Characterization of PDAT Genes in Oat (Avena sativa L.) and the Role of AsPDAT-5C in Lipid Biosynthesis and Abiotic Stress Response
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification and Sequence Analysis of PDATs in Oat
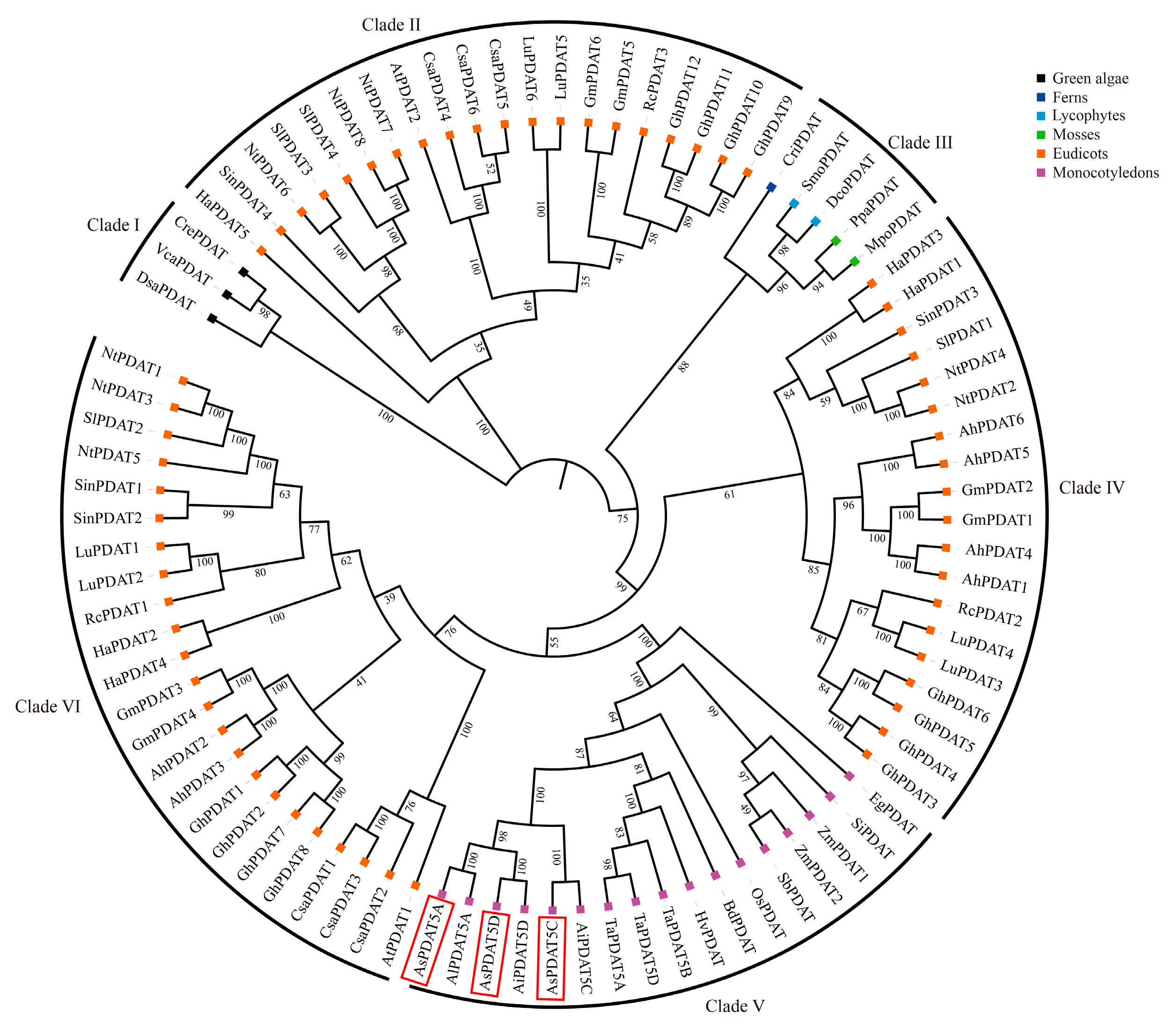
2.2. Phylogenetic Analysis of PDATs in Oat
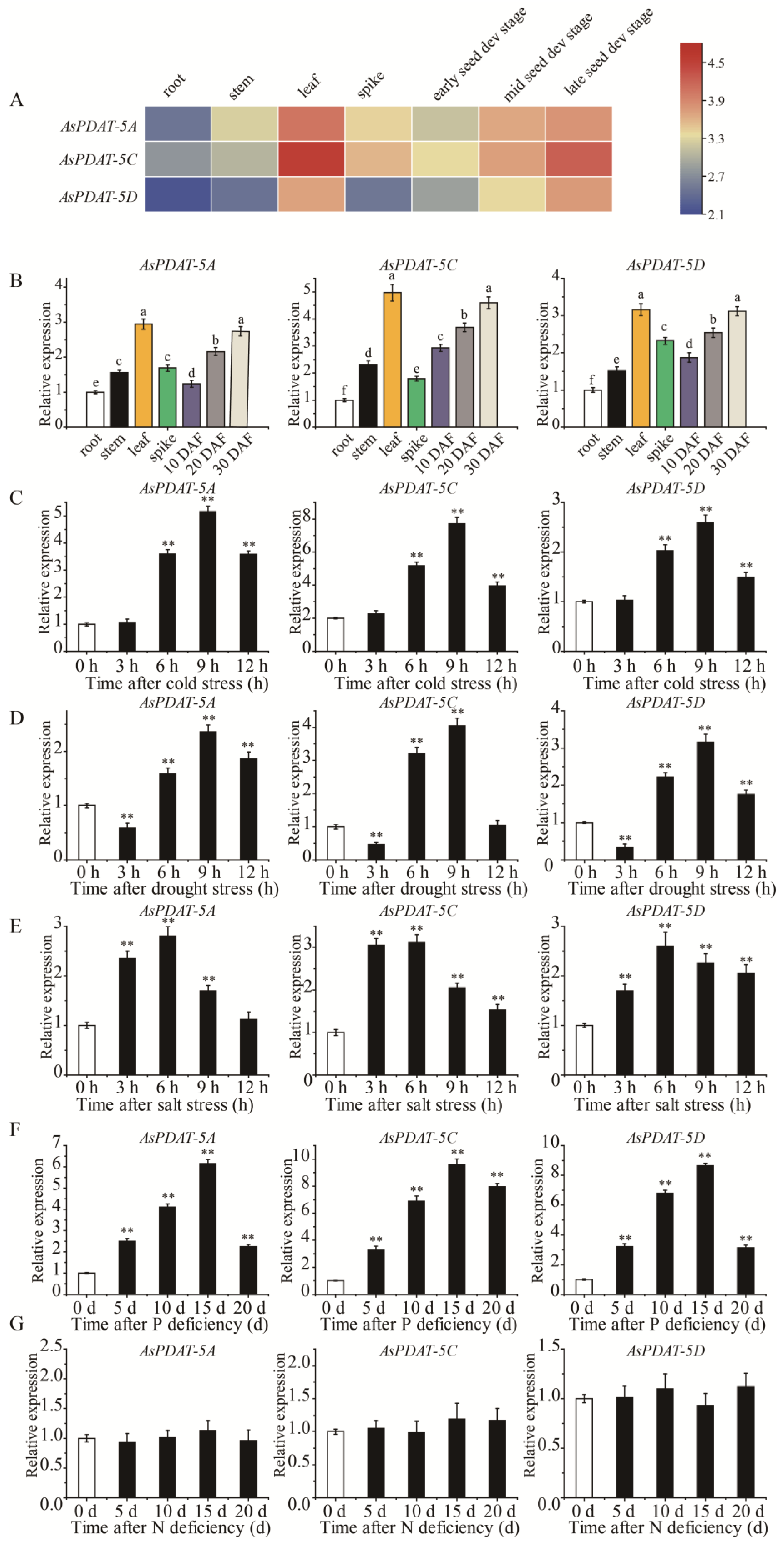
2.3. Expression Analysis of AsPDAT Genes in Oat
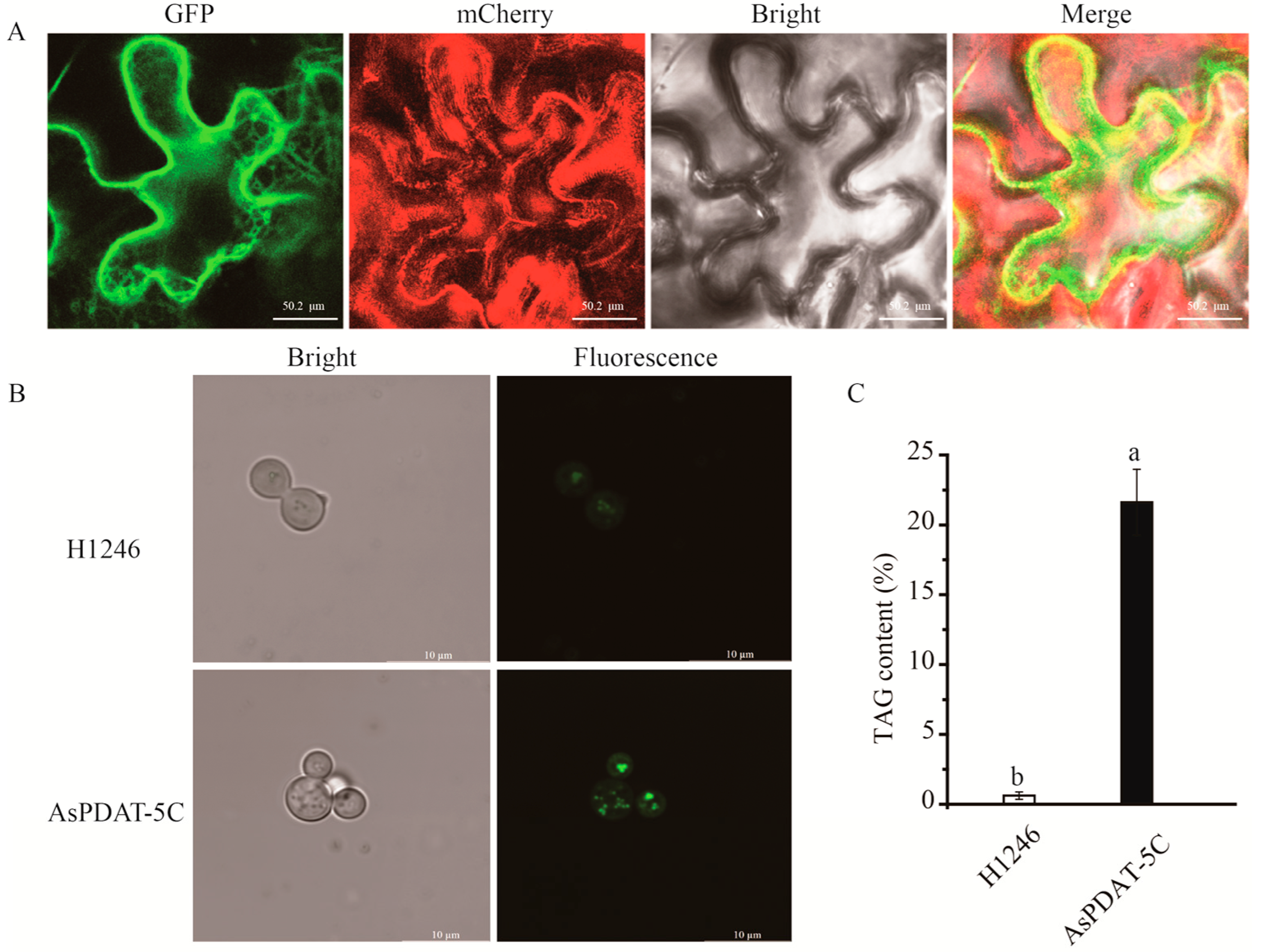
2.4. AsPDAT-5C Is Localized to Endoplasmic Reticulum and Exhibits Acyltransferase Activity
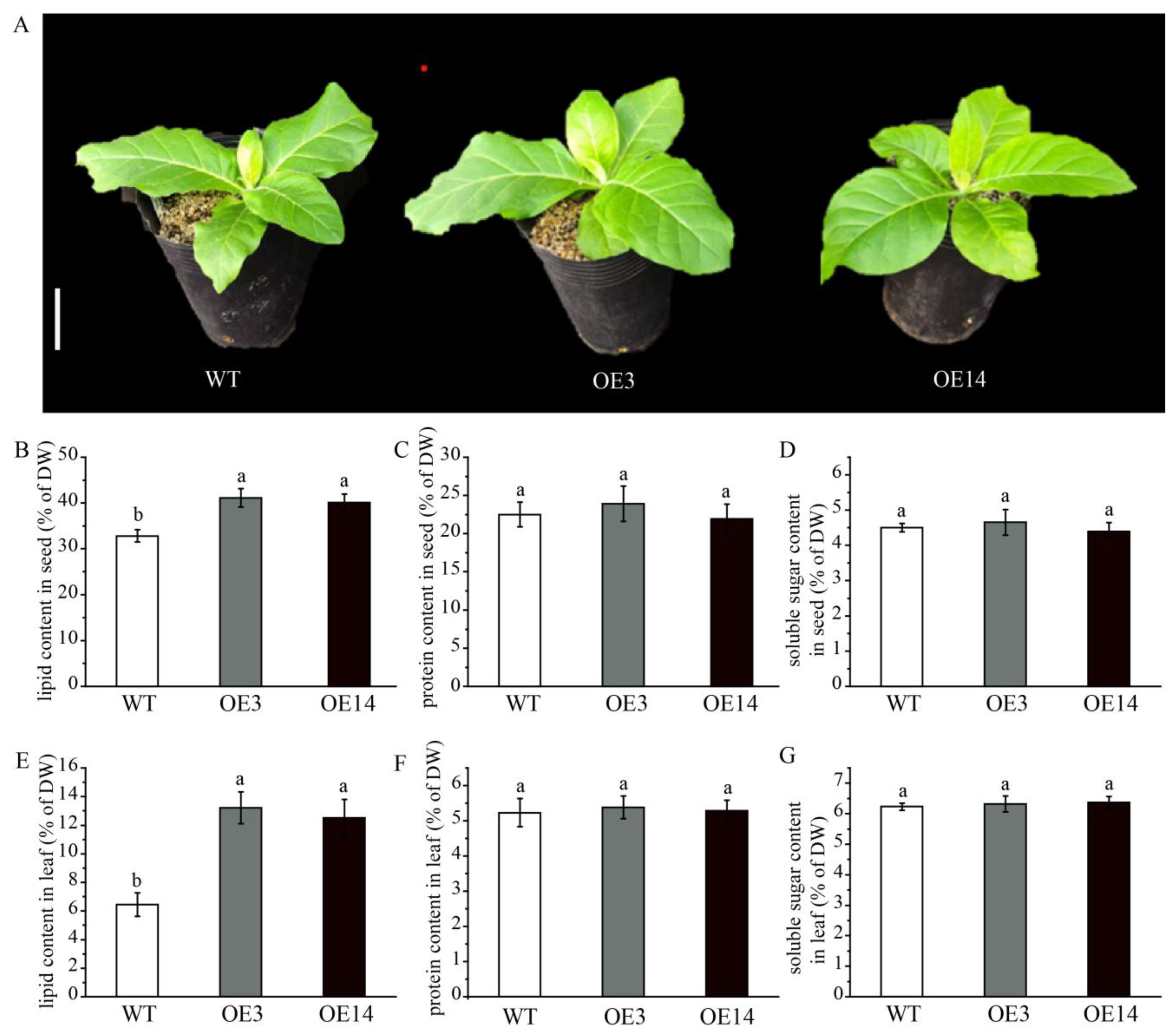
2.5. Overexpressing AsPDAT-5C Increased Total Lipid Content in Tobacco (Nicotiana tabacum)
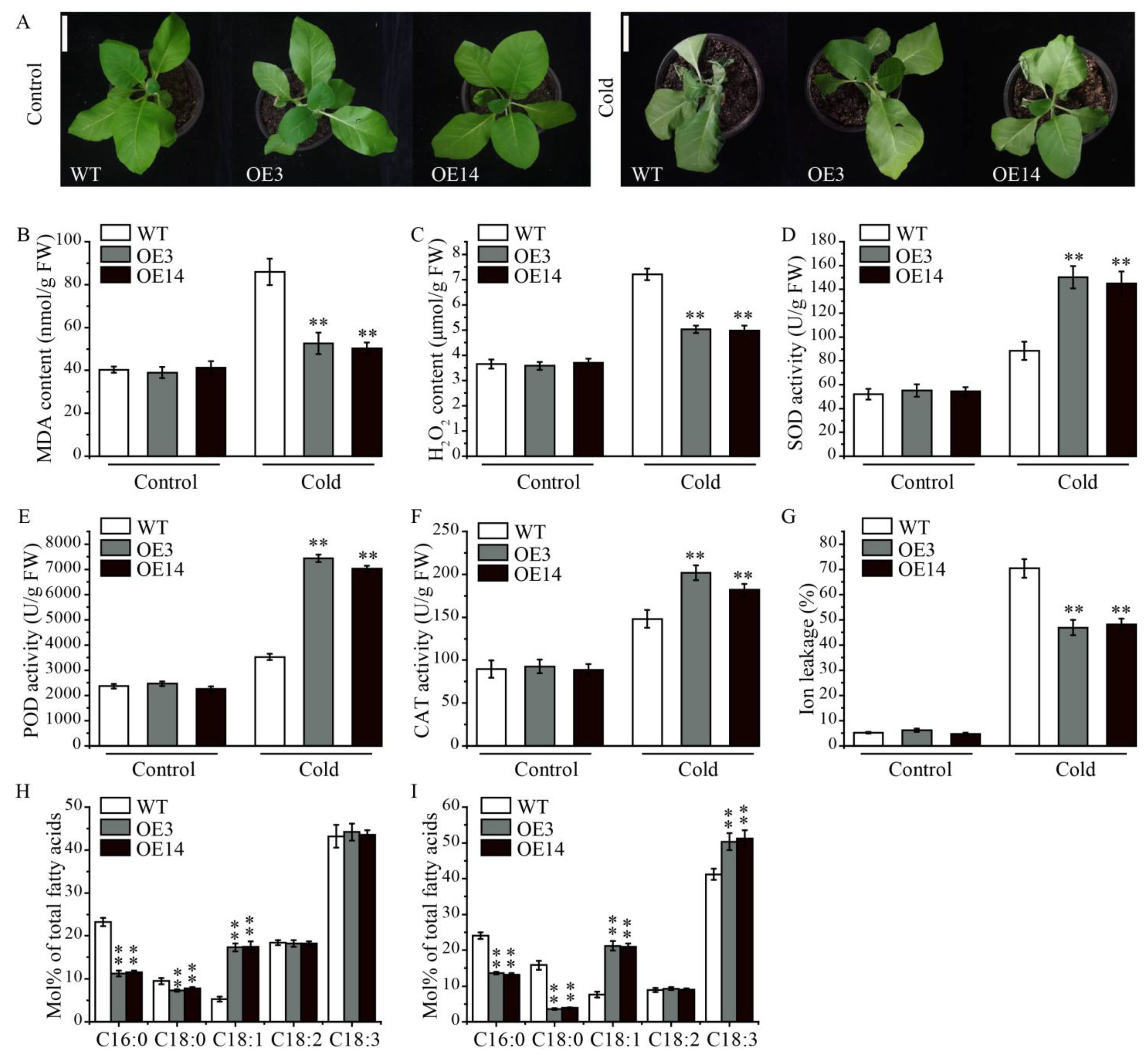
2.6. Overexpression of AsPDAT-5C Enhanced Cold Stress Tolerance
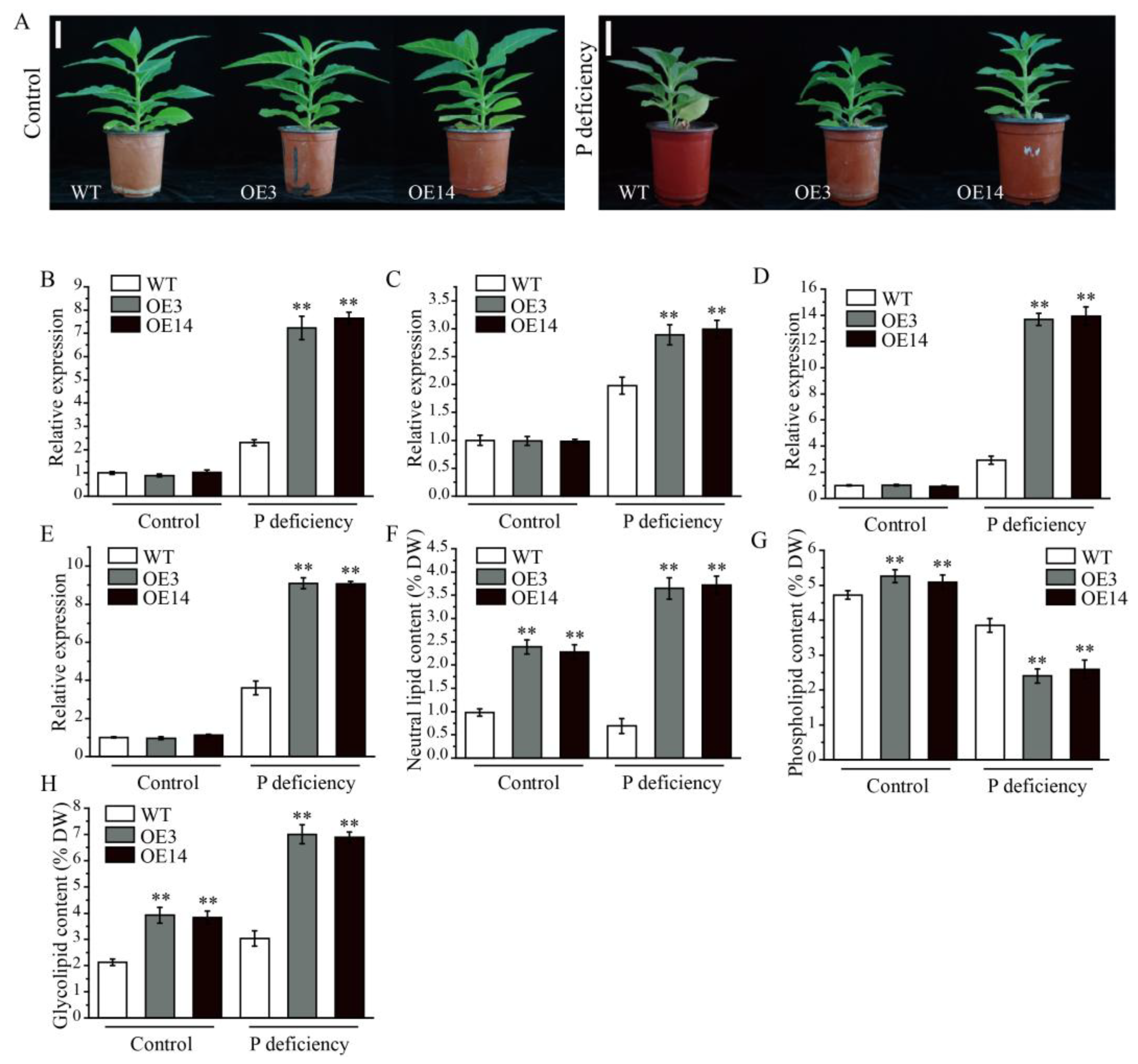
2.7. Overexpression of AsPDAT-5C Increased Tolerance to P Deficiency
3. Discussion
4. Materials and Methods
4.1. Sequence Acquisition and Bioinformatics Analysis of PDAT Family Genes
4.2. Plant Materials and Growth Conditions
4.3. RNA Isolation and Gene Expression Analysis
4.4. Subcellular Localization and AsPDAT-5C Expression in Yeast Mutant H1246
4.5. Vector Construction and Transgenic Plant Generation
4.6. Total Lipid, Soluble Sugar and Protein Measurement
4.7. Fatty Acid Profile and Lipid Composition Analysis
4.8. Analysis of Oxidative Stress Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PDAT | Phospholipid:Diacylglycerol Acyltransferase |
| DGAT | Diacylglycerol Acyltransferase |
| TAG | Triacylglycerol |
| LCAT | Lecithin:cholesterol Acyltransferase |
| TMD | Transmembrane Domain |
| DAF | Days After Flowering |
| ER | Endoplasmic Reticulum |
References
