The Mining of Candidate Genes Involved in the Camphor Biosynthesis Pathway of Cinnamomum camphora
Abstract
1. Introduction
2. Results
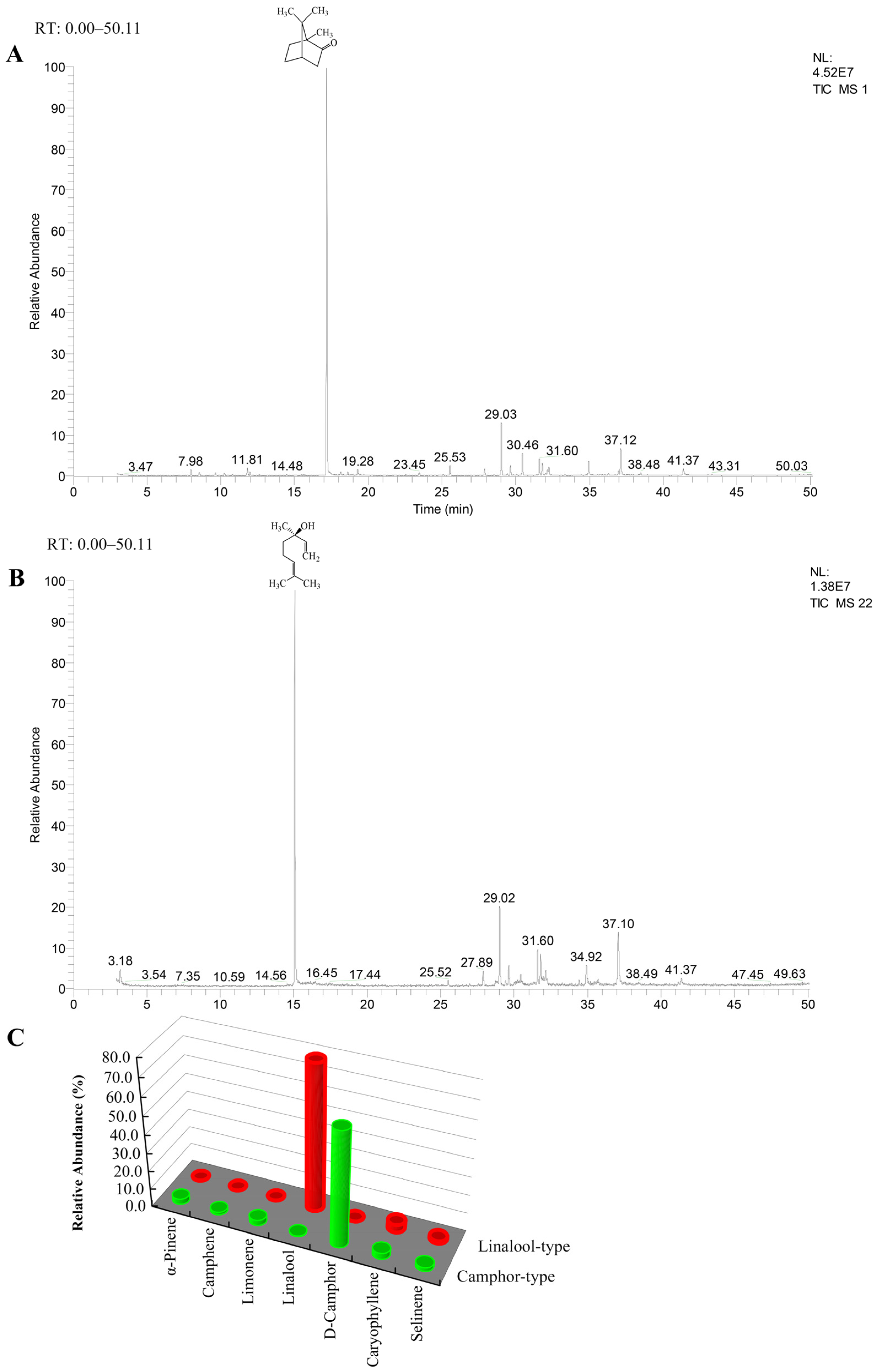
