A Multidimensional Approach to Cereal Caryopsis Development: Insights into Adlay (Coix lacryma-jobi L.) and Emerging Applications
Abstract
1. Introduction
2. Three-Dimensional Imaging Technologies for Structural Analysis of Cereal Caryopses
2.1. Micro-CT-Based Non-Destructive 3D Imaging
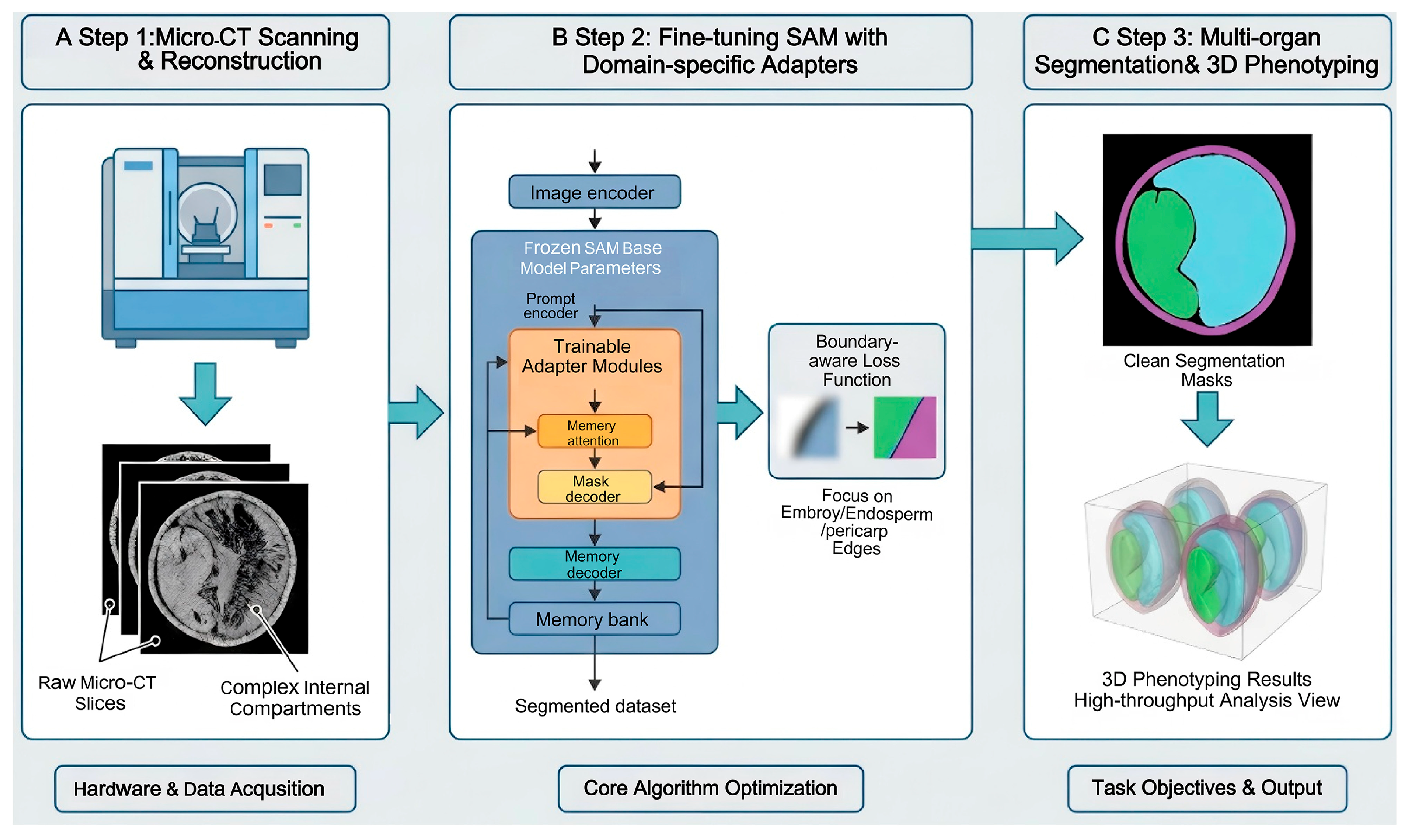
2.2. Deep Learning-Based Analysis and Phenotypic Trait Extraction
3. Genetic Regulation and Multi-Omics Analysis of Cereal Caryopsis Development
3.1. Genetic Basis of Cereal Caryopsis Development
3.1.1. Transcriptional Regulation of Storage Reserves
3.1.2. Phytohormones and Source-Sink Coordination
3.1.3. Limitations of Current Genomic Approaches: The “Position Effect” Blind Spot
3.2. Advances in Multi-Omics Studies of Caryopsis Development
3.2.1. Transcriptome-Metabolome Association Analysis
3.2.2. Spatial Omics: Bridging the Genotype-Phenotype Gap
3.3. Core Technical Challenges in Cereal Developmental Studies
4. Application Prospects of Multi-Dimensional Analytical Technologies in Adlay Research
4.1. Current Research Status and Critical Bottlenecks
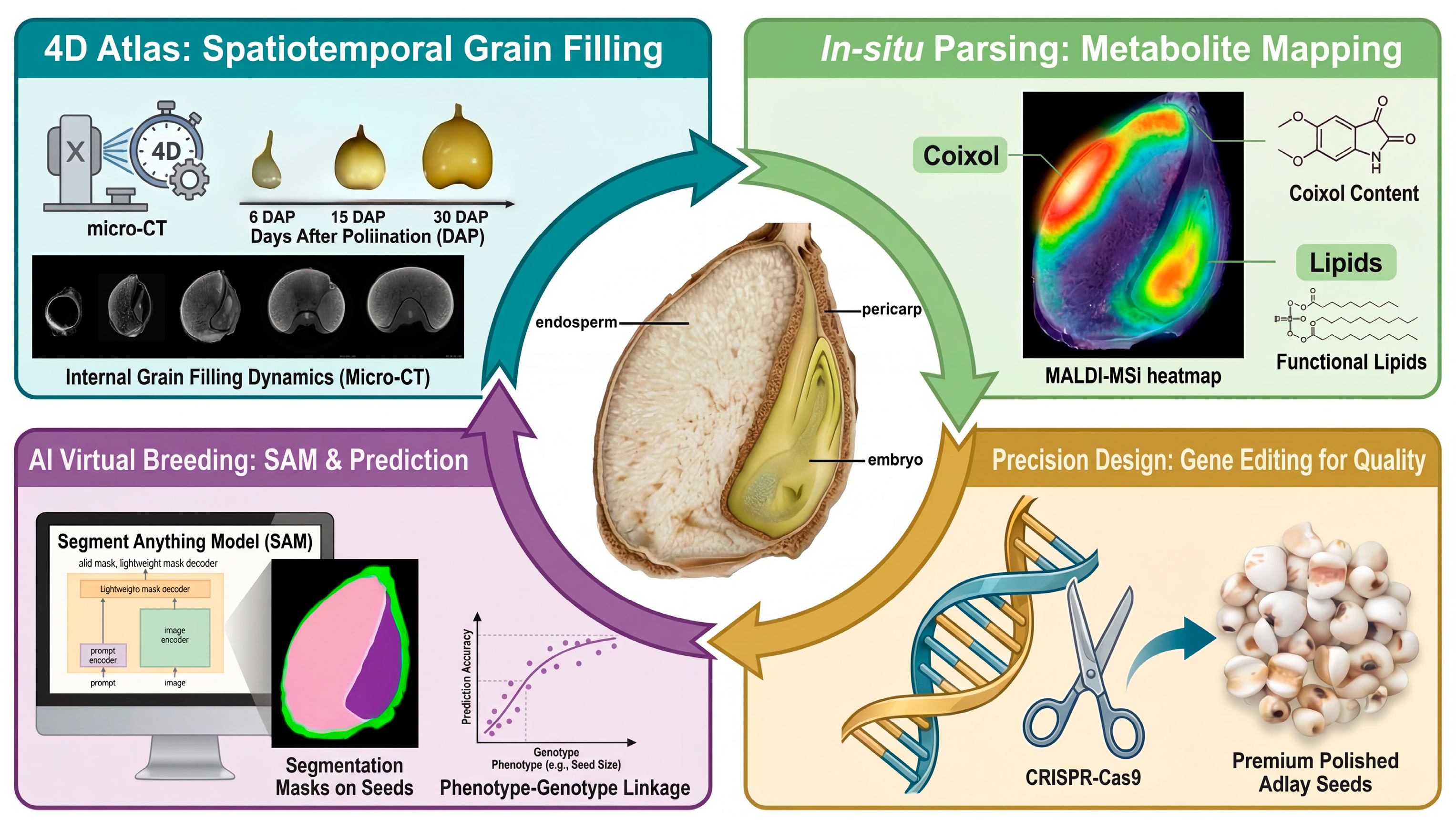
4.2. Future Directions: A Leapfrog Strategy via Multi-Dimensional Integration
- Project 1: The 4D Adlay Caryopsis Atlas (Spatiotemporal Morphogenesis)
- Project 2: In situ Pathway Parsing (Spatial Multi-Omics)
- Project 3: AI-Assisted “Virtual Breeding” (Predictive Phenomics)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yilmaz, H.; Yilmaz, A. Hidden Hunger in the Age of Abundance: The Nutritional Pitfalls of Modern Staple Crops. Food Sci. Nutr. 2025, 13, e4610. [Google Scholar] [CrossRef]
- Bao, J.; Malunga, L.N. Editorial: Compositional Diversity in Cereals in Relation to Their Nutritional Quality and Health Benefits. Front. Nutr. 2021, 8, 819923. [Google Scholar] [CrossRef]
- Spina, A.; Zingale, S. Title of the chapter: Health Benefits of Minor Cereals. In Sustainable Food Science—A Comprehensive Approach; Ferranti, P., Ed.; Elsevier: Oxford, UK, 2023; pp. 16–39. [Google Scholar] [CrossRef]
- Weng, W.F.; Peng, Y.; Pan, X.; Yan, J.; Li, X.D.; Liao, Z.Y.; Cheng, J.P.; Gao, A.J.; Yao, X.; Ruan, J.J.; et al. Adlay, an ancient functional plant with nutritional quality, improves human health. Front. Nutr. 2022, 9, 1019375. [Google Scholar] [CrossRef]
- Liu, H.; Shi, J.; Cai, Z.; Huang, Y.; Lv, M.; Du, H.; Gao, Q.; Zuo, Y.; Dong, Z.; Huang, W.; et al. Evolution and Domestication Footprints Uncovered from the Genomes of Coix. Mol. Plant 2020, 13, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.W.; Henry, A.G.; Liu, D.Y.; Allen, E.; Liu, L.; Sheng, P.F. Revealing Coix lacryma-jobi var. lacryma-jobi (Job’s tears) in Han Dynasty burials with evidence from phytolith identification. Veg. Hist. Archaeobot. 2025, 1–8. [Google Scholar] [CrossRef]
- Li, F.Y.; Shen, M.; Huang, Q.S. Coix Seed Industry Blue Book: Annual Report on China’s Coix Seed Industry Development No. 3; Social Sciences Academic Press: Beijing, China, 2019; pp. 2–5. ISBN 9787520152297. (In Chinese) [Google Scholar]
