Willats, W.G.T.; Knox, J.P. Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 2003, 338, 1797–1800. [Google Scholar]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar]
- Willats, W.G.T.; McCartney, L.; Steele-King, C.G.; Marcus, S.E.; Mort, A.; Huisman, M.; van Alebeek, G.-J.; Schols, H.A.; Voragen, A.G.J.; Le Goff, A.; et al. A xylogalacturonan epitope is specifically associated with plant cell detachment. Planta 2004, 218, 673–681. [Google Scholar]
- Jones, L.; Seymour, G.B.; Knox, J.P. Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1->4)-beta-d-galactan. Plant Physiol. 1997, 113, 1405–1412. [Google Scholar]
- Willats, W.G.T.; Marcus, S.E.; Knox, J.P. Generation of a monoclonal antibody specific to (1->5)-alpha-l-arabinan. Carbohydr. Res. 1998, 308, 149–152. [Google Scholar]
- Moller, I.; Marcus, S.E.; Haeger, A.; Verhertbruggen, Y.; Verhoef, R.; Schols, H.; Mikklesen, J.D.; Knox, J.P.; Willats, W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchial clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2008, 25, 49–58. [Google Scholar]
- Meikle, P.J.; Bonig, I.; Hoogenraad, N.J.; Clarke, A.E.; Stone, B.A. The location of (1→3)-beta-glucans in the walls of pollen tubes of Nicotiana alata using a (1→3)-beta-glucan-specific monoclonal antibody. Planta 1991, 185, 1–8. [Google Scholar]
- Matoh, T.; Takasaki, M.; Takabe, K.; Kobayashi, M. Immunocytochemistry of rhamnogalacturonan II in cell walls of higher Plants. Plant Cell Physiol. 1998, 39, 483–491. [Google Scholar]
- Johnson-Brousseau, S.A.; McCormick, S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically expressed genes. Plant J. 2004, 39, 761–775. [Google Scholar]
- Rounds, C.M.; Lubeck, E.; Hepler, P.K.; Winship, L.J. Propidium iodide competes with Ca2+ to label pectin in pollen tubes and Arabidopsis root hairs. Plant Physiol. 2011, 157, 175–187. [Google Scholar]
- Blake, A.W.; McCartney, L.; Flint, J.E.; Bolam, D.N.; Boraston, A.B.; Gilbert, H.J.; Knox, J.P. Understanding the biological rationale for the diversity of cellulose-directed carbohydrate-binding molecules in prokaryotic enzymes. J. Biol. Chem. 2006, 281, 29321–29329. [Google Scholar]
- WallMbDB. Available online: http://glycomics.ccrc.uga.edu/wall2/jsp/abIndex.jsp/ (accessed on 6 November 2012).
- Jauh, G.Y.; Lord, E.M. Localization of pectins and arabinogalactan-proteins in lily (Lilium longiflorum L.) pollen tube and style, and their possible roles in pollination. Planta 1996, 199, 251–261. [Google Scholar]
- Mollet, J.C.; Kim, S.; Jauh, G.Y.; Lord, E.M. AGPs, pollen tube growth and the reversible effects of Yariv phenylglycoside. Protoplasma 2002, 219, 89–98. [Google Scholar]
- Rubinstein, A.L.; Márque, J.; Cervera, M.S.; Bedinger, P.A. Extensin-like glycoproteins in the maize pollen tube wall. Plant Cell 1995, 7, 2211–2225. [Google Scholar]
- Chen, T.; Teng, N.J.; Wu, X.Q.; Wang, Y.H.; Tang, W.; Šamaj, J.; Baluška, F.; Lin, J.X. Disruption of actin filaments by latrunculin B affects cell wall construction in Picea meyeri pollen tube by disturbing vesicle trafficking. Plant Cell Physiol. 2007, 48, 19–30. [Google Scholar]
- Fernando, D.D.; Quinn, C.R.; Brenner, E.; Owens, J.N. Male gametophyte development and evolution in Gymnosperms. Int. J. Plant Dev. Biol. 2010, 4, 47–63. [Google Scholar]
- Parre, E.; Geitmann, A. More than a leak sealant. The mechanical properties of callose in pollen tubes. Plant Physiol. 2005, 137, 274–286. [Google Scholar] [CrossRef]
- Derksen, J.; Janssen, G.J.; Wolters-Arts, M.; Lichtscheidl, I.; Adlassnig, W.; Ovecka, M.; Doris, F.; Steer, M. Wall architecture with high porosity is established at the tip and maintained in growing pollen tubes of Nicotiana tabacum. Plant J. 2011, 68, 495–506. [Google Scholar] [CrossRef]
- Abercrombie, J.M.; O’Meara, B.C.; Moffatt, A.R.; Williams, J.H. Developmental evolution of flowering plant pollen tube cell walls: Callose synthase (CalS) gene expression patterns. EvoDevo 2011, 2, 14. [Google Scholar] [CrossRef]
- Lazzaro, M.D.; Donohue, J.M.; Soodavar, F.M. Disruption of cellulose synthesis by isoxaben causes tip swelling and disorganizes cortical microtubules in elongating conifer pollen tubes. Protoplasma 2003, 220, 201–207. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Stepka, M.; Ciampolini, F.; Charzynska, M.; Cresti, M. Localization of pectins in the pollen tube wall of Ornithogalum virens L. Does the pattern of pectin distribution depend on the growth rate of the pollen tube? Planta 2000, 210, 630–635. [Google Scholar]
- Li, Y.Q.; Chen, F.; Linskens, H.F.; Cresti, M. Distribution of unesterified and esterified pectins in cell walls of pollen tubes of flowering plants. Sex. Plant Reprod. 1994, 7, 145–152. [Google Scholar]
- Chen, K.M.; Wu, G.L.; Wang, Y.H.; Tian, C.T.; Samaj, J.; Baluska, F.; Lin, J.X. The block of intracellular calcium release affects the pollen tube development of Picea wilsonii by changing the deposition of cell wall components. Protoplasma 2008, 233, 39–49. [Google Scholar] [CrossRef]
- Willats, W.G.T.; Orfila, C.; Limberg, G.; Buchholt, H.C.; van Alebeek, G.-J.; Voragen, A.G.J.; Marcus, S.E.; Christensen, T.M.; Mikkelsen, J.D.; Murray, B.S.; et al. Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls: implications for pectin methyl esterase action, matrix properties, and cell adhesion. J. Biol. Chem. 2001, 276, 19404–19413. [Google Scholar]
- Fayant, P.; Girlanda, O.; Chebli, Y.; Aubin, C.; Villemure, I.; Geitmann, A. Finite element model of polar growth in pollen tubes. Plant Cell 2010, 22, 2579–2593. [Google Scholar] [CrossRef]
- APGIII. Angiosperm Phylogeny Group. An update of the Angiosperm phylogeny group classification for the orders and families of flowering plants. Bot. J. Linn. Soc. 2009, 161, 105–121.