- Yang, Y.; Benning, C. Functions of triacylglycerols during plant development and stress. Curr. Opin. Biotechnol. 2018, 49, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Caldo, K.M.P.; Pal-Nath, D.; Ozga, J.; Lemieux, M.J.; Weselake, R.J.; Chen, G. Properties and Biotechnological Applications of Acyl-CoA:diacylglycerol Acyltransferase and Phospholipid:diacylglycerol Acyltransferase from Terrestrial Plants and Microalgae. Lipids 2018, 53, 663–688. [Google Scholar] [CrossRef] [PubMed]
- Sah, S.K.; Fan, J.; Blanford, J.; Shanklin, J.; Xu, C. Physiological Functions of Phospholipid:Diacylglycerol Acyltransferases. Plant Cell Physiol. 2024, 65, 863–871. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Peng, F.Y.; Weselake, R.J. Genome-wide analysis of phospholipid: Diacylglycerol acyltransferase (PDAT) genes in plants reveals the eudicot-wide PDAT gene expansion and altered selective pressures acting on the core eudicot PDAT paralogs. Plant Physiol. 2015, 167, 887–904. [Google Scholar] [CrossRef]
- Dahlqvist, A.; Stahl, U.; Lenman, M.; Banas, A.; Lee, M.; Sandager, L.; Ronne, H.; Stymne, S. Phospholipid:diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. USA 2000, 97, 6487–6492. [Google Scholar] [CrossRef]
- Stahl, U.; Carlsson, A.S.; Lenman, M.; Dahlqvist, A.; Huang, B.; Banas, W.; Banas, A.; Stymne, S. Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis. Plant Physiol. 2004, 135, 1324–1335. [Google Scholar] [CrossRef]
- Mhaske, V.; Beldjilali, K.; Ohlrogge, J.; Pollard, M. Isolation and characterization of an Arabidopsis thaliana knockout line for phospholipid: Diacylglycerol transacylase gene (At5g13640). Plant Physiol. Biochem. 2005, 43, 413–417. [Google Scholar] [CrossRef]
- Li, R.; Yu, K.; Hildebrand, D.F. DGAT1, DGAT2 and PDAT expression in seeds and other organs of epoxy and hydroxy fatty acid accumulating plants. Lipids 2010, 45, 145–157. [Google Scholar] [CrossRef]