- Zeng, Y.; Yang, J.; Chen, J.; Pu, X.; Li, X.; Yang, X.; Yang, L.; Ding, Y.; Nong, M.; Zhang, S.; et al. Actional Mechanisms of Active Ingredients in Functional Food Adlay for Human Health. Molecules 2022, 27, 4808. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wan, Y.; Wang, D.Z.; Li, X.G.; Wu, J.J.; Xiao, J.; Chen, K.M.; Han, X.; Chen, Y. Spatiotemporal transcriptomics reveals key gene regulation for grain yield and quality in wheat. Genome Biol. 2025, 26, 93. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, H.; Jiang, S.; Kang, J.; Li, D.; Wang, K.; Xie, S.; Tong, C.; Liu, C.; Hu, G.; et al. A single-cell multi-omics atlas of rice. Nature 2025, 644, 722–730. [Google Scholar] [CrossRef]
- Zhu, W.; Miao, X.; Qian, J.; Chen, S.; Jin, Q.; Li, M.; Han, L.; Zhong, W.; Xie, D.; Shang, X.; et al. A translatome-transcriptome multi-omics gene regulatory network reveals the complicated functional landscape of maize. Genome Biol. 2023, 24, 60. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, M.; Wang, J.; Diao, J.; Wu, Y.; Cheng, J.; Ban, Q. Exploring the hydration promotion and cooking quality improvement of adlay seed by high hydrostatic pressure. LWT-Food Sci. Technol. 2022, 171, 114158. [Google Scholar] [CrossRef]
- Wang, L.R.; Sui, N.; Lv, H.S.; Tang, Q.; Shi, M.; Fan, H.Y.; Zhou, W.; Meng, Y.L.; Kai, G.Y. Effects of potassium foliage supplementation on Coix lacryma-jobi L. yield formation and source-sink relationship compared with those of soil supplementation. Ind. Crops Prod. 2022, 180, 114754. [Google Scholar] [CrossRef]
- Legland, D.; Alvarado, C.; Badel, E.; Guillon, F.; King, A.; Le, T.D.Q.; Rivard, C.; Paré, L.; Chateigner-Boutin, A.L.; Girousse, C. Synchrotron Based X-ray Microtomography Reveals Cellular Morphological Features of Developing Wheat Grain. Appl. Sci. 2022, 12, 3454. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Huang, G.; Wang, J.; Lv, L.; Zhao, S.; Lu, X.; Zhang, M.; Guo, M.; Zhang, C.; et al. Association analysis of maize stem vascular bundle micro-characteristics with yield components based on micro-CT and identification of related genes. Sci. Rep. 2025, 15, 13009. [Google Scholar] [CrossRef] [PubMed]
- Olsen, O.A. The Modular Control of Cereal Endosperm Development. Trends Plant Sci. 2020, 25, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, M.W.; Liu, C.M. Cereal Endosperms: Development and Storage Product Accumulation. Annu. Rev. Plant Biol. 2022, 73, 255–291. [Google Scholar] [CrossRef] [PubMed]
- Bhuvaneswari, R.; Saravanan, K.R.; Vennila, S.; Suganthi, S. Advances in Genomic Selection for Enhanced Crop Improvement: Bridging the Gap between Genomics and Plant Breeding. Plant Sci. Arch. 2020, 5, 11–16. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Coleman, K.; Schroeder, A.; Li, M. Unlocking the power of spatial omics with AI. Nat. Methods 2024, 21, 1378–1381. [Google Scholar] [CrossRef]
- Crozier, D.; Riera-Lizarazu, O.; Rooney, W.L. Application of X-ray computed tomography to analyze the structure of sorghum grain. Plant Methods 2022, 18, 3. [Google Scholar] [CrossRef]