- Fry, S.C. Cell wall polysaccharide composition and covalent crosslinking. Annu. Plant Rev. 2011, 41, 1–42. [Google Scholar]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 391–417. [Google Scholar] [CrossRef]
- Freshour, G.; Bonin, C.P.; Reiter, W.D.; Albersheim, P.; Darvill, A.G.; Hahn, M.G. Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiol. 2003, 131, 1602–1612. [Google Scholar] [CrossRef]
- Williams, J.H. Novelties of the flowering plant pollen tube underlie diversification of a key life history stage. Proc. Natl. Acad. Sci. USA 2008, 105, 11259–11263. [Google Scholar] [CrossRef]
- Laughlin, S.T.; Bertozzi, C.R. Imaging the glycome. Proc. Natl. Acad. Sci. USA 2009, 106, 12–17. [Google Scholar] [CrossRef]
- Anderson, C.T.; Wallace, I.S. Illuminating the wall: Using click chemistry to image pectins in Arabidopsis cell walls. Plant Signal. Behav. 2012, 7, 661–663. [Google Scholar] [CrossRef]
- Anderson, C.T.; Wallace, I.S.; Somerville, C.R. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proc. Natl. Acad. Sci. USA 2012, 109, 1329–1334. [Google Scholar]
- Rae, A.L.; Harris, P.J.; Bacic, A.; Clarke, A.E. Composition of the cell walls of Nicotiana alata Link et Otto pollen tubes. Planta 1985, 166, 128–133. [Google Scholar] [CrossRef]
- Nakamura, N.; Suzuki, H. Sugar composition of pollen grain and pollen tube cell walls. Phytochemistry 1981, 20, 981–984. [Google Scholar]
- Fry, S.C.; York, W.S.; Albersheim, P.; Darvill, A.; Hayashi, T.; Joseleau, J.P.; Seitz, H.U.; Kato, Y.; Pérez Lorences, E.; Maclachlan, G.A.; et al. An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol. Plant 1993, 89, 1–3. [Google Scholar] [CrossRef]
- Cavalier, D.M.; Lerouxel, O.; Neumetzler, L.; Yamauchi, K.; Reinecke, A.; Freshour, G.; Zabotina, O.A.; Hahn, M.G.; Burgert, I.; Pauly, M.; et al. Disrupting two Arabidopsis thaliana xylosyltransferase genes results in plants deficient in xyloglucan, a major primary cell wall component. Plant Cell 2008, 20, 1519–1537. [Google Scholar] [CrossRef]
- Günl, M.; Kraemer, F.; Pauly, M. Oligosaccharide Mass Profiling (OLIMP) of Cell Wall Polysaccharides by MALDI-TOF/MS. In The Plant Cell Wall: Methods and Protocols, Methods in Molecular Biology; Popper, Z.A., Ed.; Springer, Humana Press: New York, NY, USA, 2011; Volume 715, pp. 43–54. [Google Scholar]
- Lerouxel, O.; Choo, T.S.; Seveno, M.; Usadel, B.; Faye, L.; Lerouge, P.; Pauly, M. Rapid structural phenotyping of plant cell wall mutants by enzymatic oligosaccharide fingerprinting. Plant Physiol. 2002, 130, 1754–1763. [Google Scholar] [CrossRef]
- Copyright American Society of Plant Biologists. Available online: http://www.plantphysiol.org/ (accessed on 19 February 2013).