- van Erp, H.; Bates, P.D.; Burgal, J.; Shockey, J.; Browse, J. Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol. 2011, 155, 683–693. [Google Scholar] [CrossRef]
- Yoon, K.; Han, D.; Li, Y.; Sommerfeld, M.; Hu, Q. Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 2012, 24, 3708–3724. [Google Scholar] [CrossRef]
- Oelkers, P.; Cromley, D.; Padamsee, M.; Billheimer, J.T.; Sturley, S.L. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 2002, 277, 8877–8881. [Google Scholar] [CrossRef]
- Zhang, M.; Fan, J.; Taylor, D.C.; Ohlrogge, J.B. DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 2009, 21, 3885–3901. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Yan, C.; Zhang, X.; Xu, C. Dual role for phospholipid:diacylglycerol acyltransferase: Enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 2013, 25, 3506–3518. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Fei, W.; Yang, S.; Yang, F.; Qu, G.; Tang, W.; Ou, J.; Peng, D. Alteration of the fatty acid composition of Brassica napus L. via overexpression of phospholipid: Diacylglycerol acyltransferase 1 from Sapium sebiferum (L.) Roxb. Plant Sci. 2020, 298, 110562. [Google Scholar] [CrossRef] [PubMed]
- Fenyk, S.; Woodfield, H.K.; Romsdahl, T.B.; Wallington, E.J.; Bates, R.E.; Fell, D.A.; Chapman, K.D.; Fawcett, T.; Harwood, J.L. Overexpression of phospholipid: Diacylglycerol acyltransferase in Brassica napus results in changes in lipid metabolism and oil accumulation. Biochem. J. 2022, 479, 805–823. [Google Scholar] [CrossRef]
- Wang, J.; Ren, H.; Shi, Z.; Phillip, F.O.; Liu, S.; Zhang, W.; Wang, X.; Bao, X.; Guo, J. Exploring Functional Gene XsPDAT1’s Involvement in Xanthoceras sorbifolium Oil Synthesis and Its Acclimation to Cold Stress. Forests 2024, 15, 1822. [Google Scholar] [CrossRef]