- Du, Z.; Hu, Y.; Ali, B.N.; Mahmood, A. X-ray computed tomography for quality inspection of agricultural products: A review. Food Sci. Nutr. 2019, 7, 3146–3160. [Google Scholar] [CrossRef]
- Van, D.L.; Verboven, P.; Nicolai, B. Panoptic segmentation for complete labeling of fruit microstructure in 3D micro-CT images with deep learning. Plant Phenomics 2025, 7, 100087. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, J.L.; Liao, S.J.; Zhang, Y.; Lu, X.J.; Guo, X.Y.; Zhao, C.J. Study on the Micro-Phenotype of Different Types of Maize Kernels Based on Micro-CT. Smart Agric. 2021, 3, 16–28. (In Chinese) [Google Scholar] [CrossRef]
- Su, Y.; Xiao, L.T. 3D Visualization and Volume-Based Quantification of Rice Chalkiness In Vivo by Using High Resolution Micro-CT. Rice 2020, 13, 69. [Google Scholar] [CrossRef]
- Rousseau, D.; Widiez, T.; Di Tommaso, S.; Rositi, H.; Adrien, J.; Maire, E.; Langer, M.; Olivier, C.; Peyrin, F.; Rogowsky, P. Fast virtual histology using X-ray in-line phase tomography: Application to the 3D anatomy of maize developing seeds. Plant Methods 2015, 11, 55. [Google Scholar] [CrossRef]
- Xu, X.P.; Yang, X.Y.; Feng, M. A New Cereal Seed Treatment Method for Displaying Endosperm Cell Structures Under Micro CT Scanning. Chin. Bull. Bot. 2025, 60, 81–89. (In Chinese) [Google Scholar] [CrossRef]
- Guelpa, A.; Plessis, A.D.; Manley, M. A high-throughput X-ray micro-computed tomography (μCT) approach for measuring single kernel maize (Zea mays L.) volumes and densities. J. Cereal Sci. 2016, 69, 321–328. [Google Scholar] [CrossRef]
- Miao, Y.; Wang, R.; Jing, Z.; Wang, K.; Tan, M.; Li, F.; Zhang, W.; Han, J.; Han, Y. CT image segmentation of foxtail millet seeds based on semantic segmentation model VGG16-UNet. Plant Methods 2024, 20, 169. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Tan, F.; Li, C.; Jin, S.; Zhang, C.; Gao, P.; Xu, W. Stem–Leaf segmentation and phenotypic trait extraction of individual plant using a precise and efficient point cloud segmentation network. Comput. Electron. Agric. 2024, 220, 108839. [Google Scholar] [CrossRef]
- Houssein, E.H.; El-Din Helmy, B.; Oliva, D.; Elngar, A.A.; Shaban, H. Multi-Level Thresholding Image Segmentation Based on Nature-Inspired Optimization Algorithms: A Comprehensive Review. In Metaheuristics in Machine Learning: Theory and Applications; Oliva, D., Houssein, E.H., Hinojosa, S., Eds.; Studies in Computational Intelligence; Springer: Cham, Switzerland, 2021; Volume 967, pp. 311–353. [Google Scholar] [CrossRef]
- Yu, L.; Liu, L.; Yang, W.; Wu, D.; Wang, J.; He, Q.; Chen, Z.; Liu, Q. A non-destructive coconut fruit and seed traits extraction method based on Micro-CT and deeplabV3+ model. Front. Plant Sci. 2022, 13, 1069849. [Google Scholar] [CrossRef]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.-Y.; et al. Segment Anything. arXiv 2023, arXiv:2304.02643. [Google Scholar] [CrossRef]
- Uygun, T.; Ozguven, M.M. Determination of tomato leafminer: Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) damage on tomato using deep learning instance segmentation method. Eur. Food Res. Technol. 2024, 250, 1837–1852. [Google Scholar] [CrossRef]