- Sekkal, M.; Huvenne, J.-P.; Legrand, P.; Sombret, B.; Mollet, J.-C.; Mouradi-Givernaud, A.; Verdus, M.-C. Direct structural identification of polysaccharides from red algae by FTIR microspectrometry. I. Localization of agar in Gracilaria verrucosa sections. Microchim. Acta 1993, 112, 1–10. [Google Scholar] [CrossRef]
- Chen, L.; Carpita, N.C.; Reiter, W.D.; Wilson, R.H.; Jeffries, C.; McCann, M.C. A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J. 1998, 16, 385–392. [Google Scholar] [CrossRef]
- Mouille, G.; Robin, S.; Lecomte, M.; Pagant, S.; Höfte, H. Classification and identification of Arabidopsis cell wall mutants using Fourier-Transform InfraRed (FT-IR) microspectroscopy. Plant J. 2003, 35, 393–404. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, L.; Wu, X.; Li, Y.; Lin, J. Boron influences pollen germination and pollen tube growth in Picea meyeri. Tree Physiol. 2003, 23, 345–351. [Google Scholar] [CrossRef]
- Wang, Q.L.; Kong, L.A.; Hao, H.Q.; Wang, X.H.; Lin, J.X.; Samaj, J.; Baluska, F. Effects of brefeldin A on pollen germination and tube growth: Antagonistic effects on endocytosis and secretion. Plant Physiol. 2005, 139, 1692–1703. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, T.; Zhang, C.; Hao, H.; Liu, P.; Zheng, M.; Baluska, F.; Samaj, J.; Lin, J. Nitric oxide modulates the influx of extracellular Ca2+ and actin filament organization during cell wall construction in Pinus bungeana pollen tubes. New Phytol. 2009, 182, 851–862. [Google Scholar] [CrossRef]
- Mohnen, D. Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 2008, 11, 266–277. [Google Scholar] [CrossRef]
- Driouich, A.; Follet-Gueye, M.L.; Bernard, S.; Kousar, S.; Chevalier, L.; Vicré-Gibouin, M.; Lerouxel, O. Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef]
- Akita, K.; Ishimizu, T.; Tsukamoto, T.; Ando, T.; Hase, S. Successive glycosyltransfer activity and enzymatic characterization of pectic polygalacturonate 4-alpha-galacturonosyltransferase solubilized from pollen tubes of Petunia axillaris using pyridylaminated oligogalacturonates as substrates. Plant Physiol. 2002, 130, 374–379. [Google Scholar] [CrossRef]
- Caffall, K.H.; Pattathil, S.; Phillips, S.; Hahn, M.G.; Mohnen, D. Arabidopsis thaliana T-DNA mutants implicate GAUT genes in the biosynthesis of pectin and xylan in cell walls and seed testa. Mol. Plant 2009, 2, 1000–1014. [Google Scholar]
- Kong, Y.; Zhou, G.; Yin, Y.; Xu, Y.; Pattathil, S.; Hahn, M.G. Molecular analysis of a family of Arabidopsis genes related to galacturonosyltransferases. Plant Physiol. 2011, 155, 1791–1805. [Google Scholar] [CrossRef]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar]
- Bouton, S.; Leboeuf, E.; Mouille, G.; Leydecker, M.T.; Talbotec, J.; Granier, F.; Lahaye, M.; Höfte, H.; Truong, H.N. QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 2002, 14, 2577–2590. [Google Scholar] [CrossRef]
- Qin, Y.; Leydon, A.R.; Manziello, A.; Pandey, R.; Mount, D.; Denic, S.; Vasic, B.; Johnson, M.A.; Palanivelu, R. Penetration of the stigma and style elicits a novel transcriptome in pollen tubes, Pointing to genes critical for growth in a pistil. PLoS Genet. 2009, 5, e1000621. [Google Scholar] [CrossRef]
- Jensen, J.K.; Sørensen, S.O.; Harholt, J.; Geshi, N.; Sakuragi, Y.; Møller, I.; Zandleven, J.; Bernal, A.J.; Jensen, N.B.; Sørensen, C.; et al. Identification of a xylogalacturonan xylosyltransferase involved in pectin biosynthesis in Arabidopsis. Plant Cell 2008, 20, 1289–1302. [Google Scholar] [CrossRef]
- Harholt, J.; Suttangkakul, A.; Scheller, H.V. Biosynthesis of pectin. Plant Physiol. 2010, 153, 384–395. [Google Scholar] [CrossRef]
- Konishi, T.; Takeda, T.; Miyazaki, Y.; Ohnishi-Kameyama, M.; Hayashi, T.; O'Neill, M.A.; Ishii, T. A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology 2007, 17, 345–354. [Google Scholar]
- Drakakaki, G.; Zabotina, O.; Delgado, I.; Robert, S.; Keegstra, K.; Raikhel, N. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 2006, 142, 1480–1492. [Google Scholar] [CrossRef]
- Rautengarten, C.; Ebert, B.; Herter, T.; Petzold, C.J.; Ishii, T.; Mukhopadhyay, A.; Usadel, B.; Scheller, H.V. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell 2011, 23, 1373–1390. [Google Scholar] [CrossRef]
- Iwai, H.; Hokura, A.; Oishi, M.; Chida, H.; Ishii, T.; Sakai, S.; Satoh, S. The gene responsible for borate cross-linking of pectin Rhamnogalacturonan-II is required for plant reproductive tissue development and fertilization. Proc. Natl. Acad. Sci. USA 2006, 103, 16592–16597. [Google Scholar]
- Wu, A.M.; Rihouey, C.; Seveno, M.; Hörnblad, E.; Singh, S.; Matsunaga, T.; Ishii, T.; Lerouge, P.; Marchant, A. The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 2009, 57, 718–731. [Google Scholar] [CrossRef]
- Brown, D.M.; Zhang, Z.; Stephens, E.; Dupree, P.; Turner, S.R. Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 2009, 57, 732–746. [Google Scholar] [CrossRef]
- Delmas, F.; Séveno, M.; Northey, J.G.; Hernould, M.; Lerouge, P.; McCourt, P.; Chevalier, C. The synthesis of the rhamnogalacturonan II component 3-deoxy-d-manno-2-octulosonic acid (Kdo) is required for pollen tube growth and elongation. J. Exp. Bot. 2008, 59, 2639–2647. [Google Scholar] [CrossRef]