2.1. The Composition of Leaf Extracts from Two Chemotypes of C. camphora
2.2. The Statistics of RNA Sequencing in Two Chemotypes of C. camphora
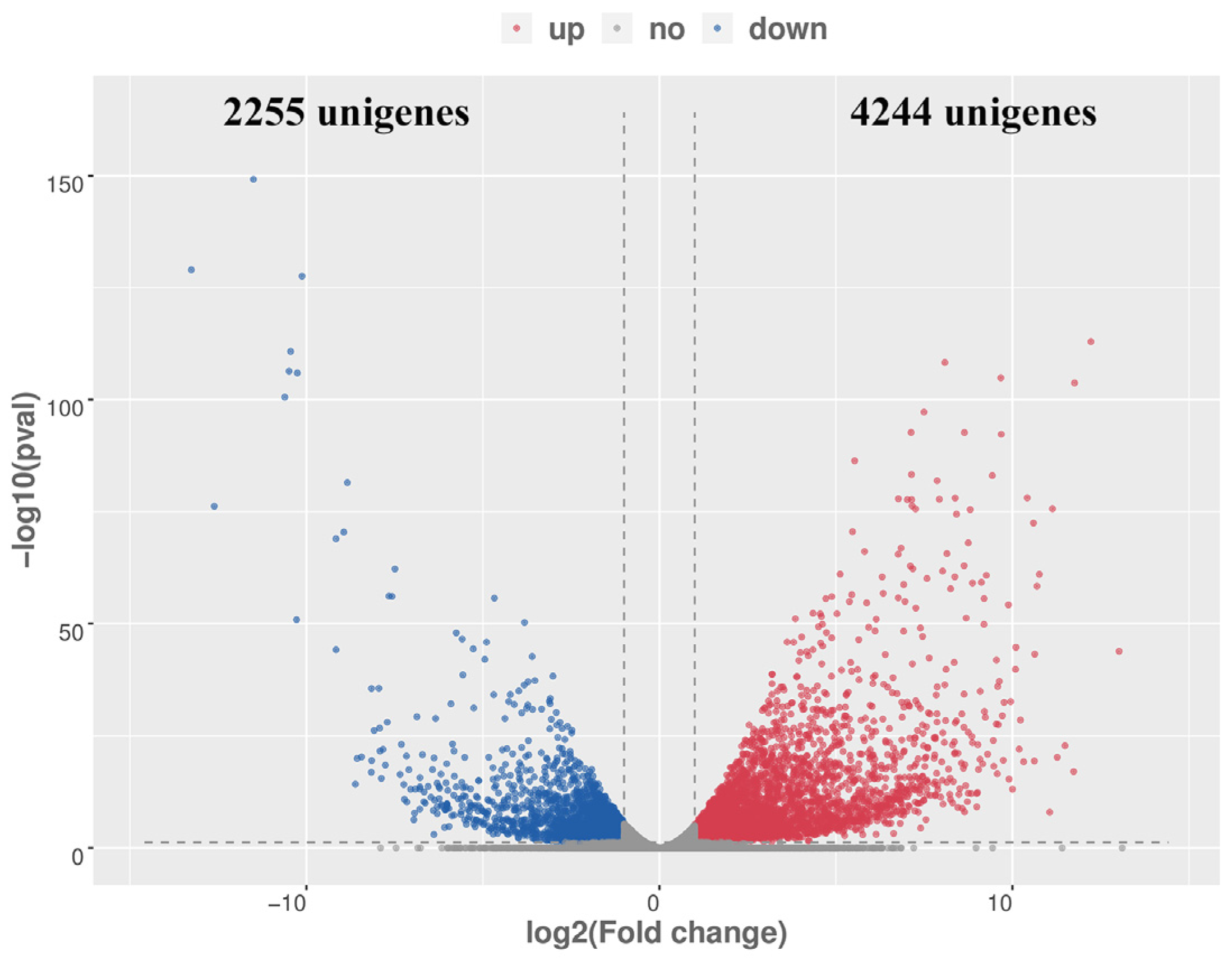