- Zhang, W.; Dang, L.M.; Nguyen, L.Q.; Alam, N.; Bui, N.D.; Park, H.Y.; Moon, H. Adapting the Segment Anything Model for Plant Recognition and Automated Phenotypic Parameter Measurement. Horticulturae 2024, 10, 398. [Google Scholar] [CrossRef]
- Luna, M.; Chikontwe, P.; Park, S.H. Enhanced Nuclei Segmentation and Classification via Category Descriptors in the SAM Model. Bioengineering 2024, 11, 294. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Yuan, C.; Li, H.; Hu, J. Enhancing Agricultural Image Segmentation with an Agricultural Segment Anything Model Adapter. Sensors 2023, 23, 7884. [Google Scholar] [CrossRef]
- Sun, J.; Yan, S.; Yao, X.; Gao, B.; Yang, J. A Segment Anything Model based weakly supervised learning method for crop mapping using Sentinel-2 time series images. Int. J. Appl. Earth Obs. Geoinf. 2024, 133, 104085. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, H.; Yang, X.; Lu, X.; Guo, Z.; Jiang, L.; He, Y.; Cen, H. Biomass phenotyping of oilseed rape through UAV multi-view oblique imaging with 3DGS and SAM model. Comput. Electron. Agric. 2025, 235, 110320. [Google Scholar] [CrossRef]
- Dong, Q.; Xu, Q.; Kong, J.; Peng, X.; Zhou, W.; Chen, L.; Wu, J.; Xiang, Y.; Jiang, H.; Cheng, B. Overexpression of ZmbZIP22 gene alters endosperm starch content and composition in maize and rice. Plant Sci. 2019, 283, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wu, C.; Li, Q.; Du, M.; Li, Y.; Cheng, B.; Li, X.; Wu, J. ZmSSRP1, transactivated by OPAQUE11, positivelyregulates starch biosynthesis in maize endosperm. Plant Biotechnol. J. 2025, 23, 3770–3782. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, F.; Nie, X.; Xiong, Y.; Bao, Y.; Ling, S.; Sun, X.; Wang, X.; Yao, J. Rice endosperm starch regulator1 controls starch granule formation and amylopectin synthesis in rice endosperm. Plant Sci. 2025, 359, 112620. [Google Scholar] [CrossRef]
- Satoh-Cruz, M.; Crofts, A.J.; Takemoto-Kuno, Y.; Sugino, A.; Washida, H.; Crofts, N.; Okita, T.W.; Ogawa, M.; Satoh, H.; Kumamaru, T. Protein Disulfide Isomerase Like 1-1 Participates in the Maturation of Proglutelin Within the Endoplasmic Reticulum in Rice Endosperm. Plant Cell Physiol. 2010, 51, 1581–1593. [Google Scholar] [CrossRef]
- Yasuda, H.; Hirose, S.; Kawakatsu, T.; Wakasa, Y.; Takaiwa, F. Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol. 2009, 50, 1532–1543. [Google Scholar] [CrossRef]
- Wang, Q.; Feng, F.; Zhang, K.; He, Y.; Qi, W.; Ma, Z.; Song, R. ZmICE1a regulates the defence-storage trade-off in maize endosperm. Nat. Plants 2024, 10, 1999–2013. [Google Scholar] [CrossRef]
- Kavi Kishor, P.B.; Tiozon, R.N., Jr.; Fernie, A.R.; Sreenivasulu, N. Abscisic acid and its role in the modulation of plant growth, development, and yield stability. Trends Plant Sci. 2022, 27, 1283–1295. [Google Scholar] [CrossRef]
- Nonhebel, H.M.; Griffin, K. Production and roles of IAA and ABA during development of superior and inferior rice grains. Funct. Plant Biol. 2020, 47, 716–726. [Google Scholar] [CrossRef] [PubMed]