- Boyle, N.R.; Page, M.D.; Liu, B.; Blaby, I.K.; Casero, D.; Kropat, J.; Cokus, S.J.; Hong-Hermesdorf, A.; Shaw, J.; Karpowicz, S.J.; et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [Google Scholar] [CrossRef]
- Mueller, S.P.; Unger, M.; Guender, L.; Fekete, A.; Mueller, M.J. Phospholipid:Diacylglycerol Acyltransferase-Mediated Triacylglyerol Synthesis Augments Basal Thermotolerance. Plant Physiol. 2017, 175, 486–497. [Google Scholar] [CrossRef]
- Demski, K.; Losiewska, A.; Jasieniecka-Gazarkiewicz, K.; Klinska, S.; Banas, A. Phospholipid:Diacylglycerol Acyltransferase1 Overexpression Delays Senescence and Enhances Post-heat and Cold Exposure Fitness. Front. Plant Sci. 2020, 11, 611897. [Google Scholar] [CrossRef]
- Yuan, L.; Mao, X.; Zhao, K.; Ji, X.; Ji, C.; Xue, J.; Li, R. Characterisation of phospholipid: Diacylglycerol acyltransferases (PDATs) from Camelina sativa and their roles in stress responses. Biol. Open 2017, 6, 1024–1034. [Google Scholar] [CrossRef]
- Hernandez, M.L.; Moretti, S.; Sicardo, M.D.; Garcia, U.; Perez, A.; Sebastiani, L.; Martinez-Rivas, J.M. Distinct Physiological Roles of Three Phospholipid:Diacylglycerol Acyltransferase Genes in Olive Fruit with Respect to Oil Accumulation and the Response to Abiotic Stress. Front. Plant Sci. 2021, 12, 751959. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Lei, X.; Liu, T.; Xiong, Y.; Wu, J.; Xiong, Y.; You, M.; Zhao, J.; Zhang, J.; Ma, X. Integration of machine learning and genome-wide association study to explore the genomic prediction accuracy of agronomic trait in oats (Avena sativa L.). Plant Genome 2025, 18, e20549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Kaur, R.; Donoso, T.; Ohm, J.B.; Gupta, R.; Lefsrud, M.; Singh, J. Metabolic engineering-induced transcriptome reprogramming of lipid biosynthesis enhances oil composition in oat. Plant Biotechnol. J. 2024, 22, 3459–3472. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, X.; Li, N.; Du, C.; Wang, K.; Zhao, C.; Wang, Z.; Hu, Y.; Zhang, M. WRINKLED1 homologs highly and functionally express in oil-rich endosperms of oat and castor. Plant Sci. 2019, 287, 110193. [Google Scholar] [CrossRef]
