Abstract
Rice (Oryza sativa L.) root system plays a critical role in water and nutrient uptake, influencing overall plant growth and crop yield. In this study, we characterized the Osdrp2b mutant, which exhibits a short-root phenotype and was identified through map-based cloning. The Osdrp2b mutation was traced to the gene encoding a dynamin-related protein, and the mutant displayed reduced cell elongation and impaired cell division in the root tip. Further analysis revealed that ROS (reactive oxygen species) accumulation was elevated in the mutant roots, and treatment with ROS inhibitors restored root elongation in the Osdrp2b mutant, indicating that altered ROS homeostasis is associated with the phenotype. Transcriptomic analysis highlighted the differential expression of genes involved in cell wall organization and hydrogen peroxide catabolism. Agronomic evaluations of the Osdrp2b mutant demonstrated compromised shoot growth, reduced tiller number, and lower seed setting rates, indicating the impact of the mutation on rice yield. Overall, these results suggest that OsDRP2B is involved in regulating root growth, potentially through effects on ROS homeostasis and associated signaling networks. These findings provide a basis for future studies on improving rice root development and agronomic performance.
1. Introduction
Rice (Oryza sativa L.), one of the most important staple crops, supports over half the global population [1]. Economic models predict that by 2050, crop yields will be insufficient to meet the demands of the growing global population, emphasizing the urgent need to improve grain production [2]. The root system is a critical organ responsible for water and nutrient uptake, as well as providing anchorage. Serving as the primary interface between plants and the soil, root system architecture is closely linked to shoot morphology and crop yield [3]. Therefore, breeding strategies targeting root traits have garnered considerable attention from researchers. As a model monocot plant, rice has a typical fibrous root system comprising seminal roots, crown roots, lateral roots, and root hairs [4]. Numerous studies have demonstrated the indispensable role of the rice root system in yield formation [5,6]. Given the crucial role of rice roots in yield formation, understanding the molecular mechanisms that govern root development is essential for improving rice production.
Reactive oxygen species (ROS), including free radical species such as superoxide (O2−) and non-radical species like hydrogen peroxide (H2O2), are now recognized as crucial signaling molecules involved in various developmental processes, including root development [7]. Appropriate ROS distribution and homeostasis are required for maintaining root meristem activity, root elongation, and differentiation [8,9,10,11]. The regulatory function of ROS requires finely tuned homeostasis, maintained by a balance between ROS production and scavenging. In Arabidopsis thaliana, a highly dynamic and complementary network of over 150 genes has been identified as being involved in ROS homeostasis, some of which are also implicated in root development [12]. The protein phosphatase LIKE SEX FOUR2 (LSF2), identified as a key regulator of ROS homeostasis under oxidative stress, influenced root development [13]. Several studies in rice have also revealed the role of ROS homeostasis in root development. For example, altered ROS levels caused by overexpression or loss of function of specific genes result in defective root elongation and development [14,15]. Despite these advances, the precise mechanisms governing ROS homeostasis regulation in rice roots remain largely unclear.
Dynamin-related proteins (DRPs) are large multidomain GTPases that mediate membrane remodeling events essential for intracellular trafficking. In animal cells, dynamins assemble into helical structures at the necks of budding vesicles, facilitating vesicle scission via GTP hydrolysis [16]. In plants, two major DRP subfamilies-DRP1 and DRP2-have been identified and shown to function in various post-Golgi trafficking pathways, including clathrin-mediated endocytosis and cell plate formation. In A. thaliana, DRP family members have been shown to play critical roles in plant cytokinesis, polar cell expansion, and vacuolar trafficking [17,18,19]. In rice, DRPs have been implicated in cell wall biosynthesis, plant height regulation, and plant immunity [20,21,22]. A DRP2 family member was shown to participate in membrane trafficking related to cell wall biosynthesis, as mutants exhibited altered mechanical strength and secondary wall structure [20]. Additionally, DRPs also regulate lateral root development. The drp mutants exhibited increased LR number and diameter, which are associated with reduced endocytic activity and altered auxin signaling [23]. Despite these advances, the function of DRPs in regulating root growth in rice remains poorly characterized. Notably, although both membrane trafficking and ROS homeostasis are essential for root development, whether DRP family proteins are involved in the regulation of ROS homeostasis during rice root growth has not yet been reported. In the present study, a rice mutant exhibiting a short root phenotype was identified and named Osdrp2b through map-based cloning. Treatment of the Osdrp2b mutant with a ROS inhibitor partially restored the root phenotype to wild-type levels, suggesting that altered ROS homeostasis is associated with the mutant phenotype. Our findings indicate that OsDRP2B may regulate rice root development, potentially through effects on ROS homeostasis.
2. Results
2.1. Characterization of Root Growth Defects in the Osdrp2b Mutant
To investigate genes involved in rice root development, we screened a mutant library of Oryza sativa L. indica cv. Kasalath generated through EMS (ethylmethane sulfonate) mutagenesis. Among these, a mutant with a severe short-root phenotype attracted our attention. Subsequent map-based cloning identified the causative mutation as Osdrp2b. As shown in Figure 1a, both primary and adventitious root lengths were markedly reduced in the Osdrp2b mutant compared to the wild type (WT). Moreover, shoot growth was also compromised. Stereomicroscopic analysis further confirmed the short-root phenotype of the Osdrp2b mutant compared with the WT (Figure 1b). A time-course analysis was subsequently conducted to confirm these observations. As shown in Figure 1c,d, the primary root, adventitious roots, and shoot of the Osdrp2b mutant exhibited severe growth retardation compared to the WT. Collectively, these results suggest that the Osdrp2b mutation impairs root development in rice.
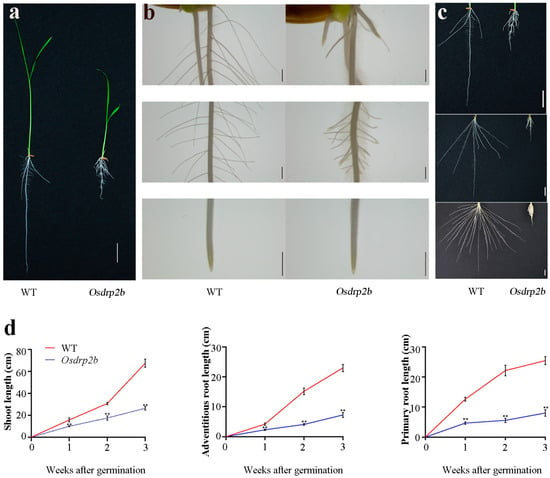
Figure 1.
Phenotypic characterization of the Osdrp2b mutant. (a) Seedling phenotypes of WT and Osdrp2b grown in rice solution culture for 7 days. bar = 2 cm. (b) Stereomicroscope images of WT and Osdrp2b roots. bars = 100 μm. (c) Root phenotype of WT and Osdrp2b from 1 to 3 weeks after germination. bars = 2 cm. (d) Quantification of shoot length, adventitious root length, and primary root length of WT and Osdrp2b from 1 to 3 weeks after germination. Values are means ± SE. Significant differences were determined using Student’s t-test, and asterisks (**) indicating a significant difference (p < 0.01) from WT.
To elucidate the cellular basis of the short-root phenotype in Osdrp2b, we performed a root longitudinal section analysis. As shown in Figure 2a,b, the average cell length in the elongation and maturation zones was significantly reduced in the Osdrp2b mutant compared to the WT. This observation indicates that cell elongation is compromised in the root tips of Osdrp2b mutants. To explore potential changes in radial growth, we also conducted a root cross-section analysis. As shown in Supplementary Figure S1a, Osdrp2b roots exhibited an increased diameter in both the elongation and maturation zones compared to the WT. Quantitative analysis further revealed that the width of cortical cells in these zones was significantly greater in Osdrp2b than in the WT (Supplementary Figure S1b), indicating that Osdrp2b roots undergo altered radial expansion of root cells.