- Lima, R.B.; Pankaj, R.; Ehlert, S.T.; Finger, P.; Fröhlich, A.; Bayle, V.; Landrein, B.; Sampathkumar, A.; Figueiredo, D.D. Seed coat-derived brassinosteroid signaling regulates endosperm development. Nat. Commun. 2024, 15, 9352. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Shi, C.; Wang, L.; Sun, M.X. The parental contributions to early plant embryogenesis and the concept of maternal-to-zygotic transition in plants. Curr. Opin. Plant Biol. 2022, 65, 102144. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, R.; Xu, Z.; Zhang, Y.; Li, L.; Fu, J.; Wang, G.; Wang, J.; Du, X.; Gu, R. Label-Free Comparative Proteomic Analysis Combined with Laser-Capture Microdissection Suggests Important Roles of Stress Responses in the Black Layer of Maize Kernels. Int. J. Mol. Sci. 2020, 21, 1369–1383. [Google Scholar] [CrossRef]
- Li, Y.; Huang, K.; Zhang, L.; Zhang, B.; Duan, P.; Zhang, G.; Huang, X.; Zhou, C.; Han, N.; Zheng, L.; et al. A molecular framework for the GS2-SUG1 module-mediated control of grain size and weight in rice. Nat. Commun. 2025, 16, 3944. [Google Scholar] [CrossRef]
- Diao, Z.; Lu, L.; Wang, X.; Kong, F.; Wang, S.; Sui, J.; Qi, C.; Li, S. A module involving the cyclin-dependent kinase OsCDKF3 regulates grain size in rice. Plant Physiol. 2025, 199, 418–431. [Google Scholar] [CrossRef]
- Achary, V.M.M.; Reddy, M.K. CRISPR-Cas9 mediated mutation in GRAIN WIDTH and WEIGHT2 (GW2) locus improves aleurone layer and grain nutritional quality in rice. Sci. Rep. 2021, 11, 21941. [Google Scholar] [CrossRef] [PubMed]
- Hachiken, T.; Masunaga, Y.; Ishii, Y.; Ohta, T.; Ichitani, K.; Fukunaga, K. Deletion commonly found in Waxy gene of Japanese and Korean cultivars of Job’s tears (Coix lacryma-jobi L.). Mol. Breed. 2012, 30, 1747–1756. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Yang, M.; Yao, Y.; Cui, Y.; Li, X.; Ding, B.; Yao, X.; Wu, K. Transcriptomic insights into grain size development in naked barley (Hordeum vulgare L. var. nudum Hook. f): Based on weighted gene co-expression network analysis. PeerJ 2025, 13, e19856. [Google Scholar] [CrossRef]
- Yang, L.F.; Yang, Y.; Huang, L.Q.; Cui, X.M.; Liu, Y. From single- to multi-omics: Future research trends in medicinal plants. Brief. Bioinf. 2022, 24, bbac485. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, J.; Zhou, X.; Chen, X.; Li, Q.; Tan, H.; Dong, X.; Xiao, Y.; Chen, L.; Chen, W. Dynamic metabolic and transcriptomic profiling of methyl jasmonate-treated hairy roots reveals synthetic characters and regulators of lignan biosynthesis in Isatis indigotica Fort. Plant Biotechnol. J. 2016, 14, 2217–2227. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, H.; Song, Y.; Chen, J.; Bai, J.; Tang, J.; Wang, Q.; Fotopoulos, V.; Zhu, Q.H.; Yang, R.; et al. Transcription factor OsbZIP10 modulates rice grain quality by regulating OsGIF1. Plant J. 2024, 119, 2181–2198. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Ma, S.; Chen, X.M.; Yi, F.; Li, B.B.; Liang, X.G.; Liao, S.J.; Gao, L.H.; Zhou, S.L.; Ruan, Y.L. A transcriptional landscape underlying sugar import for grain set in maize. Plant J. 2022, 110, 228–242. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, W.; Shi, Y.; Wei, H.; Zhou, C.; Huang, X.; Chen, Z.; Xiang, T.; Wang, L.; Han, N.; et al. Coordinated Transcriptome and Metabolome Analyses of a Barley hvhggt Mutant Reveal a Critical Role of Tocotrienols in Endosperm Starch Accumulation. J. Agric. Food Chem. 2024, 72, 1146–1161. [Google Scholar] [CrossRef]