- Hu, H.; Gutierrez-Gonzalez, J.J.; Liu, X.; Yeats, T.H.; Garvin, D.F.; Hoekenga, O.A.; Sorrells, M.E.; Gore, M.A.; Jannink, J.L. Heritable temporal gene expression patterns correlate with metabolomic seed content in developing hexaploid oat seed. Plant Biotechnol. J. 2020, 18, 1211–1222. [Google Scholar] [CrossRef]
- Hayden, D.M.; Rolletschek, H.; Borisjuk, L.; Corwin, J.; Kliebenstein, D.J.; Grimberg, A.; Stymne, S.; Dehesh, K. Cofactome analyses reveal enhanced flux of carbon into oil for potential biofuel production. Plant J. 2011, 67, 1018–1028. [Google Scholar] [CrossRef]
- Vanhercke, T.; Dyer, J.M.; Mullen, R.T.; Kilaru, A.; Rahman, M.M.; Petrie, J.R.; Green, A.G.; Yurchenko, O.; Singh, S.P. Metabolic engineering for enhanced oil in biomass. Prog. Lipid Res. 2019, 74, 103–129. [Google Scholar] [CrossRef]
- Falarz, L.J.; Xu, Y.; Caldo, K.M.P.; Garroway, C.J.; Singer, S.D.; Chen, G. Characterization of the diversification of phospholipid:diacylglycerol acyltransferases in the green lineage. Plant J. 2020, 103, 2025–2038. [Google Scholar] [CrossRef]
- Pan, X.; Siloto, R.M.; Wickramarathna, A.D.; Mietkiewska, E.; Weselake, R.J. Identification of a pair of phospholipid:diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J. Biol. Chem. 2013, 288, 24173–24188. [Google Scholar] [CrossRef]
- Kim, H.U.; Lee, K.R.; Go, Y.S.; Jung, J.H.; Suh, M.C.; Kim, J.B. Endoplasmic reticulum-located PDAT1-2 from castor bean enhances hydroxy fatty acid accumulation in transgenic plants. Plant Cell Physiol. 2011, 52, 983–993. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, H.; Guo, L.; Deng, C.; Wang, C.; Wang, Y.; Kang, L.; Zhou, P.; Yu, K.; Dong, X.; et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 2022, 54, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Arora, A.; Singh, V. Biodiesel from oil produced in vegetative organs of biomass—A review. Bioresour. Technol. 2021, 326, 124772. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.W.; Lee, H.G.; Do, H.; Kim, H.U.; Seo, P.J. Transcriptional regulation of triacylglycerol accumulation in plants under environmental stress conditions. J. Exp. Bot. 2022, 73, 2905–2917. [Google Scholar] [CrossRef] [PubMed]
- Klinska-Bachor, S.; Kedzierska, S.; Demski, K.; Banas, A. Phospholipid:diacylglycerol acyltransferase1-overexpression stimulates lipid turnover, oil production and fitness in cold-grown plants. BMC Plant Biol. 2023, 23, 370. [Google Scholar] [CrossRef]
- Shomo, Z.D.; Mahboub, S.; Vanviratikul, H.; McCormick, M.; Tulyananda, T.; Roston, R.L.; Warakanont, J. All members of the Arabidopsis DGAT and PDAT acyltransferase families operate during high and low temperatures. Plant Physiol. 2024, 195, 685–697. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, S.; Bai, B.; Chen, Y.; Xiang, Z.; Chen, C.; Kuang, X.; Yang, Y.; Fu, J.; Chen, L.; et al. OsKASI-2 is required for the regulation of unsaturation levels of membrane lipids and chilling tolerance in rice. Plant Biotechnol. J. 2024, 22, 2157–2172. [Google Scholar] [CrossRef]
- Ghosal, A.; Banas, A.; Ståhl, U.; Dahlqvist, A.; Lindqvist, Y.; Stymne, S. Saccharomyces cerevisiae phospholipid:diacylglycerol acyl transferase (PDAT) devoid of its membrane anchor region is a soluble and active enzyme retaining its substrate specificities. Biochim. Biophys. Acta 2007, 1771, 1457–1463. [Google Scholar] [CrossRef]