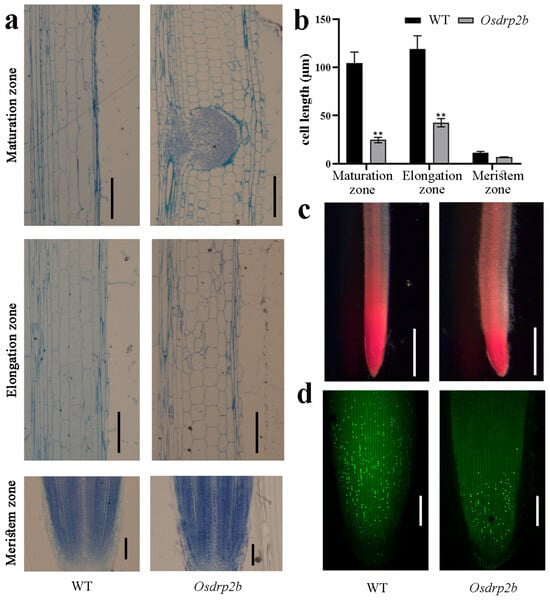
Figure 2.
Root cell division and elongation analysis in the Osdrp2b mutant. (a) Longitudinal sections of the maturation zone (top), elongation zone (middle), and meristem zone (bottom) in 3-day-old WT and Osdrp2b roots. bars = 100 μm. (b) Quantification of cell length in roots of 3-day-old WT and Osdrp2b plants. Values are means ± SE. Statistical significance was determined using Student’s t-test, and asterisks (**) indicate a significant difference from WT (p < 0.01). (c) Acetocarmine staining of root tips in WT and Osdrp2b. Scale bars = 500 μm. (d) EdU staining of 4-day-old WT and Osdrp2b root tips to visualize S-phase entry. Scale bars = 100 μm.
Meristematic activity in root tips was further evaluated using acetocarmine and EdU staining. As shown in Figure 2c, acetocarmine staining demonstrated a markedly smaller stained region in the mutant compared to WT, indicating reduced cell division activity. Consistently, EdU incorporation was substantially lower in Osdrp2b than in the WT (Figure 2d). These findings suggest that the short-root phenotype in Osdrp2b mutants may result from defects in both cell elongation and cell division activity at the root tip, accompanied by abnormal radial cell expansion.
2.2. Map-Based Cloning of the OsDRP2B
The causal gene responsible for the root development defect in Osdrp2b was identified through a map-based cloning approach. Crossing the Osdrp2b mutant with the japonica rice variety Nipponbare produced F1 progeny exhibiting wild-type phenotypes, indicating that the mutation is recessive. Self-pollination of the F1 plants resulted in an F2 population with phenotypic segregation: 1412 individuals exhibited normal root development, while 519 individuals displayed the short-root phenotype. Chi-square analysis (χ2 = 3.53, p > 0.05) confirmed a 3:1 segregation ratio. These results suggest that the Osdrp2b phenotype is controlled by a single recessive gene.
For preliminary mapping, we employed 98 pairs of SSR markers distributed across the 12 rice chromosomes and identified a linkage with the molecular marker RM5472 on chromosome 2. To further refine the mapping, we expanded the population size and developed InDel-specific markers, which localized the gene to a 109 kb region between markers In1 and In4 (Figure 3a). This region contains 15 candidate genes, as annotated by the National Rice Data Center. Sequencing of these candidate genes revealed that the causative mutation lies within the OsDRP2B (LOC_Os02g50550). OsDRP2B encodes a dynamin-related protein consisting of 923 amino acids. The Osdrp2b mutation results from a deletion of an adenine (A) at nucleotide position 1164 in the coding sequence, causing a premature stop codon and truncation of the protein at amino acid 414 (Figure 3a).
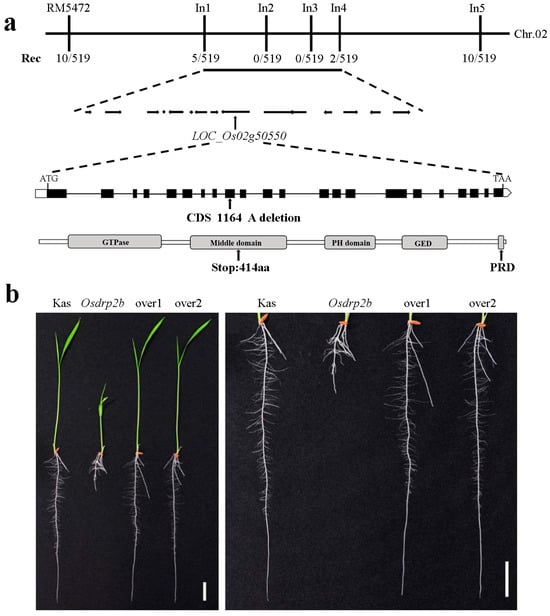
Figure 3.
Map-based cloning of OsDRP2B. (a) The OsDRP2B locus was mapped to chromosome 2 between the markers In1 and In4. Rec represents recombination frequency. Black boxes and lines indicate exons and introns, respectively. The arrow marks the adenine deletion. (b) Complementation analysis of Osdrp2b. Two independent lines of OsDRP2B-overexpressing transgenic plants (over1 and over2) in the Osdrp2b mutant background are shown. Scale bars = 2 cm.
To confirm that the short-root phenotype was caused by the OsDRP2B mutation, we performed a transgenic complementation assay. Transgenic plants carrying 35S::CDS-OsDRP2B exhibited root lengths comparable to those of the wild-type plants (Figure 3b). These results demonstrate that the short-root phenotype in Osdrp2b is attributable to the mutation in the OsDRP2B.
To further characterize OsDRP2B, its tissue-specific expression pattern was examined using a GUS reporter driven by the OsDRP2B promoter. GUS staining revealed that OsDRP2B was expressed in the primary root tip and adventitious root tip of seedlings (Figure 4a,b). GUS signals were also detected in the maturation zones of both primary and adventitious roots, which are regions characterized by root hair differentiation and vascular tissue development (Figure 4c,d). In mature plants at the reproductive stage, GUS expression was observed in multiple aerial and reproductive tissues, including stems, leaves, leaf sheaths, glumes, stamens, and pistils (Figure 4e–j). Together, these results indicate that OsDRP2B is broadly expressed in rice during both vegetative and reproductive development.
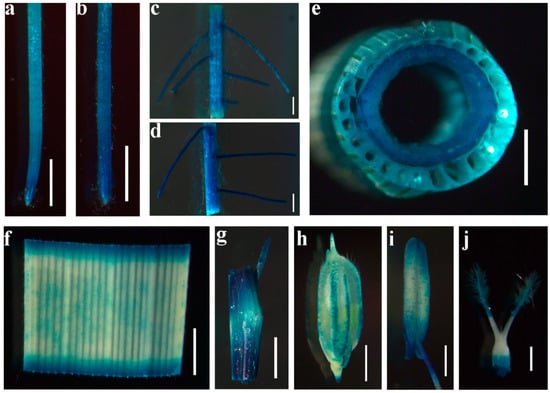
Figure 4.
Expression pattern of OsDRP2B. Expression pattern of OsDRP2B in transgenic rice lines expressing a GUS reporter driven by the OsDRP2B promoter. Histochemical GUS staining revealed expression in (a) primary root tip, (b) adventitious root tip, (c) maturation zone of the primary root, (d) maturation zone of the adventitious root, (e) stem, (f) leaves, (g) leaf sheath, (h) glume, (i) stamen, (j) pistil. bars: (a,b,e–g) = 1 mm; (c,d,i,j) = 500 μm; (h) = 2 mm.