- Chen, C.; Ge, Y.; Lu, L. Opportunities and challenges in the application of single-cell and spatial transcriptomics in plants. Front. Plant Sci. 2023, 14, 1185377. [Google Scholar] [CrossRef]
- Yao, J.; Marand, A.P.; Bai, Y.; Schmitz, R.J.; Fan, L. Advances in plant spatial multi-omics data analysis. Trends Plant Sci. 2025. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Vidkjær, N.H.; Massange-Sanchez, J.A.; Laursen, B.B.; Gislum, R.; Borg, S.; Jiang, D.; Hebelstrup, K.H. Changes in spatiotemporal protein and amino acid gradients in wheat caryopsis after N-topdressing. Plant Sci. 2020, 291, 110336. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Liu, J.; Li, D.; Liu, C.M. Rice caryopsis development II: Dynamic changes in the endosperm. J. Integr. Plant Biol. 2016, 58, 786–798. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Jiang, S.J.; Kong, S.X.; Zhang, X.; Yue, J.H.; Zhao, W.Q.; Li, L.; Lin, S.Y. Advances in Single-Cell Transcriptome Sequencing and Spatial Transcriptome Sequencing in Plants. Plants 2024, 13, 1679. [Google Scholar] [CrossRef]
- Yuan, Y.; Huo, Q.; Zhang, Z.; Wang, Q.; Wang, J.; Chang, S.; Cai, P.; Song, K.M.; Galbraith, D.W.; Zhang, W.; et al. Decoding the gene regulatory network of endosperm differentiation in maize. Nat. Commun. 2024, 15, 34. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Fan, X.; Li, X.; Zhang, M.; Liu, Y.; Ni, Y.; Li, Y.; Jiang, Y.; Gao, X.; Zhao, Y.; et al. Uncovering chromatin accessibility dynamics in early maize endosperm and seed coat differentiation. Plant J. 2025, 123, e70316. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Xiao, W.; Tian, L.; Guo, L.; Ma, G.; Ji, C.; Huang, Y.; Wang, H.; Wu, X.; Yang, T.; et al. Spatial transcriptomics uncover sucrose post-phloem transport during maize kernel development. Nat. Commun. 2023, 14, 7191. [Google Scholar] [CrossRef]
- Wu, H.; Scanlon, M.J. Single-Cell and Spatial Transcriptomic Analysis of Maize Embryo Development. Cold Spring Harb. Protocols 2025. [Google Scholar] [CrossRef]
- Huang, Q.; Xu, M.; Zhang, H.; He, D.; Kong, Y.; Chen, L.; Song, H. Transcriptome and proteome analyses of the molecular mechanisms associated with coix seed nutritional quality in the process of breeding. Food Chem. 2019, 272, 549–558. [Google Scholar] [CrossRef]
- Huda, M.N.; Li, X.; Jahan, T.; He, Y.; Guan, C.; Zhang, K.; Gao, A.; Georgiev, M.I.; Zhou, M. Acceleration of the genetic gain for nutraceutical improvement of adlay (Coix L.) through genomic approaches: Current status and future prospects. Food Rev. Int. 2022, 39, 5377–5401. [Google Scholar] [CrossRef]