- Liu, X.L.; Liu, L.; Niu, Q.K.; Xia, C.; Yang, K.Z.; Li, R.; Chen, L.Q.; Zhang, X.Q.; Zhou, Y.; Ye, D. Male gametophyte defective 4 encodes a rhamnogalacturonan II xylosyltransferase and is important for growth of pollen tubes and roots in Arabidopsis. Plant J. 2011, 65, 647–660. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, W.; Li, W.Q.; Xia, C.; Liao, H.Z.; Zhang, X.Q.; Ye, D. MALE GAMETOPHYTE DEFECTIVE 2, encoding a sialyltransferase-like protein, is required for normal pollen germination and pollen tube growth in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 829–843. [Google Scholar] [CrossRef]
- Kobayashi, M.; Kouzu, N.; Inami, A.; Toyooka, K.; Konishi, Y.; Matsuoka, K.; Matoh, T. Characterization of Arabidopsis CTP:3-deoxy-d-manno-2-octulosonate cytidylyltransferase (CMP-KDO synthetase), the enzyme that activates KDO during rhamnogalacturonan II biosynthesis. Plant Cell Physiol. 2011, 52, 1832–1843. [Google Scholar] [CrossRef]
- Goubet, F.; Mohnen, D. Solubilization and partial characterization of homogalacturonan-methyltransferase from microsomal membranes of suspension-cultured tobacco cells. Plant Physiol. 1999, 121, 281–290. [Google Scholar] [CrossRef]
- Pauly, M.; Scheller, H.V. O-Acetylation of plant cell wall polysaccharides: Identification and partial characterization of a rhamnogalacturonan O-acetyl-transferase from potato suspension-cultured cells. Planta 2000, 210, 659–667. [Google Scholar] [CrossRef]
- Mouille, G.; Ralet, M.C.; Cavelier, C.; Eland, C.; Effroy, D.; Hématy, K.; McCartney, L.; Truong, H.N.; Gaudon, V.; Thibault, J.F.; et al. Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 2007, 50, 605–614. [Google Scholar]
- Miao, Y.; Li, H.Y.; Shen, J.; Wang, J.; Jiang, L. QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J. Exp. Bot. 2011, 62, 5063–5078. [Google Scholar] [CrossRef]
- Manabe, Y.; Nafisi, M.; Verhertbruggen, Y.; Orfila, C.; Gille, S.; Rautengarten, C.; Cherk, C.; Marcus, S.E.; Somerville, S.; Pauly, M.; et al. Loss-of-function mutation of REDUCED WALL ACETYLATION2 in Arabidopsis leads to reduced cell wall acetylation and increased resistance to Botrytis cinerea. Plant Physiol. 2011, 155, 1068–1078. [Google Scholar] [CrossRef]
- Zabotina, O.A. Xyloglucan and its biosynthesis. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Lerouxel, O.; Drakakaki, G.; Alonso, A.P.; Liepman, A.H.; Keegstra, K.; Raikhel, N.; Wilkerson, C.G. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 8550–8555. [Google Scholar] [CrossRef]
- Cavalier, D.M.; Keegstra, K. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. J. Biol. Chem. 2006, 281, 34197–34207. [Google Scholar] [CrossRef]
- Vuttipongchaikij, S.; Brocklehurst, D.; Steele-King, C.; Ashford, D.A.; Gomez, L.D.; McQueen-Mason, S.J. Arabidopsis GT34 family contains five xyloglucan α-1,6-xylosyltransferases. New Phytol. 2012, 195, 585–595. [Google Scholar] [CrossRef]
- Madson, M.; Dunand, C.; Li, X.; Verma, R.; Vanzin, G.F.; Caplan, J.; Shoue, D.A.; Carpita, N.C.; Reiter, W.D. The MUR3 gene of Arabidopsis encodes a xyloglucan galactosyltransferase that is evolutionarily related to animal exostosins. Plant Cell 2003, 15, 1662–1670. [Google Scholar] [CrossRef]
- Jensen, J.K.; Schultink, A.; Keegstra, K.; Wilkerson, C.G.; Pauly, M. RNA-Seq analysis of developing nasturtium seeds (Tropaeolum majus): Identification and characterization of an additional galactosyltransferase involved in xyloglucan biosynthesis. Mol. Plant 2012, 5, 984–992. [Google Scholar]
- Perrin, R.M.; DeRocher, A.E.; Bar-Peled, M.; Zeng, W.; Norambuena, L.; Orellana, A.; Raikhel, N.V.; Keegstra, K. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 1999, 284, 1976–1979. [Google Scholar] [CrossRef]
- Vanzin, G.F.; Madson, M.; Carpita, N.C.; Raikhel, N.V.; Keegstra, K.; Reiter, W.D. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proc. Natl. Acad. Sci. USA 2002, 99, 3340–3345. [Google Scholar]
- Guerriero, G.; Fugelstad, J.; Bulone, V. What do we really know about cellulose biosynthesis in Higher Plants? J. Integr. Plant Biol. 2010, 52, 161–175. [Google Scholar] [CrossRef]
- Richmond, T. Higher plant cellulose synthases. Genome Biol. 2000, 1, reviews3001.1–reviews3001.6. [Google Scholar] [CrossRef] [Green Version]
- Doblin, M.S.; Kurek, I.; Jacob-Wilk, D.; Delmer, D.P. Cellulose biosynthesis in plants: From genes to rosettes. Plant Cell Physiol. 2002, 43, 1407–1420. [Google Scholar] [CrossRef]
- Richmond, T.A.; Somerville, C.R. Integrative approaches to determining Csl function. Plant Mol. Biol. 2001, 47, 131–143. [Google Scholar] [CrossRef]
- Bernal, A.J.; Yoo, C.M.; Mutwil, M.; Jensen, J.K.; Hou, G.; Blaukopf, C.; Sørensen, I.; Blancaflor, E.B.; Scheller, H.V.; Willats, W.G. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 2008, 148, 1238–1253. [Google Scholar] [CrossRef]
- Wang, L.; Guo, K.; Li, Y.; Tu, Y.; Hu, H.; Wang, B.; Cui, X.; Peng, L. Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 282. [Google Scholar] [CrossRef]
- Doblin, M.S.; de Melis, L.; Newbigin, E.; Bacic, A.; Read, S.M. Pollen tubes of Nicotiana alata express two genes from different β-Glucan synthase families. Plant Physiol. 2001, 125, 2040–2052. [Google Scholar]