2.3. Analysis of Agronomic Traits of Osdrp2b
Due to the significant impact of root systems on shoot morphology and rice yield, we analyzed the effects of the Osdrp2b mutation on agronomic traits. As shown in Figure 5a, the mutant plants grown under field conditions exhibited a significantly reduced plant height compared to WT. Similarly, both root length and panicle length in mature Osdrp2b mutants were notably shorter than those in WT (Figure 5b,c). In addition, the mutant displayed a significantly lower seed-setting rate and reduced tiller number than WT (Figure 5d). These findings suggest that OsDRP2B plays a role in regulating rice yield.
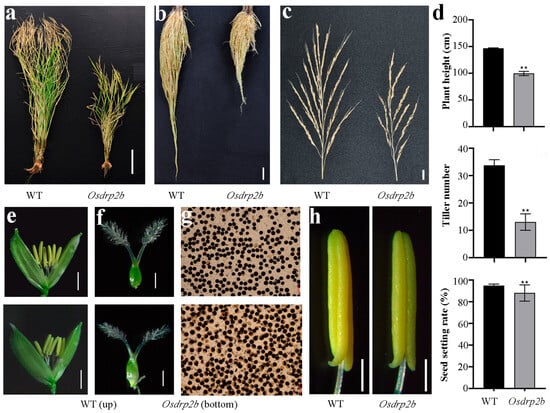
Figure 5.
Agronomic traits and reproductive organ phenotypes of the Osdrp2b mutant. (a) Mature WT and Osdrp2b plants grown under field conditions. Scale bar = 20 cm. (b) Roots of mature WT and Osdrp2b. Scale bar = 2 cm. (c) Spikes of WT and Osdrp2b from field planting. Scale bar = 2 cm. (d) Agronomic traits at the maturation stage, including plant height, effective tiller number, stem thickness, and seed setting rate of WT and Osdrp2b. Values are means ± SE. Statistical significance was determined using Student’s t-test, and asterisks (**) indicate a significant difference from WT (p < 0.01). (e) Florets of WT (top) and Osdrp2b (bottom). Scale bar = 1 mm. (f) Pistils of WT (top) and Osdrp2b (bottom). Scale bar = 500 μm. (g) I2-KI staining of pollen from WT (top) and Osdrp2b (bottom). (h) Stamens of WT and Osdrp2b. Scale bars = 100 μm.
To further assess the effects of the mutation at the heading stage, we examined the florets and found no significant morphological differences between the Osdrp2b mutant and the WT (Figure 5e). Pollen viability analysis using I2-KI staining showed that most mutant pollen grains were spherical but exhibited weaker staining and appeared largely unfilled compared to WT (Figure 5g). In contrast, no obvious morphological differences were observed in the stamens (Figure 5h). These results suggest that the reduced seed-setting rate in Osdrp2b may be attributed to decreased pollen viability.
2.4. Analysis of ROS Levels and Their Impact on Root Growth in the Osdrp2b Mutant
Given the important role of ROS in regulating plant growth, we assessed ROS levels in Osdrp2b. DAB staining revealed a markedly deeper brown coloration in primary roots of the mutant compared with WT, indicating increased hydrogen peroxide accumulation (Figure 6a). Similarly, NBT staining showed much stronger signals in the mutant root tips (Figure 6b), suggesting elevated superoxide anion levels.
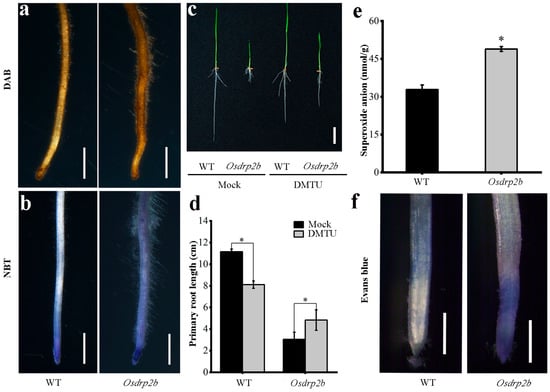
Figure 6.
ROS analysis of the primary root in the Osdrp2b mutant. (a) DAB staining of the primary root tip of Osdrp2b and wild-type (WT) plants. Bars = 100 μm. (b) NBT staining of the primary root tip of Osdrp2b and WT plants. Bars = 100 μm. (c) Primary root length of Osdrp2b and WT after DMTU treatment. Bars = 2 cm. (d) Quantification of primary root length in Osdrp2b and WT after DMTU treatment in (c). Values are means ± SE. Statistical significance was determined using Student’s t-test, and asterisks (*) indicate a significant difference from WT (p < 0.05). (e) Measurement of endogenous superoxide anion content in the primary roots of Osdrp2b and WT. Values are means ± SE. Statistical significance was determined using Student’s t-test, and asterisks (*) indicate a significant difference from WT (p < 0.05). (f) Evans Blue staining of the primary root tip of Osdrp2b and WT plants. Bars = 1 mm.
To examine the relationship between the short-root phenotype and ROS levels in Osdrp2b, we treated the rice plants with the ROS inhibitor DMTU. After DMTU treatment, the primary root length of the WT decreased, whereas that of the Osdrp2b mutant increased, nearly restoring WT levels (Figure 6c,d). This result suggests that the excessive accumulation of ROS in Osdrp2b inhibits root elongation. Consistently, quantification of endogenous superoxide anion content confirmed significantly higher levels in Osdrp2b compared with WT (Figure 6e).
To further assess the impact of ROS on cell viability, Evans Blue staining was performed. Mutant root tips accumulated significantly more dye than WT, indicating increased cell death (Figure 6f). Together, these results suggest that the elevated ROS levels in the mutant impair root growth.
2.5. Transcriptome Analysis of Osdrp2b
We conducted transcriptome sequencing on both WT and Osdrp2b plants to elucidate the molecular basis of the shortened root phenotype in Osdrp2b. As shown in Figure 7a, compared to the WT, 305 genes were upregulated, and 968 genes were downregulated in Osdrp2b (q < 0.05, FC > 2). Further enrichment analysis of the differentially expressed genes (DEGs) identified 106 significantly enriched GO terms (Table S2). In the biological process (BP) category, DEGs were primarily enriched in processes such as the hydrogen peroxide catabolic process, response to oxidative stress, xyloglucan metabolic process, and cell wall biogenesis. In the cellular components (CC) category, significant enrichment was observed in the cell wall, apoplast, and plasma membrane. Additionally, in molecular functions (MF), the DEGs were enriched in peroxidase activity, calcium ion binding, and electron transfer activity. These results suggest that the Osdrp2b phenotype may be associated with dysregulated expression of genes involved in these GO terms.
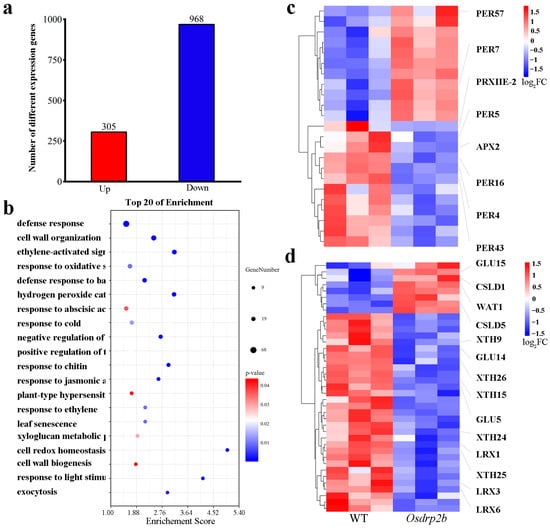
Figure 7.