- Wei, X.; Wang, J.; Tan, Y.; Zhu, H.; Wang, Y.; Zhou, S.; Guo, J.; Wang, Y.; Huang, L. Analysis of Coix Seed Oil Biosynthesis Facilitates the Identification of Lysophosphatidic Acid Acyltransferase. J. Agric. Food Chem. 2025, 73, 12093–12104. [Google Scholar] [CrossRef]
- Wei, X.; Li, Y.; Zhou, S.; Guo, C.; Dong, X.; Li, Q.; Guo, J.; Wang, Y.; Huang, L. The Differences of Nutrient Components in Edible and Feeding Coix Seed at Different Developmental Stages Based on a Combined Analysis of Metabolomics. Molecules 2023, 28, 3759. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Bolger, A.M.; Poorter, H.; Dumschott, K.; Bolger, M.E.; Arend, D.; Osorio, S.; Gundlach, H.; Mayer, K.F.X.; Lange, M.; Scholz, U.; et al. Computational aspects underlying genome to phenome analysis in plants. Plant J. 2019, 97, 182–198. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Hu, X.; Song, J.; Guo, J.; Lu, B. An Overview of High-Throughput Crop Phenotyping: Platform, Image Analysis, Data Mining, and Data Management. In Plant Functional Genomics: Methods and Protocols; Maghuly, F., Ed.; Springer: New York, NY, USA, 2024; Volume 1, pp. 3–38. [Google Scholar] [CrossRef]
- Li, X.D.; Lu, X.J.; Wei, X.Y.; Fan, H.; Zeng, T.; Shi, M. Traits Inheritance of Female Sterile Line FS2106 in Adlay (Coix L.). J. Plant Genet. Resour. 2024, 25, 1083–1091. (In Chinese) [Google Scholar] [CrossRef]
- Qin, Y.M.; Liu, Y.L.; Sun, K.; Xiao, S.L.; Jin, B.Z.; Yao, J.W.; Yu, H.F. Cloning and Expression Analysis of ramosa2 Gene in Coix lacryma-jobi L. Mol. Plant Breed. 2023, 21, 6292–6299. (In Chinese) [Google Scholar] [CrossRef]
- Zeng, T.; Pan, H.; Wei, X.Y.; Lu, X.J.; Shi, M.; Long, S.W.; Li, G.; Li, X.D. Effects of Nitrogen Application Rate on Dry Matter Accumulation and Nitrogen Utilization Characteristics of Different Adlay Varieties. Shandong Agric. Sci. 2025, 57, 119–125. (In Chinese) [Google Scholar] [CrossRef]
- Ao, M.H.; Song, Z.Q.; Yang, X.Y. Effects of Different Sowing Dates on Starch Accumulation, Yield and Medicinal Quality of Coix lacryma-jobi seeds. Mol. Plant Breed. 2024, 22, 3306–3311. (In Chinese) [Google Scholar] [CrossRef]
- Hang, S.; Liu, M.; Xia, Y.Q.; Cheng, Y.; Liu, D.L.; Song, B. Effects of Altitude on Photosynthetic Characteristics and Grain Filling of Coix lacryma-jobi L. Shandong Agric. Sci. 2025, 57, 57–65. (In Chinese) [Google Scholar] [CrossRef]
- Wang, W.; Zhao, Y.; Bai, Z.; Zhang, J.; Liu, D. Extraction, purification, structural characterization and bioactivities of polysaccharides from coix seed: A review. Food Chem. 2025, 492, 145396. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, F.M.; Wang, M.; Yang, T.M.; Sun, D.L. The dynamic accumulation pattern of coixol in different parts and growth periods of Coix lacryma-jobi L. Hubei Agric. Sci. 2025, 64, 114–119. (In Chinese) [Google Scholar] [CrossRef]
- Zhu, F. Coix: Chemical composition and health effects. Trends Food Sci. Technol. 2017, 61, 160–175. [Google Scholar] [CrossRef]
- Xi, X.Y.; Ye, B.X. Studies on endosperm development and deposition of storage reserves in Coix lacryma-jobi. Acta Bot. Sin. 1995, 37, 118–124. (In Chinese) [Google Scholar]