- Cai, G.; Faleri, C.; Del Casino, C.; Emons, A.M.C.; Cresti, M. Distribution of callose synthase, cellulose synthase, and sucrose synthase in tobacco pollen tube is controlled in dissimilar ways by actin filaments and microtubules. Plant Physiol. 2011, 155, 1169–1190. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Chen, C.; Xiong, G.; Tan, X.Y.; Yang, K.Z.; Wang, Z.C.; Zhou, Y.; Ye, D.; Chen, L.Q. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 2011, 62, 5161–5177. [Google Scholar] [CrossRef]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef]
- Boavida, L.C.; Shuai, B.; Yu, H.J.; Pagnussat, G.C.; Sundaresan, V.; McCormick, S. A collection of Ds insertional mutants associated with defects in male gametophyte development and function in Arabidopsis thaliana. Genetics 2009, 181, 1369–1385. [Google Scholar]
- Verma, D.P.; Hong, Z. Plant callose synthase complexes. Plant Mol. Biol. 2001, 47, 693–701. [Google Scholar] [CrossRef]
- Nishikawa, S.; Zinkl, G.M.; Swanson, R.J.; Maruyama, D.; Preuss, D. Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol. 2005, 5, 15. [Google Scholar] [CrossRef]
- Dong, X.; Hong, Z.; Sivaramakrishnan, M.; Mahfouz, M.; Verma, D.P. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005, 42, 315–328. [Google Scholar] [CrossRef]
- Brownfield, L.; Ford, K.; Doblin, M.S.; Newbigin, E.; Read, S.; Bacic, A. Proteomic and biochemical evidence links the callose synthase in Nicotiana alata pollen tubes to the product of the NaGSL1 gene. Plant J. 2007, 52, 147–156. [Google Scholar] [CrossRef]
- Brownfield, L.; Wilson, S.; Newbigin, E.; Bacic, A.; Read, S. Molecular control of the glucan synthase-like protein NaGSL1 and callose synthesis during growth of Nicotiana alata pollen tubes. Biochem. J. 2008, 414, 43–52. [Google Scholar] [CrossRef]
- Pacini, E.; Franchi, G.G.; Ripaccioli, M. Ripe pollen structure and histochemistry of some Gymnosperms. Plant Syst. Evol. 1999, 217, 81–99. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Durachko, D.M.; Cosgrove, D.J. Two endogenous proteins that induce cell wall extension in plants. Plant Cell 1992, 4, 1425–1433. [Google Scholar]
- Li, Z.-C.; Durachko, D.M.; Cosgrove, D.J. An oat coleoptile wall protein that induces wall extension in vitro and that is antigenically related to a similar protein from cucumber hypocotyls. Planta 1993, 191, 349–356. [Google Scholar]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 1–11. [Google Scholar]
- McQueen-Mason, S.; Cosgrove, D.J. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. USA 1994, 91, 6574–6578. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Cosgrove, D.J. Expansin mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 1995, 107, 87–100. [Google Scholar]
- Sharova, E.I. Expansins: Proteins involved in cell wall softening during plant growth and morphogenesis. Russ. J. Plant Physiol. 2007, 54, 713–727. [Google Scholar]
- Arabidopsis genes. Available online: https://homes.bio.psu.edu/expansins/arabidopsis.htm/ (accessed on 6 November 2012).
- Dai, S.; Li, L.; Chen, T.; Chong, K.; Xue, Y.; Wang, T. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics 2006, 6, 2504–2529. [Google Scholar]
- Li, L.C.; Bedinger, P.A.; Volk, C.; Jones, A.D.; Cosgrove, D.J. Purification and characterization of four β-expansins (Zea m 1 Isoforms) from maize pollen. Plant Physiol. 2003, 132, 2073–2085. [Google Scholar]
- Jin, Y.; Tashpulatov, A.S.; Katholnigg, H.; Heberle-Bors, E.; Touraev, A. Isolation and characterization of two wheat β-expansin genes expressed during male gametophyte development. Protoplasma 2006, 228, 13–19. [Google Scholar]
- Zaidi, M.A.; O’Leary, S.; Wu, S.; Gleddie, S.C.; Eudes, F.; Laroche, A.; Robert, L.S. A molecular and proteomic investigation of proteins rapidly released from triticale pollen upon hydration. Plant Mol Biol. 2012, 79, 101–121. [Google Scholar]
- Cosgrove, D.J.; Bedinger, P.; Durachko, D.M. Group 1 allergens of grass pollen as cell wall-loosening agents. Proc. Natl. Acad. Sci.USA 1997, 94, 6559–6564. [Google Scholar]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2007, 2, e718. [Google Scholar]
- Magrane, M.; UniProt Consortium. UniProt Knowledgebase: A hub of integrated protein data. Database 2011. [Google Scholar] [CrossRef]
- Hende, H.; Bradford, K.J.; Brummel, D.A.; Cho, H.T.; Cosgrove, D.J.; Fleming, A.J.; Gehring, C.; Lee, Y.; McQuenn-Mason, S.; Rose, J.K.C.; et al. Nomenclature for members of the expansin superfamily of genes and proteins. Plant Mol. Biol. 2004, 55, 311–314. [Google Scholar]
- Swanson, R.; Clark, T.; Preuss, D. Expression profiling of Arabidopsis stigma tissue identifies stigma-specific genes. Sex. Plant Reprod. 2005, 18, 163–171. [Google Scholar]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: Current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar]
- Fry, S.C.; Smith, R.C.; Renwick, K.F.; Martin, D.J.; Hodge, S.K.; Matthews, K.J. Xyloglucan endotransglycosylase, a new wall-loosening enzyme activity from plants. Biochem. J. 1992, 282, 821–828. [Google Scholar]