Transcriptome comparative analysis of WT and Osdrp2b mutant. (a) Number of differentially expressed genes (DEGs) that were up-regulated or down-regulated in WT versus Osdrp2b as determined by RNA-seq analysis. DEGs were identified with a q-value < 0.05 and a fold change > 2. (b) The Top20 GO terms in BP category by Gene Ontology (GO) functional enrichment analysis of differentially expressed genes (DEGs) in Osdrp2b vs. WT. The size of each circle corresponds to the number of genes in each pathway, while the color gradient represents the p-value for the enrichment of each gene set. (c) Heatmap showing expression profiles of DEGs involved in hydrogen peroxide catabolic process. (d) Heatmap showing expression profiles of DEGs involved in cell wall biogenesis and organization.
We ranked the GO terms within the BP category by p-value, as shown in Figure 7b. Among the top 20 enriched terms, processes related to ROS, such as hydrogen peroxide catabolic process and response to oxidative stress, were particularly prominent, indicating a potential role of ROS homeostasis in the Osdrp2b mutant. Furthermore, GO terms related to cell wall organization, cell wall biogenesis, and xyloglucan metabolic process were also enriched, indicating a potential involvement of OsDRP2B in cell wall structure, which is essential for cell division and elongation. We further analyzed DEGs involved in ROS metabolism and cell wall remodeling. As shown in Figure 7c, the expression of several peroxidase genes was significantly altered in the Osdrp2b mutant. Specifically, PER7 and PER57 were significantly upregulated. In contrast, PER5, PER4, PER16, PER43, PRXIIE, and APX2 were significantly downregulated (fold change > 2, q < 0.05). These results suggest a potential dysregulation of hydrogen peroxide metabolism in the Osdrp2b mutant. Regarding cell wall biogenesis and organization, GLU15, CSLD1, and WAT1 were upregulated in the Osdrp2b mutant, while XTH9, XTH15, XTH24, XTH25, XTH26, LRX1, LRX3, LRX6, CSLD5, GLU5, and GLU14 were downregulated (Figure 7d). These differential expression patterns suggest that Osdrp2b disrupts ROS homeostasis and alters the balance of cell wall biosynthesis and remodeling processes, potentially contributing to its defective root phenotype.
To validate the RNA-seq results, qRT-PCR was performed on nine representative genes: OsPER57, OsPER7, OsPER43, OsPER2 (ROS homeostasis), OsXTH15, OsLRX6, OsXTH26, OsPGL32 (cell wall remodeling), and OsCYCB1-2 (cell division). Except for OsCYCB1-2, which showed downregulation in the mutant contrary to RNA-seq results, qPCR results for the other eight genes were consistent with transcriptome trends (Supplementary Figure S2), supporting the reliability of the sequencing results.
GO enrichment also revealed hormone-related processes involving auxin, abscisic acid (ABA), ethylene, and jasmonic acid (JA) signaling (list hits > 3) (Supplementary Figure S3a). However, exogenous treatments with these hormones failed to rescue the short-root phenotype (Supplementary Figure S3b), suggesting that hormone-related transcriptional changes alone may not fully account for the observed phenotype.
To complement the conventional GO enrichment analysis, we performed Gene Set Enrichment Analysis (GSEA) based on the full gene expression dataset. A total of 124 gene sets were significantly enriched (|NES| > 1, p < 0.05, FDR < 0.25) (Table S5). Notably, gene sets related to the response to reactive oxygen species (NES = −2.01, FDR = 0.001), cell wall organization (NES = −1.55, FDR = 0.128), and positive regulation of the cell cycle (NES = −1.48, FDR = 0.192) were suppressed in the Osdrp2b mutant (Supplementary Figure S4). To examine whether these transcriptomic alterations were reflected at the physiological and biochemical levels, we analyzed ROS-related enzyme activities and cell wall components in rice roots. Under control conditions, catalase (CAT) activity was higher and peroxidase (POD) activity was lower in the Osdrp2b mutant than in the WT. After DMTU treatment, CAT activity decreased in the mutant, whereas POD activity increased in both genotypes (Supplementary Figure S5a,b). In addition, cell wall composition was altered in Osdrp2b, with reduced cellulose and increased hemicellulose contents (Supplementary Figure S5c,d). These findings indicate global dysregulation of ROS metabolism, cell wall remodeling, and cell cycle progression, consistent with the observed cellular and developmental defects in root growth.
3. Discussion
ROS homeostasis is essential for root development. Our findings support a model in which OsDRP2B may regulate rice root growth primarily through maintaining ROS homeostasis.
3.1. ROS Homeostasis Regulates Root Growth
ROS play a crucial role in plant root growth and development by modulating processes such as cell division, expansion, hormonal signaling pathways, and cell wall remodeling [7,24]. Impaired root tip growth in Osdrp2b is associated with disturbed ROS homeostasis, suggesting that redox imbalance contributes to the short-root phenotype (Figure 2 and Figure 6). Similar observations have been reported in A. thaliana. Knockdown of SYNTAXIN OF PLANTS81 (AtSYP81) resulted in short roots with a severe reduction in root meristem activity and altered ROS levels [25]. Restoration of ROS levels in Atsyp81 reversed these root growth defects. In contrast, Osdrp2b root growth was partially restored upon treatment with a ROS inhibitor (Figure 6d), suggesting that both excessive and insufficient ROS may impact root development. Likewise, Arabidopsis scr mutants also exhibit short roots and reduced epidermal cell length, which could be partially rescued by modifying redox homeostasis via UPB1 mutation [26]. These findings align with the observed defects in cell elongation in Osdrp2b. Taken together, these observations reinforce the concept that maintaining ROS homeostasis, rather than a fixed ROS level, is essential for proper rice root development, although other pathways may also contribute. Future studies could explore the impact of the UPB1 mutation in the Osdrp2b background to further elucidate how ROS homeostasis regulates rice root development.
ROS homeostasis is controlled by the balance between ROS production and scavenging, involving various enzymes, such as respiratory burst oxidase homologs (RBOHs), peroxidases (PER), and catalase (CAT) [7]. In Osdrp2b mutants, the expression of PERs was significantly affected, with most being downregulated (Figure 7c), reminiscent of the UPB1-PER regulatory network in Arabidopsis [27]. In a previous study, transcriptomic analyses of the elongation zone in Arabidopsis roots also revealed coordinated regulation of peroxidases and xyloglucan endo-transglycosylase/hydrolase (XTH) genes, essential for cell wall remodeling. Silencing XTH genes resulted in shorter root lengths [28], similarly to suppressed expression of XTH genes in Osdrp2b (Figure 7d). Consistent with these transcriptomic observations, the activities of key ROS-related enzymes were altered in Osdrp2b roots and partially normalized following DMTU treatment. Under control conditions, CAT activity was elevated while POD activity was reduced in the mutant, likely reflecting a compensatory response to excessive ROS accumulation. After DMTU treatment, CAT activity decreased and POD activity increased in the mutant, suggesting an improved cellular redox environment. These biochemical changes, together with the altered cell wall composition observed in Osdrp2b (Supplementary Figure S5c,d), provide further support that disruption of ROS homeostasis contributes to the short-root phenotype.