- Liu, C.; Tai, Y.; Luo, J.; Wu, Y.; Zhao, X.; Dong, R.; Ding, X.; Zhao, S.; Luo, L.; Liu, P.; et al. Integrated multi-omics analysis provides insights into genome evolution and phosphorus deficiency adaptation in pigeonpea (Cajanus cajan). Hortic. Res. 2022, 9, uhac107. [Google Scholar] [CrossRef]
- Wen, J.; Chai, X.; Huang, X.; Yang, H.; Lei, T.; Dong, S.; Li, R.; Wang, J.; Zhou, Y. PfPAH1-1 gene enhances plant tolerance to low phosphate stress by modulating cell membrane lipid remodeling. Plant Physiol. Biochem. 2025, 221, 109593. [Google Scholar] [CrossRef]
- Pant, B.D.; Burgos, A.; Pant, P.; Cuadros-Inostroza, A.; Willmitzer, L.; Scheible, W.R. The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 2015, 66, 1907–1918. [Google Scholar] [CrossRef]
- Kobayashi, K.; Awai, K.; Nakamura, M.; Nagatani, A.; Masuda, T.; Ohta, H. Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 2009, 57, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary geneticsanalysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, X.; Sun, Y.; Chai, X.; Wen, J.; Yang, Z.; Xue, J.; Zhang, X.; Jia, X.; Wang, J.; et al. Three Distinctive Diacylglycerol Acyltransferases (DGAT1, DGAT2, and DGAT3) from Perilla frutescens and Their Potential in Metabolic Engineering for Designed Oil Production. J. Agric. Food Chem. 2025, 73, 20254–20272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.L.; Su, Y.T.; Wang, L.L.; Ma, L.; Sun, Y.; Li, R.Z.; Ge, L.P. Characterization of GPAT gene family in Euphorbia lathyris L. and elucidating the role of ElGPAT9 in the biosynthesis of oils and pollen viability. Ind. Crops Prod. 2024, 213, 118473. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in isolated chloroplasts. I. Kinetics and stoichiometry of fatty acid peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Velikova, V.; Yordanov, I.; Edreva, A. Oxidative stress and some antioxidant systems in acid rain-treated bean plants. Plant Sci. 2000, 151, 59–66. [Google Scholar] [CrossRef]