- Nishitani, K.; Tominaga, R. Endo-xyloglucan transferase, a novel class of glycosyltransferase that catalyzes transfer of a segment of xyloglucan molecule to another xyloglucan molecule. J. Biol. Chem. 1992, 267, 21058–21064. [Google Scholar]
- Yokoyama, R.; Nishitani, K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001, 42, 1025–1033. [Google Scholar]
- Yokoyama, R.; Rose, J.K.C.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar]
- Becnel, J.; Natarajan, M.; Kipp, A.; Braam, J. Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol. Biol. 2006, 61, 451–467. [Google Scholar]
- Kurasawa, K.; Matsui, A.; Yokoyama, R.; Kuriyama, T.; Yoshizumi, T.; Matsui, M.; Suwabe, K.; Watanabe, M.; Nishitani, K. The AtXTH28 gene, a xyloglucan endotransglucosylase/hydrolase, is involved in automatic self-pollination in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 413–422. [Google Scholar]
- Zhang, G.F.; Staehelin, L.A. Functional compartmentation of the Golgi apparatus of plant cells: Immunocytochemical analysis of high-pressure frozen- and freeze-substituted sycamore maple suspension culture cells. Plant Physiol. 1992, 99, 1070–1083. [Google Scholar]
- Gaffe, J.; Tieman, D.M.; Handa, A.K. Pectin methylesterase isoforms in tomato (Lycopersicon esculentum) tissues (effects of expression of a pectin methylesterase antisense gene). Plant Physiol. 1994, 105, 199–203. [Google Scholar]
- Rhee, S.Y.; Somerville, C.R. Tetrad pollen formation in quartet mutants of Arabidopsis thaliana is associated with persistence of pectic polysaccharides of the pollen mother cell wall. Plant J. 1998, 15, 79–88. [Google Scholar]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell-wall degradation. Plant Physiol. 2003, 133, 1170–1180. [Google Scholar]
- Francis, K.E.; Lam, S.Y.; Copenhaver, G.P. Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methyl-esterase gene. Plant Physiol. 2006, 142, 1004–1013. [Google Scholar]
- Wolf, S.; Mouille, G.; Pelloux, J. Homogalacturonan methyl-esterification and plant development. Mol. Plant 2009, 2, 851–860. [Google Scholar]
- Zhu, Y.; Zhao, P.; Wu, X.; Wang, W.; Scali, M.; Cresti, M. Proteomic identification of differentially expressed proteins in mature and germinated maize pollen. Acta Physiol. Plant 2011, 33, 1467–1474. [Google Scholar]
- Ge, W.; Song, Y.; Zhang, C.; Zhang, Y.; Burlingame, A.L.; Guo, Y. Proteomic analyses of apoplastic proteins from germinating Arabidopsis thaliana pollen. Biochim. Biophys. Acta 2011, 1814, 1964–1973. [Google Scholar]
- Li, Y.Q.; Mareck, A.; Faleri, C.; Moscatelli, A.; Liu, Q.; Cresti, M. Detection and localization of pectin methylesterase isoforms in pollen tubes of Nicotiana tabacum L. Planta 2002, 214, 734–740. [Google Scholar]
- Jiang, L.; Yang, S.L.; Xie, L.F.; Puah, C.S.; Zhang, X.Q.; Yang, W.C.; Sundaresan, V.; Ye, D. VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 2005, 17, 584–596. [Google Scholar]
- Tian, G.W.; Chen, M.H.; Zaltsman, A.; Citovsky, V. Pollen-specific pectin methylesterase involved in pollen tube growth. Dev. Biol. 2006, 294, 83–91. [Google Scholar]
- Bosch, M.; Cheung, A.Y.; Hepler, P.K. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005, 138, 1334–1346. [Google Scholar]
- Wolf, S.; Grsic-Rausch, S.; Rausch, T.; Greiner, S. Identification of pollen-expressed pectin methylesterase inhibitors in Arabidopsis. FEBS Lett. 2003, 555, 551–555. [Google Scholar]
- Raiola, A.; Camardella, L.; Giovane, A.; Mattei, B.; de Lorenzo, G.; Cervone, F.; Bellincampi, D. Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors. FEBS Lett. 2004, 557, 199–203. [Google Scholar]
- Pina, C.; Pinto, F.; Feijó, J.A.; Becker, J.D. Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol. 2005, 138, 744–756. [Google Scholar]
- Zhang, G.Y.; Feng, J.; Wu, J.; Wang, X.W. BoPMEI1, a pollen-specific pectin methylesterase inhibitor, has an essential role in pollen tube growth. Planta 2010, 231, 1323–1334. [Google Scholar]
- Lehner, A.; Leroux, C.; Mollet, J.C. Unpublished work. University of Rouen: Mont Saint-Aignan, France, 2013. [Google Scholar]
- Röckel, N.; Wolf, S.; Kost, B.; Rausch, T.; Greiner, S. Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 2008, 53, 133–143. [Google Scholar]
- Leboeuf, E.; Guillon, F.; Thoiron, S.; Lahaye, M. Biochemical and immunohistochemical analysis of pectic polysaccharides in the cell walls of Arabidopsis mutant QUASIMODO 1 suspension-cultured cells: Implications for cell adhesion. J. Exp. Bot. 2005, 56, 3171–3182. [Google Scholar]
- Durand, C.; Vicre-Gibouin, M.; Follet-Gueye, M.L.; Duponchel, L.; Moreau, M.; Lerouge, P.; Driouich, A. The organization pattern of root border-likes cells of Arabidopsis thaliana is dependent on cell wall homogalacturonan. Plant Physiol. 2009, 150, 1411–1421. [Google Scholar]
- Mollet, J.C.; Park, S.Y.; Nothnagel, E.A.; Lord, E.M. A lily stylar pectin is necessary for pollen tube adhesion to an in vitro stylar matrix. Plant Cell 2000, 12, 1737–1749. [Google Scholar]
- Park, S.Y.; Jauh, G.Y.; Mollet, J.C.; Eckard, K.J.; Nothnagel, E.A.; Walling, L.L.; Lord, E.M. A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 2000, 12, 151–164. [Google Scholar]
- Tung, C.W.; Dwyer, K.G.; Nasrallah, M.E.; Nasrallah, J.B. Genome-wide identification of genes expressed in Arabidopsis pistils specifically along the path of pollen tube growth. Plant Physiol. 2005, 138, 977–989. [Google Scholar]