Transcriptome analysis revealed that multiple hormone-related signaling pathways were affected in Osdrp2b. However, exogenous hormone treatments failed to rescue the short-root phenotype, suggesting that the defect is not due to a simple hormone deficiency and may involve disrupted crosstalk between hormone and ROS signaling. Exogenous phytohormones, such as auxin and ABA, can induce ROS production to mediate their effects on root growth [9,29]. In Osdrp2b, excessive ROS accumulation may mask or override hormone effects and influence feedback regulation of hormone-related genes. Consistently, treatment with the ROS inhibitor DMTU partially restored root growth, indicating that the abnormal expression of hormone-related genes likely reflects disturbed ROS homeostasis. The precise molecular mechanisms underlying the interaction between OsDRP2B-mediated ROS homeostasis and hormone signaling remain to be elucidated.
3.2. The Conserved Role of DRP2 in Plant Development
In Arabidopsis, the DRP family participates in endocytosis, plasma membrane formation, pollen grain development, and cytoskeleton organization [17,30]. The adl1A mutant, encoding a dynamin-like protein, exhibits severely stunted roots and reduced seed set [30], phenotypes reminiscent of Osdrp2b (Figure 1a and Figure 5d). Furthermore, AtDRP2, the homolog of OsDRP2B, affects gametophyte development [31]. This is also consistent with our findings, where the pollen viability was reduced (Figure 5g). These parallels suggest that the DRP2 genes may play a conserved role in regulating both root and reproductive development.
Although DRP family proteins share conserved molecular features, accumulating evidence indicates functional diversification among different DRP members in rice. For example, OsDRP1C has been shown to regulate lateral root diameter and the formation of L-type lateral roots by modulating auxin distribution through clathrin-mediated endocytosis [23]. DBC1, an allelic gene of OsDRP2B, primarily controls plant height by regulating cell division [21]. Moreover, BC3/OsDRP2B was previously reported to participate in membrane trafficking and endocytosis, thereby affecting the plasma membrane abundance of cellulose synthase and secondary wall structure [20]. Building on these studies, our work suggests that OsDRP2B is also involved in rice root development, contributing to root growth, at least in part, through the regulation of ROS homeostasis. This observation expands the current understanding of the functional diversity of DRP family members.
DRPs are involved in cell plate formation via clathrin-mediated endocytosis, influencing cytokinesis [32]. For instance, the Atdrp1 mutant exhibited defects in cell plate assembly, ultimately leading to abnormal cell division, and showed hypersensitivity to the membrane trafficking inhibitor, resulting in restricted root elongation [33]. These findings align with our results that OsDRP2B affects cell division.
Based on studies in Arabidopsis and other rice DRP homologs, we speculate that OsDRP2B may be involved in regulating ROS homeostasis through clathrin-mediated endocytosis. Our transcriptome analysis revealed a significant enrichment of endocytosis-related genes among the differentially expressed genes in the Osdrp2b mutant (Figure 7b), suggesting a possible link between OsDRP2B and endocytosis. However, direct experimental evidence is still lacking. In Arabidopsis, DRP2-mediated clathrin-dependent endocytosis is critical for the spatial regulation of RBOHD, affecting ROS production [19,34,35]. Similarly, in rice, the Osdrp1c mutant exhibits reduced endocytosis and altered auxin transporter localization [23], suggesting a conserved role of DRPs in vesicle trafficking. Furthermore, OsDRP2B has been previously implicated in membrane trafficking and cellulose biosynthesis by regulating the plasma membrane abundance of OsCESA4 [20]. The involvement of OsDRP2B in ROS regulation via endocytosis remains hypothetical and requires future experimental validation, such as endocytic activity assays or subcellular localization of ROS-related proteins.
In conclusion, OsDRP2B is a key regulator of rice root development, as the Osdrp2b mutant exhibits short roots due to reduced cell elongation and impaired cell division in the root tip. Elevated ROS levels in the mutant and restoration of root growth by ROS inhibitors indicate that OsDRP2B contributes to root development via maintaining ROS homeostasis. Mechanistically, OsDRP2B may act via clathrin-mediated endocytosis to modulate ROS balance, which in turn interacts with hormone signaling to regulate cell division and elongation, ultimately controlling root growth (Figure 8). These findings provide a conceptual framework for the working model of OsDRP2B and highlight its potential as a target for optimizing rice root architecture and improving yield.
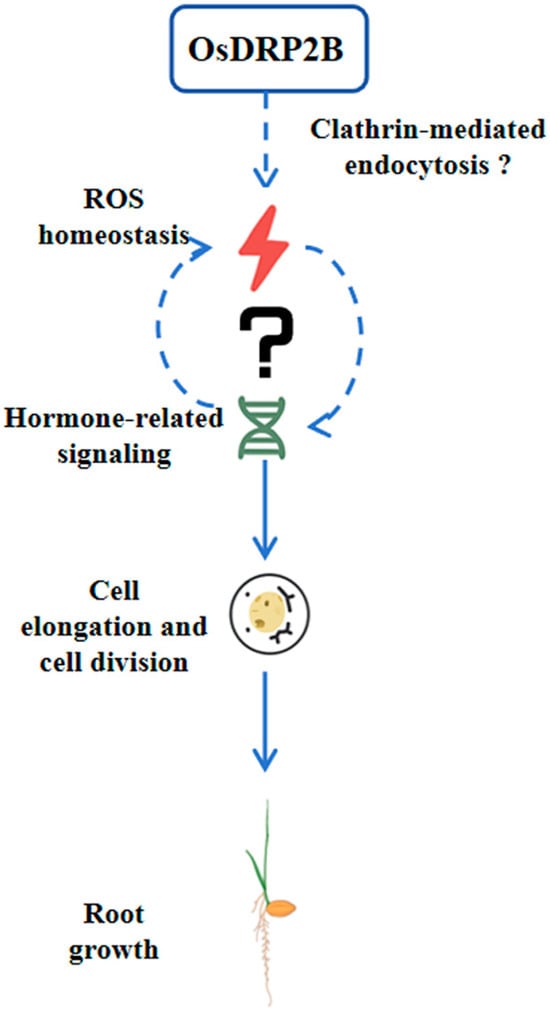
Figure 8.
A proposed working model of OsDRP2B in regulating rice root growth. OsDRP2B is proposed to modulate ROS homeostasis, possibly via clathrin-mediated endocytosis. Altered ROS levels may directly affect cell division and elongation in the root tip and/or indirectly influence these processes through crosstalk with hormone signaling pathways. Through these mechanisms, OsDRP2B ultimately regulates rice root growth.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
The Osdrp2b mutant was isolated from an ethyl methane sulfonate (EMS)-mutagenized rice (Oryza sativa L. India cv. Kasalath) mutant library. Wild-type (WT) plants of the same genetic background (Kasalath) were used as controls in all experiments.
Seeds were surface-sterilized and germinated in distilled water for 2 days, after which uniform seedlings were transferred to a greenhouse and cultivated in standard rice culture medium (pH 5.5), following the protocol described by Zhu et al. [36]. The climate chamber conditions were maintained at 32/22 °C (day/night), with 60–70% relative humidity, a light intensity of 16,000 lux, and a 12 h photoperiod. The rice culture medium was renewed weekly.
To investigate the impact of reactive oxygen species (ROS) on root growth and development in Osdrp2b, seedlings were cultured in a solution supplemented with 1 mM N, N′-dimethylthiourea (DMTU). To evaluate the effect of phytohormones on the root growth of Osdrp2b, seedlings were treated with culture solutions supplemented with 0.2 μM 2,4-D, 0.5 μM ABA, 4 mg/L ethephon, or 0.5 μM JA. Seedlings grown in hormone-free culture solution were used as controls. All treatments were performed for 7 days unless otherwise indicated. At least three independent biological replicates were conducted for each treatment.