- Ahmar, S.; Usman, B.; Hensel, G.; Jung, K.H.; Gruszka, D. CRISPR enables sustainable cereal production for a greener future. Trends Plant Sci. 2024, 29, 179–195. [Google Scholar] [CrossRef] [PubMed]
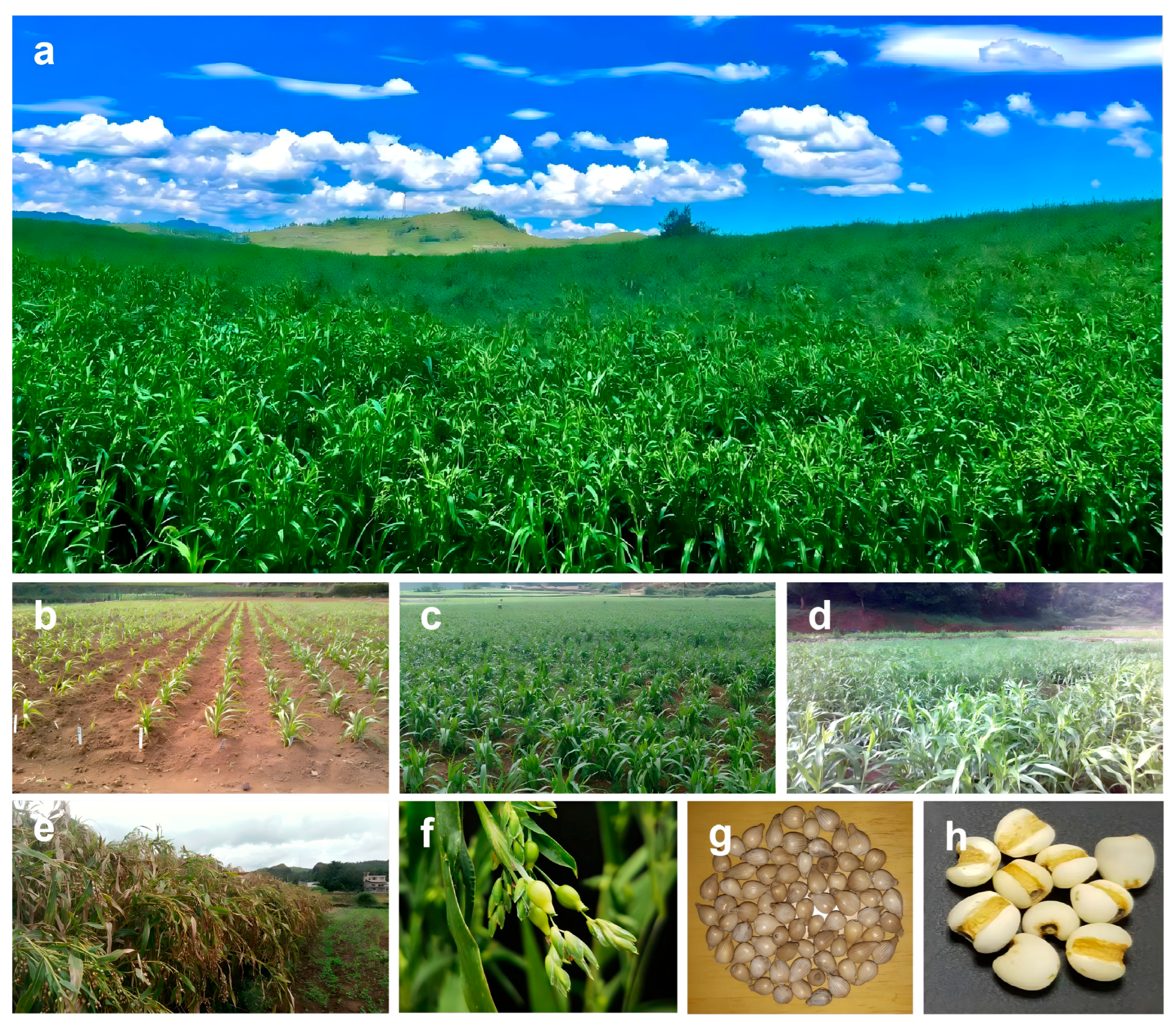
| Resolution Tier (Voxel Size) | Typical Applications and Traits | Example Crops and Studies | Advantages | Limitations | Relevance to Adlay (Leapfrog Strategy) |
|---|---|---|---|---|---|
| Low/Macro (~20–80 µm) | Whole-kernel morphometry: volume, density, husk/pericarp thickness, overall shape. | Maize (high-throughput single-kernel volumes/densities [28]); Sorghum grain structure [21]; Wheat grain morphology [14]. | High throughput; batch scanning possible; suitable for large samples. | Cannot resolve internal tissues or fine defects. | Yield traits: Rapid, non-destructive screening of husk thickness and kernel-to-husk ratio for improved edible yield. |
| Medium (~5–15 µm) | Quality and internal defects: chalkiness, fissures, embryo morphology, insect damage, plumpness. | Rice chalkiness (optimal ~5 µm [25]); Maize plumpness and internal cracks [24]; Pest detection in rice [22]. | Good balance of detail and speed; effective for mature grain quality assessment. | Limited for cellular-level structures or early development. | Seed quality/health: Detection of internal defects or pests without dehulling; embryo vigor screening. |
| High/Sub-micron (<5 µm, often phase-contrast) | Cellular microstructure: starch granule packing, aleurone layer, cell wall formation, early endosperm cellularization. | Maize developing seeds (phase-contrast [26]); Foxtail millet grain structure [29]; Wheat microstructural evolution [14]. | Resolves fine cellular boundaries and developmental dynamics. | Small field of view; long scan times; high data volume. | Developmental mechanisms: Essential for visualizing early endosperm cellularization and nutrient pathways—directly adopting advanced protocols to bypass lower-resolution stages used in other cereals. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Yang, X.; Zhang, J.; Ao, M.; Lei, J.; Yang, C. A Multidimensional Approach to Cereal Caryopsis Development: Insights into Adlay (Coix lacryma-jobi L.) and Emerging Applications. Plants 2026, 15, 320. https://doi.org/10.3390/plants15020320
Yang X, Zhang J, Ao M, Lei J, Yang C. A Multidimensional Approach to Cereal Caryopsis Development: Insights into Adlay (Coix lacryma-jobi L.) and Emerging Applications. Plants. 2026; 15(2):320. https://doi.org/10.3390/plants15020320
Chicago/Turabian StyleYang, Xiaoyu, Jian Zhang, Maohong Ao, Jing Lei, and Chenglong Yang. 2026. "A Multidimensional Approach to Cereal Caryopsis Development: Insights into Adlay (Coix lacryma-jobi L.) and Emerging Applications" Plants 15, no. 2: 320. https://doi.org/10.3390/plants15020320
APA StyleYang, X., Zhang, J., Ao, M., Lei, J., & Yang, C. (2026). A Multidimensional Approach to Cereal Caryopsis Development: Insights into Adlay (Coix lacryma-jobi L.) and Emerging Applications. Plants, 15(2), 320. https://doi.org/10.3390/plants15020320