- Nieuwland, J.; Feron, R.; Huisman, B.A.H.; Fasolino, A.; Hilbers, C.W.; Derksen, J.; Mariani, C. Lipid transfer proteins enhance cell wall extension in tobacco. Plant Cell 2005, 17, 2009–2019. [Google Scholar]
- Chae, K.; Kieslich, C.A.; Morikis, D.; Kim, S.C.; Lord, E.M. A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 2009, 21, 3902–3914. [Google Scholar]
- Gou, J.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination and Plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar]
- Wing, R.A.; Yamaguchi, J.; Larabell, S.K.; Ursin, V.M.; McCormick, S. Molecular and genetic characterization of two pollen-expressed genes that have sequence similarity to pectate lyases of the plant pathogen Erwinia. Plant Mol. Biol. 1989, 14, 17–28. [Google Scholar]
- Wu, Y.; Qiu, X.; Du, S.; Erickson, L. PO149, a new member of pollen pectate lyase-like gene family from alfalfa. Plant Mol. Biol. 1996, 32, 1037–1042. [Google Scholar]
- Kulikauskas, R.; McCormick, S. Identification of the tobacco and Arabidopsis homologues of the pollen-expressed LAT59 gene of tomato. Plant Mol. Biol. 1997, 34, 809–814. [Google Scholar]
- Marin-Rodriguez, M.V.; Orchard, J.; Seymour, G.B. Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 2002, 53, 2115–2119. [Google Scholar] [CrossRef]
- Palusa, S.G.; Golovkin, M.; Shin, S.B.; Richardson, D.N.; Reddy, A.S. Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 2007, 174, 537–550. [Google Scholar] [CrossRef]
- Sun, L.; van Nocker, S. Analysis of promoter activity of members of the PECTATE LYASE-LIKE (PLL) gene family in cell separation in Arabidopsis. BMC Plant Biol. 2010, 10, 152. [Google Scholar] [CrossRef]
- Dai, S.; Chen, T.; Chong, K.; Xue, Y.; Liu, S.; Wang, T. Proteomics identification of differentially expressed proteins associated with pollen germination and tube growth reveals characteristics of germinated Oryza sativa pollen. Mol. Cell. Proteomics 2007, 6, 207–230. [Google Scholar]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1991, 280, 309–316. [Google Scholar]
- Glycoside Hydrolase Family Classification. Available online: http://www.cazy.org/Glycoside-Hydrolases.html/ (accessed on 23 October 2012).
- Del Campillo, E.; Lewis, L.N. Occurrence of 9.5 cellulase and other hydrolases in flower reproductive organs undergoing major cell wall disruption. Plant Physiol. 1992, 99, 1015–1020. [Google Scholar] [CrossRef]
- Aouar, L.; Chebli, Y.; Geitmann, A. Morphogenesis of complex plant cell shapes: The mechanical role of crystalline cellulose in growing pollen tubes. Sex. Plant Reprod. 2010, 23, 15–27. [Google Scholar] [CrossRef]
- Roggen, H.P.J.; Stanley, R.G. Cell-wall-hydrolysing enzymes in wall formation as measured by pollen-tube extension. Planta 1969, 84, 295–303. [Google Scholar] [CrossRef]
- Takeda, H.; Yoshikawa, T.; Liu, X.Z.; Nakagawa, N.; Li, Y.Q.; Sakurai, N. Molecular cloning of two exo-beta-glucanases and their in vivo substrates in the cell walls of lily pollen tubes. Plant Cell Physiol. 2004, 45, 436–444. [Google Scholar] [CrossRef]
- Kotake, T.; Li, Y.Q.; Takahashi, M.; Sakurai, N. Characterization and function of wall-bound exo-beta-glucanases of Lilium longiflorum pollen tubes. Sex. Plant Reprod. 2000, 13, 1–9. [Google Scholar] [CrossRef]
- Hruba, P.; Honys, D.; Twell, D.; Capkova, V.; Tupy, J. Expression of beta-galactosidase and beta-xylosidase genes during microspore and pollen development. Planta 2005, 220, 931–940. [Google Scholar] [CrossRef]
- Pressey, R.; Reger, B.J. Polygalacturonase in pollen from corn and other grasses. Plant Sci. 1969, 59, 57–62. [Google Scholar] [CrossRef]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef]
- Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815.
- González-Carranza, Z.H.; Elliott, K.A.; Roberts, J.A. Expression of polygalacturonases and evidence to support their role during cell separation processes in Arabidopsis thaliana. J. Exp. Bot. 2007, 58, 3719–3730. [Google Scholar] [CrossRef]
- Kim, J.; Shiu, S.H.; Thoma, S.; Li, W.H.; Patterson, S.E. Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 2007, 7, R87. [Google Scholar]
- Torki, M.; Mandaron, P.; Mache, R.; Falconet, D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 2000, 242, 427–436. [Google Scholar] [CrossRef]
- Huang, L.; Ye, Y.; Zhang, Y.; Zhang, A.; Liu, T.; Cao, J. BcMF9, a novel polygalacturonase gene, is required for both Brassica campestris intine and exine formation. Ann. Bot. 2009, 104, 1339–1351. [Google Scholar] [CrossRef]
- Huang, L.; Cao, J.; Zhang, A.; Ye, Y.; Zhang, Y.; Liu, T. The polygalacturonase gene BcMF2 from Brassica campestris is associated with intine development. J. Exp. Bot. 2009, 60, 301–313. [Google Scholar]
- Tamari, F.; Shore, J.S. Allelic variation for a short-specific polygalacturonase in Turnera subulata: Is it associated with the degree of self-compatibility? Int. J. Plant Sci. 2006, 167, 125–133. [Google Scholar] [CrossRef]
- Dearnaley, J.D.W.; Daggard, G.A. Expression of a polygalacturonase enzyme in germinating pollen of Brassica napus. Sex. Plant Reprod. 2001, 13, 265–271. [Google Scholar] [CrossRef]