4.2. Mapping and Cloning of the OsDRP2B
To map the OsDRP2B locus, the Osdrp2b homozygous mutant was crossed with Nipponbare, a japonica rice wild-type to generate an F1 mapping population. From the F2 generation derived by selfing the F1, 30 and 519 short-root lines exhibiting a phenotype consistent with Osdrp2b were selected for coarse and fine mapping of OsDRP2B, respectively. Genomic DNA was extracted from seedlings using a standard CTAB method. PCR-based InDel markers polymorphic between Kasalath and Nipponbare were developed and used for linkage analysis. The primers used for mapping are listed in Supplementary Data Table S1.
OsDRP2B was mapped to a 109 kb region on chromosome 2, between the sequence marker sites In1 and In4. Candidate genes within this interval were identified based on the Rice Genome Annotation Project database. Coding sequences of candidate genes were amplified and sequenced to identify potential EMS-induced mutations.
4.3. Vector Construction and Plant Transformation
For genetic complementation, the full-length coding sequence of OsDRP2B was amplified from wild-type rice cDNA and cloned into the binary vector pCAMBIA1300 under the control of the CaMV 35S promoter. For tissue expression patterns analysis, a 2456 bp OsDRP2B promoter was amplified by PCR and cloned into pCAMBIA11300NHGUS. All constructs were confirmed by sequencing. The resulting vectors were introduced into wild-type or mutant rice via Agrobacterium tumefaciens-mediated transformation. Transgenic seedlings were selected on hygromycin-containing medium and positive plants were further confirmed by PCR amplification of the hygromycin resistance (hpt) gene. The primers used for genotyping are listed in Supplementary Data Table S1.
4.4. Histochemical Analysis and GUS Assay
Histochemical GUS analysis was performed following the protocol described by Ding et al. [37]. Transgenic plant samples at the indicated developmental stages were incubated in GUS staining solution (100 mM NaH2PO4 buffer pH 7.0, 0.5% Triton X-100, 0.5 mg mL−1 X-Gluc and 20% methanol). Samples were vacuum-infiltrated for 1 min and then incubated overnight at 37 °C. After staining, tissues were destained with 70% ethanol, then were mounted on slides and photographed using a stereo microscope (Leica MZ95, Nussloch, Germany).
4.5. Acetocarmine and EdU Staining
Acetocarmine and EdU staining were performed as previously described by Ding et al. [37]. Briefly, for acetocarmine staining, root tips of 7-day-old WT and Osdrp2b seedlings were stained with 1% acetocarmine for 10 min in the dark, washed with 45% acetic acid, and observed under a stereo microscope (Leica MZ95). For EdU staining, roots of 4-day-old WT and Osdrp2b seedlings were incubated in 20 μM EdU medium for 2 h, fixed for 30 min, and washed three times with phosphate buffer (pH 7.2). Samples were then incubated in EdU detection solution for 30 min in the dark and imaged using a confocal microscope (Zeiss LSM 510, Jena, Germany).
4.6. Histological Observation
Histological observation was conducted as previously described by Ye et al. [38]. Root tips from 3-day-old rice seedlings were excised and fixed overnight at 4 °C in 0.1 M sodium phosphate buffer (pH 7.2) containing 2.5% glutaraldehyde. Samples were washed three times for 30 min each in the same buffer and then post-fixed in 1% osmium tetroxide (OsO4) for 4 h at room temperature, followed by a 30 min wash in the same buffer. Samples were dehydrated through a graded ethanol series and embedded in pure Spurr resin, which was polymerized overnight at 70 °C. Semithin sections (2 μm thick) were cut with a diamond knife on a Power Tome XL microtome (RMC-Boeckeler Instruments, Tucson, AZ, USA) and stained with 0.1% methylene blue for 3–5 min at 70 °C. After rinsing with distilled water, images were captured using a Nikon 90i microscope (Nikon, Tokyo, Japan). At least three independent biological replicates were analyzed.
4.7. Detection of Reactive Oxygen Species (ROS), Antioxidant Enzyme Activities and Cell Death
Hydrogen peroxide (H2O2) accumulation in rice roots was detected using an endogenous peroxidase-dependent staining method with 3,3′-diaminobenzidine (DAB; Sigma-Aldrich, St. Louis, MO, USA). Roots were incubated in 1 mg/mL DAB prepared in double-distilled water for 20 min at room temperature (22–25 °C) in the dark. Superoxide (O2−) production was examined by staining root tissues with 2 mM nitroblue tetrazolium chloride (NBT; Sigma-Aldrich) in 10 mM phosphate buffer (pH 7.0) for 10 min at room temperature under ambient light. Cell death was assessed using 1 mg/mL Evans Blue (Sigma-Aldrich) in double-distilled water for 10 min at room temperature, followed by three washes with distilled water to remove excess dye. Stained tissues were mounted on glass slides and imaged using a Nikon 90i microscope (Nikon, Japan). More than ten tissue samples were examined for each genotype, and all experiments were repeated at least twice. Representative images are shown.
Quantitative measurements of superoxide anion (O2−) levels and the activities of antioxidant enzymes, peroxidase (POD) and catalase (CAT), in rice seedlings were performed using commercial kits (Suzhou Grace Biotechnology Co., Ltd., Suzhou, China), following the manufacturer’s instructions. At least three independent biological replicates were analyzed.
4.8. Transcriptome Analysis and qRT-PCR
Transcriptome analysis by RNA sequencing was conducted as previously described by Ma et al. [39]. Roots of 7-day-old WT and Osdrp2b plants, grown in solution culture were harvested for RNA-seq experiments. Three biological replicates for each sample were collected. Total RNA extraction, cDNA library construction, and sequencing were conducted by OE Biotech Co., Ltd. (Shanghai, China) using the Illumina NovaSeq 6000 platform. GSEA was performed using the GSEA software (version V2.0.1.8, www.oebiotech.com). A predefined gene set was used, and all genes were ranked based on their differential expression between the two sample types. The analysis tested whether members of the predefined gene set were significantly enriched at the top or bottom of the ranked gene list.
For qRT-PCR analysis, total RNA was extracted from 7-day-old WT and Osdrp2b seedlings using RNAiso plus (Takara, Kyoto, Japan) and quantified by NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA). cDNA synthesis was performed with the ABScript II First-Strand Synthesis Kit (ABclonal, Wuhan, China). qRT-PCR was carried out on a LightCycler 96 System (Roche, Basel, Switzerland) using ChamQ SYBR qPCR Master Mix (Vazyme Biotech, Nanjing, China). Primers are listed in Table S1.
4.9. Pollen Activity Staining Assay
Spikelets about to flower from WT and Osdrp2b plants were carefully selected for examination. After gently opening the hull, anthers were placed in several drops of 1% iodine-potassium iodide (I2-KI; Sigma-Aldrich) solution on glass slides and gently crumbled to release pollen grains. Samples were stained at room temperature (22–25 °C) for 5 min before observation. Pollen viability was assessed under a Nikon Eclipse 90i light microscope (Nikon, Japan). More than five plants per genotype were analyzed, and the experiment was repeated at least twice. Representative images are shown.