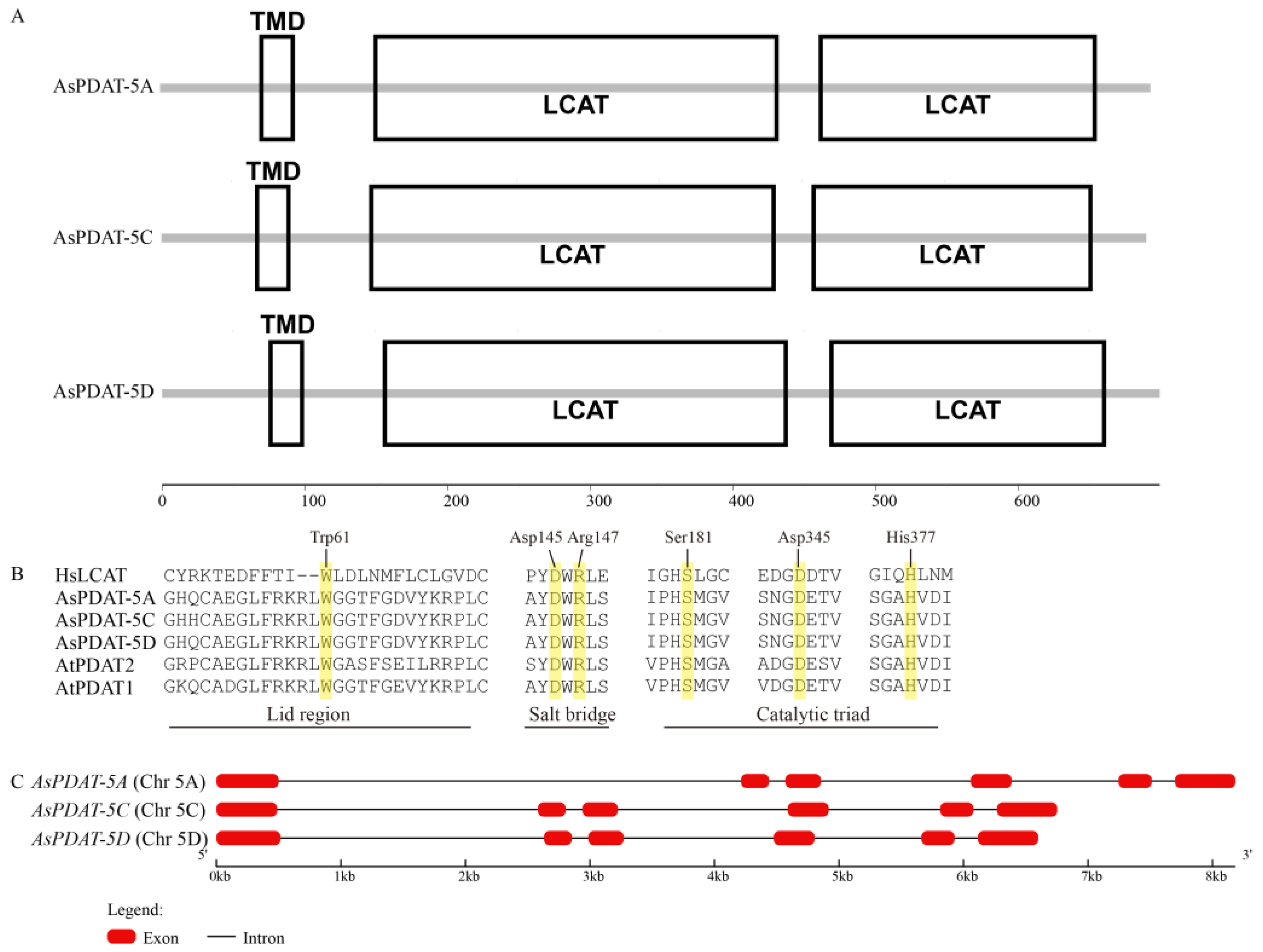
| Gene ID (OatOmics) | Gene Name | Chromosome | Amino Acid Number | Molecular Weight (Da) | pI | Instability Index | Aliphatic Index | Grand Average of Hydropathicity | Predictions for Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| 1059_5A004970 | AsPDAT-5A | 5A | 694 | 76,474.89 | 5.8 | 44.04 | 73.89 | −0.323 | ER |
| 1059_5C004228 | AsPDAT-5C | 5C | 690 | 76,019.45 | 5.91 | 42.93 | 73.9 | −0.325 | ER |
| 1059_5D004221 | AsPDAT-5D | 5D | 699 | 76,875.18 | 5.8 | 43 | 73.22 | −0.324 | ER |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Sun, Y.; Yang, J.; Hu, R.; Li, C.; Yang, Q.; Sun, X.; Zhang, Z.; Li, R.; Xue, J. Characterization of PDAT Genes in Oat (Avena sativa L.) and the Role of AsPDAT-5C in Lipid Biosynthesis and Abiotic Stress Response. Plants 2026, 15, 35. https://doi.org/10.3390/plants15010035
Sun Y, Yang J, Hu R, Li C, Yang Q, Sun X, Zhang Z, Li R, Xue J. Characterization of PDAT Genes in Oat (Avena sativa L.) and the Role of AsPDAT-5C in Lipid Biosynthesis and Abiotic Stress Response. Plants. 2026; 15(1):35. https://doi.org/10.3390/plants15010035
Chicago/Turabian StyleSun, Yan, Jinzhou Yang, Ruirui Hu, Chen Li, Qian Yang, Xiping Sun, Zhiwei Zhang, Runzhi Li, and Jinai Xue. 2026. "Characterization of PDAT Genes in Oat (Avena sativa L.) and the Role of AsPDAT-5C in Lipid Biosynthesis and Abiotic Stress Response" Plants 15, no. 1: 35. https://doi.org/10.3390/plants15010035
APA StyleSun, Y., Yang, J., Hu, R., Li, C., Yang, Q., Sun, X., Zhang, Z., Li, R., & Xue, J. (2026). Characterization of PDAT Genes in Oat (Avena sativa L.) and the Role of AsPDAT-5C in Lipid Biosynthesis and Abiotic Stress Response. Plants, 15(1), 35. https://doi.org/10.3390/plants15010035