- Bosch, M.; Hepler, P.K. Pectin methylesterases and pectin dynamics in pollen tubes. Plant Cell 2005, 17, 3219–3226. [Google Scholar] [CrossRef]
- Dey, P.M.; del Campillo, E. Biochemistry of the multiple forms of glycosidases in plants. Adv. Enzymol. Relat. Areas Mol. Biol. 1984, 56, 141–249. [Google Scholar]
- Ahn, Y.O.; Zheng, M.; Bevan, D.R.; Esen, A.; Shiu, S.H.; Benson, J.; Peng, H.P.; Miller, J.T.; Cheng, C.L.; Poulton, J.E.; et al. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry 2007, 68, 1510–1520. [Google Scholar] [CrossRef]
- Tanthanuch, W.; Chantarangsee, M.; Maneesan, J.; Ketudat-Cairns, J. Genomic and expression analysis of glycosyl hydrolase family 35 genes from rice (Oryza sativa L.). BMC Plant Biol. 2008, 8, 84. [Google Scholar] [CrossRef]
- Singh, M.B.; Knox, R.B. Grass pollen allergens: Antigenic relationships detected using monoclonal antibodies and dot blotting immunoassay. Int. Arch. Allergy Appl. Immunol. 1985, 78, 300–304. [Google Scholar] [CrossRef]
- Cheung, A.Y.; Wang, H.; Wu, H.M. A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 1995, 82, 383–393. [Google Scholar] [CrossRef]
- Wu, H.M.; Wang, H.; Cheung, A.Y. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell 1995, 82, 395–403. [Google Scholar] [CrossRef]
- Rogers, H.J.; Maund, S.L.; Johnson, L.H. A β-galactosidase-like gene is expressed during tobacco pollen development. J. Exp. Bot. 2001, 52, 67–75. [Google Scholar] [CrossRef]
- Walker, M.; Tehseen, M.; Doblin, M.S.; Pettolino, F.A.; Wilson, S.M.; Bacic, A.; Golz, J.F. The transcriptional regulator LEUNIG_HOMOLOG regulates mucilage release from the Arabidopsis testa. Plant Physiol. 2011, 156, 46–60. [Google Scholar] [CrossRef]
- Western, T.L.; Burn, J.; Tan, W.L.; Skinner, D.J.; Martin-McCaffrey, L.; Moffatt, B.A.; Haughn, G.W. Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 2001, 127, 998–1011. [Google Scholar] [CrossRef]
- Sampedro, J.; Pardo, B.; Gianzo, C.; Guitian, E.; Revilla, G.; Zarra, I. Lack of alpha-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol. 2010, 154, 1105–1115. [Google Scholar] [CrossRef]
- Lehner, A.; Dardelle, F.; Soret-Morvan, O.; Lerouge, P.; Driouich, A.; Mollet, J.C. Pectins in the cell wall of Arabidopsis thaliana pollen tube and pistil. Plant Signal. Behav. 2010, 5, 1282–1285. [Google Scholar] [CrossRef]
- Anderson, J.R.; Barnes, W.S.; Bedinger, P. 2,6-Dichlorobenzonitrile, a cellulose biosynthesis inhibitor, affects morphology and structural integrity of petunia and lily pollen tubes. Plant Physiol. 2002, 159, 61–67. [Google Scholar] [CrossRef]
- Whitney, S.E.C.; Wilson, E.; Webster, J.; Bacic, A.; Reid, J.S.G.; Gidley, M.J. Effects of structural variation in xyloglucan polymers on interactions with bacterial cellulose. Am. J. Bot. 2006, 93, 1402–1414. [Google Scholar] [CrossRef]
- Pena, M.J.; Ryden, P.; Madson, M.; Smith, A.C.; Carpita, N.C. The galactose residues of xyloglucan are essential to maintain mechanical strength of the primary cell walls in Arabidopsis during growth. Plant Physiol. 2004, 134, 443–451. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Andeme-Onzighi, C.; Aboughe-Angone, S.; Bardor, M.; Ishii, T.; Lerouge, P.; Driouich, A. The reb1-1 mutation of Arabidopsis: Effect on the structure and localization of galactose-containing cell wall polysaccharides. Plant Physiol. 2006, 140, 1406–1417. [Google Scholar] [CrossRef]
- Zabotina, O.A.; van de Ven, W.T.; Freshour, G.; Drakakaki, G.; Cavalier, D.; Mouille, G.; Hahn, M.G.; Keegstra, K.; Raikhel, N.V. Arabidopsis XXT5 gene encodes a putative alpha-1,6-xylosyltransferase that is involved in xyloglucan biosynthesis. Plant J. 2008, 56, 101–115. [Google Scholar] [CrossRef]
- Gille, S.; Pauly, M. O-acetylation of plant cell wall polysaccharides. Front. Plant Sci. 2012, 3. [Google Scholar] [CrossRef]
- Pena, P.J.; Kong, Y.; York, W.S.; O’Neill, M.A. A galacturonic acid-containing xyloglucan is involved in Arabidopsis root hair tip growth. Plant Cell 2012, 24, 1–14. [Google Scholar] [CrossRef]
- Wang, T.; Zabotina, O.; Hong, M. Pectin-cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry 2012, 51, 9846–9856. [Google Scholar] [CrossRef]
- Vogler, H.; Draeger, C.; Weber, A.; Felekis, D.; Eichenberger, C.; Routier-Kierzkowska, A.L.; Boisson-Dernier, A.; Ringli, C.; Nelson, B.J.; Smith, R.S.; et al. The pollen tube: A soft shell with a hard core. Plant J. 2012. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mollet, J.-C.; Leroux, C.; Dardelle, F.; Lehner, A. Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth. Plants 2013, 2, 107-147. https://doi.org/10.3390/plants2010107
Mollet J-C, Leroux C, Dardelle F, Lehner A. Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth. Plants. 2013; 2(1):107-147. https://doi.org/10.3390/plants2010107
Chicago/Turabian StyleMollet, Jean-Claude, Christelle Leroux, Flavien Dardelle, and Arnaud Lehner. 2013. "Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth" Plants 2, no. 1: 107-147. https://doi.org/10.3390/plants2010107
APA StyleMollet, J.-C., Leroux, C., Dardelle, F., & Lehner, A. (2013). Cell Wall Composition, Biosynthesis and Remodeling during Pollen Tube Growth. Plants, 2(1), 107-147. https://doi.org/10.3390/plants2010107