4.10. Determination of Cellulose and Hemicellulose Contents
The cellulose and hemicellulose contents in the roots of 7-day-old rice seedlings were measured using commercial kits (Cellulose content determination kit, G0715W; Hemicellulose content determination kit, G0716W; Suzhou Grace Biotechnology Co., Ltd., Suzhou, China) according to the manufacturer’s instructions. At least three independent biological replicates were analyzed.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants15020313/s1, Figure S1: Root radial expansion analysis in the Osdrp2b mutant. (a) Cross sections of the maturation zone (top), elongation zone (middle), and meristematic zone (bottom) in 3-day-old WT and Osdrp2b roots. Scale bars = 100 μm. (b) Quantification of cortical cell width in roots of 3-day-old WT and Osdrp2b seedlings. Values represent means ± SE. Significant differences were determined using Student’s t-test (p < 0.05); Figure S2: Validation of RNA-seq results by qRT-PCR. Relative expression levels of nine selected genes involved in ROS homeostasis (OsPER57, OsPER7, OsPER43, OsPER2), cell wall remodeling (OsXTH15, OsLRX6, OsXTH26, OsPGL32), and cell division (OsCYCB1–2) in 7-day-old WT and Osdrp2b seedlings as determined by qRT-PCR. Gene expression levels were normalized to OsACTIN1. Values represent means ± SE. Significant differences were determined using Student’s t-test (p < 0.05); Figure S3: Hormone-related GO term enrichment and hormone treatment assays. (a) GO enrichment analysis of differentially expressed genes (DEGs) revealed significant enrichment of hormone-related biological processes. Hormone-related GO terms are ranked by the number of DEGs (list hits) associated with each term. (b) Phenotypes of WT and Osdrp2b seedlings grown in nutrient solution with or without 0.2 μM 2,4-D (auxin), 0.5 μM ABA, 4 mg/L ethephon (ethylene-releasing compound), and 0.5 μM JA for 7 days. WT and Osdrp2b seedlings grown without hormones serve as control groups. Bar = 2 cm; Figure S4: Gene Set Enrichment Analysis (GSEA) in Osdrp2b. GSEA was performed using the full gene expression dataset to identify significantly enriched biological processes. Representative suppressed gene sets in the Osdrp2b mutant are shown, including response to reactive oxygen species (a), cell wall organization (b), and positive regulation of the cell cycle (c). Significance thresholds were set at |NES| > 1, p < 0.05, and FDR < 0.25. Figure S5: ROS-related enzyme activities and cell wall composition in Osdrp2b. (a,b) Catalase (CAT) and peroxidase (POD) activities in 7-day-old WT and Osdrp2b seedlings under control conditions and after DMTU treatment. Different letters (a, b, c) indicate significant differences among groups (one-way ANOVA, p < 0.05). (c,d) Cellulose and hemicellulose contents in roots of 7-day-old WT and Osdrp2b seedlings. Significant differences between genotypes were assessed by Student’s t-test and are indicated by asterisks (* p < 0.05); Table S1. Primers used in the study; Table S2. Enriched GO terms in Osdrp2b compared WT (p < 0.05); Table S3. The expression of the DEGs enriched to hydrogen peroxide catabolic process (q < 0.05 and FC > 2); Table S4. The expression of the DEGs enriched to cell wall biogenesis and cell wall organization (q < 0.05 and FC > 2); Table S5. Significantly enriched gene sets identified by Gene Set Enrichment Analysis (GSEA) (|NES| > 1, p < 0.05, FDR < 0.25).
Author Contributions
Conceptualization: L.J., Z.G., Q.Z., M.L., M.W., T.M. and W.D.; Methodology: L.J., Z.G., Y.H., Q.Z., Q.L., X.L. and Y.C.; Software: Y.H.; Validation: Z.G.; Formal analysis: L.J., Z.G., F.Z., Q.L., X.L. and X.W.; Investigation: L.J., Z.G., Y.H., F.Z., Q.L., X.L. and Y.C.; Resources: Z.G., X.L. and X.W. Data curation: L.J.; Writing—original draft preparation: L.J. and T.M.; Writing—review and editing: M.L., M.W., T.M. and W.D.; Visualization: L.J.; Supervision: X.W., T.M. and W.D.; Project administration: W.D.; Funding acquisition: W.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 32071981), The Youth Science and Technology Innovation Leading Talent Project of Ningbo, China (Grant No. 2024QL061), and Chongqing Natural Science Foundation (Grant No. CSTB2022NSCQ-MSX0460).
Data Availability Statement
The RNA-seq data generated in this study have been deposited in the NCBI BioProject database under accession number PRJNA1383587. Other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
We would like to thank the developers of the Large Language Model-ChatGPT (version GPT-4) for assisting with grammar and language refinement during the drafting of this paper.
Conflicts of Interest
The authors have no conflicts of interest to declare.
References
- Shelden, M.C.; Munns, R. Crop root system plasticity for improved yields in saline soils. Front. Plant Sci. 2023, 14, 1120583. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef]
- Rogers, E.D.; Benfey, P.N. Regulation of plant root system architecture: Implications for crop advancement. Curr. Opin. Biotechnol. 2015, 32, 93–98. [Google Scholar] [CrossRef]
- Meng, F.; Xiang, D.; Zhu, J.; Li, Y.; Mao, C. Molecular Mechanisms of Root Development in Rice. Rice 2019, 12, 1. [Google Scholar] [CrossRef]
- Guan, X.J.; Chen, J.; Chen, X.M.; Xie, J.; Deng, G.Q.; Hu, L.Z.; Li, Y.; Qian, Y.F.; Qiu, C.F.; Peng, C.R. Root characteristics and yield of rice as affected by the cultivation pattern of strong seedlings with increased planting density and reduced nitrogen application. J. Integr. Agric. 2022, 21, 1278–1289. [Google Scholar] [CrossRef]
- Xie, X.; Quintana, M.R.; Sandhu, N.; Subedi, S.R.; Zou, Y.; Rutkoski, J.E.; Henry, A. Establishment method affects rice root plasticity in response to drought and its relationship with grain yield stability. J. Exp. Bot. 2021, 72, 5208–5220. [Google Scholar] [CrossRef]
- Eljebbawi, A.; Guerrero, Y.D.R.; Dunand, C.; Estevez, J.M. Highlighting reactive oxygen species as multitaskers in root development. iScience 2021, 24, 101978. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Han, X.; Benfey, P.N. RGF1 controls root meristem size through ROS signalling. Nature 2020, 577, 85–88. [Google Scholar] [CrossRef]
- Ivanchenko, M.G.; den Os, D.; Monshausen, G.B.; Dubrovsky, J.G.; Bednarova, A.; Krishnan, N. Auxin increases the hydrogen peroxide (H2O2) concentration in tomato (Solanum lycopersicum) root tips while inhibiting root growth. Ann. Bot. 2013, 112, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Gayomba, S.R.; Muday, G.K. Flavonols regulate root hair development by modulating accumulation of reactive oxygen species in the root epidermis. Development 2020, 147, dev185819. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, N.; Xu, J.; Li, Y.; Lin, X.; Sun, C.; Zhu, Y. Pseudomonas chlororaphis subsp. aurantiaca Stimulates Lateral Root Development by Integrating Auxin and Reactive Oxygen Species Signaling in Arabidopsis. J. Agric. Food Chem. 2024, 72, 23776–23789. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Sokolov, L.N.; Ye, J.; Tang, C.Y.; Shi, J.; Zhen, Y.; Lan, W.; Hong, Z.; Qi, J.; Lu, G.H.; et al. The LIKE SEX FOUR2 regulates root development by modulating reactive oxygen species homeostasis in Arabidopsis. Sci. Rep. 2016, 6, 28683. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guo, Y.; Wu, Q.; Xie, Z.; Yu, B.; Zeng, R.; Min, Q.; Huang, J. OsFPFL4 is Involved in the Root and Flower Development by Affecting Auxin Levels and ROS Accumulation in Rice (Oryza sativa). Rice 2020, 13, 2. [Google Scholar] [CrossRef]
- Yang, S.; Chen, N.; Qi, J.; Salam, A.; Khan, A.R.; Azhar, W.; Yang, C.; Xu, N.; Wu, J.; Liu, Y.; et al. OsUGE2 Regulates Plant Growth through Affecting ROS Homeostasis and Iron Level in Rice. Rice 2024, 17, 6. [Google Scholar] [CrossRef]
- Fujimoto, M.; Tsutsumi, N. Dynamin-related proteins in plant post-Golgi traffic. Front. Plant Sci. 2014, 5, 408. [Google Scholar] [CrossRef]
- Kang, B.H.; Busse, J.S.; Bednarek, S.Y. Members of the Arabidopsis dynamin-like gene family, ADL1, are essential for plant cytokinesis and polarized cell growth. Plant Cell 2003, 15, 899–913. [Google Scholar] [CrossRef]
- Jin, J.B.; Kim, Y.A.; Kim, S.J.; Lee, S.H.; Kim, D.H.; Cheong, G.W.; Hwang, I. A new dynamin-like protein, ADL6, is involved in trafficking from the trans-Golgi network to the central vacuole in Arabidopsis. Plant Cell 2001, 13, 1511–1525. [Google Scholar] [CrossRef]
- Fujimoto, M.; Arimura, S.; Ueda, T.; Takanashi, H.; Hayashi, Y.; Nakano, A.; Tsutsumi, N. Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. USA 2010, 107, 6094–6099. [Google Scholar] [CrossRef]
- Xiong, G.Y.; Li, R.; Qian, Q.A.; Song, X.Q.; Liu, X.L.; Yu, Y.C.; Zeng, D.L.; Wan, J.M.; Li, J.Y.; Zhou, Y.H. The rice dynamin-related protein DRP2B mediates membrane trafficking, and thereby plays a critical role in secondary cell wall cellulose biosynthesis. Plant J. 2010, 64, 56–70. [Google Scholar] [CrossRef]
- Sang, X.; Zhang, X.; Xiong, Y.; Xie, J.; Shi, L.; Tian, W.; Wang, X.; Li, Y.; Sun, Y.; He, G. DBC1, an allelic OsDRP2B gene, regulating plant height by controlling cell division in Oryza sativa L. Mol. Breed. 2020, 40, 37. [Google Scholar] [CrossRef]
- Xu, G.; Zhong, X.; Shi, Y.; Liu, Z.; Jiang, N.; Liu, J.; Ding, B.; Li, Z.; Kang, H.; Ning, Y.; et al. A fungal effector targets a heat shock-dynamin protein complex to modulate mitochondrial dynamics and reduce plant immunity. Sci. Adv. 2020, 6, eabb7719. [Google Scholar] [CrossRef]
- Kawai, T.; Akahoshi, R.; Shelley, I.J.; Kojima, T.; Sato, M.; Tsuji, H.; Inukai, Y. Auxin Distribution in Lateral Root Primordium Development Affects the Size and Lateral Root Diameter of Rice. Front. Plant Sci. 2022, 13, 834378. [Google Scholar] [CrossRef] [PubMed]
- Su, J.L.; Liu, Y.M.; Han, F.Q.; Gao, F.X.; Gan, F.Y.; Huang, K.; Li, Z.S. ROS, an Important Plant Growth Regulator in Root Growth and Development: Functional Genes and Mechanism. Biology 2024, 13, 1033. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, H.; Zhao, X.; Zhou, J.; Qin, G.; Liu, Y.; Kou, X.; Zhao, Z.; Wu, T.; Zhu, J.K.; et al. SYNTAXIN OF PLANTS81 regulates root meristem activity and stem cell niche maintenance via ROS signaling. Plant Physiol. 2023, 191, 1365–1382. [Google Scholar] [CrossRef]
- Fu, J.; Zhang, X.; Liu, J.; Gao, X.; Bai, J.; Hao, Y.; Cui, H. A mechanism coordinating root elongation, endodermal differentiation, redox homeostasis and stress response. Plant J. 2021, 107, 1029–1039. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional Regulation of ROS Controls Transition from Proliferation to Differentiation in the Root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.H.; Holman, T.J.; Sorensen, I.; Cancho-Sanchez, E.; Wells, D.M.; Swarup, R.; Knox, J.P.; Willats, W.G.T.; Ubeda-Tomas, S.; Holdsworth, M.; et al. Multi-omics analysis identifies genes mediating the extension of cell walls in the Arabidopsis thaliana root elongation zone. Front. Cell Dev. Biol. 2015, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, J.; He, J.; Qin, Y.; Hua, D.; Duan, Y.; Chen, Z.; Gong, Z. ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 2014, 10, e1004791. [Google Scholar] [CrossRef]
- Kang, B.H.; Busse, J.S.; Dickey, C.; Rancour, D.M.; Bednarek, S.Y. The arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001, 126, 47–68. [Google Scholar] [CrossRef][Green Version]
- Backues, S.K.; Korasick, D.A.; Heese, A.; Bednarek, S.Y. The Arabidopsis dynamin-related protein2 family is essential for gametophyte development. Plant Cell 2010, 22, 3218–3231. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.L.; Geisler-Lee, C.J.; Zhang, Z.M.; Verma, D.P.S. Phragmoplastin dynamics: Multiple forms, microtubule association and their roles in cell plate formation in plants. Plant Mol. Biol. 2003, 53, 297–312. [Google Scholar] [CrossRef]
- Collings, D.A.; Gebbie, L.K.; Howles, P.A.; Hurley, U.A.; Birch, R.J.; Cork, A.H.; Hocart, C.H.; Arioli, T.; Williamson, R.E. Arabidopsis dynamin-like protein DRP1A: A null mutant with widespread defects in endocytosis, cellulose synthesis, cytokinesis, and cell expansion. J. Exp. Bot. 2008, 59, 361–376. [Google Scholar] [CrossRef]
- Menzel, W.; Menzel, W.; Stenzel, I.; Helbig, L.M.; Krishnamoorthy, P.; Neumann, S.; Eschen-Lippold, L.; Heilmann, M.; Lee, J.; Heilmann, I. A PAMP-triggered MAPK cascade inhibits phosphatidylinositol 4,5-bisphosphate production by PIP5K6 in Arabidopsis thaliana. New Phytol. 2019, 224, 833–847. [Google Scholar] [CrossRef]
- Lee, J.; Nguyen, H.H.; Park, Y.; Lin, J.X.; Hwang, I. Spatial regulation of RBOHD via AtECA4-mediated recycling and clathrin-mediated endocytosis contributes to ROS accumulation during salt stress response but not flg22-induced immune response. Plant J. 2022, 109, 816–830. [Google Scholar] [CrossRef]
- Zhu, Z.X.; Liu, Y.; Liu, S.J.; Mao, C.Z.; Wu, Y.R.; Wu, P. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol. Plant 2012, 5, 154–161. [Google Scholar] [CrossRef]
- Ding, W.N.; Wu, J.; Ye, J.; Zheng, W.J.; Wang, S.S.; Zhu, X.N.; Zhou, J.Q.; Pan, Z.C.; Zhang, B.T.; Zhu, S.H. A Pelota-like gene regulates root development and defence responses in rice. Ann. Bot. 2018, 122, 359–371. [Google Scholar] [CrossRef]
- Ye, J.; Ding, W.; Chen, Y.; Zhu, X.; Sun, J.; Zheng, W.; Zhang, B.; Zhu, S. A nucleoside diphosphate kinase gene OsNDPK4 is involved in root development and defense responses in rice (Oryza sativa L.). Planta 2020, 251, 77. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Zhang, X.F.; Jia, L.H.Y.; Hua, Y.Y.; Li, X.; Qiu, S.Y.; Chen, Y.J.; Wang, X.F.; Zhu, S.H.; Mao, C.Z.; et al. OsCYP22 Interacts with OsCSN5 to Affect Rice Root Growth and Auxin Signalling. Plant Cell Environ. 2025, 48, 3955–3967. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.