2.3. Characterization of the Differentially Expressed Genes
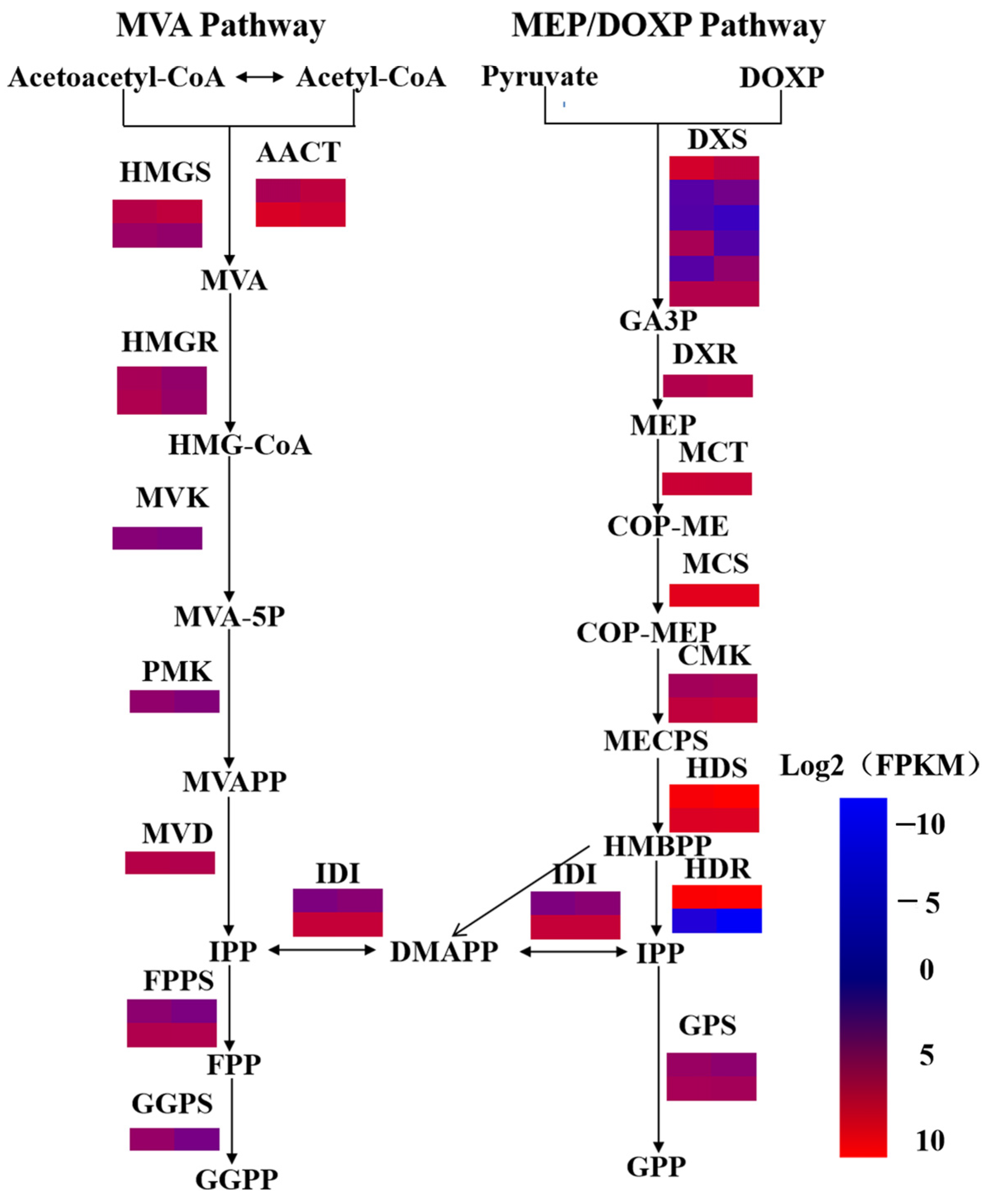
2.4. Identification of Genes Involved in Terpenoid Biosynthesis
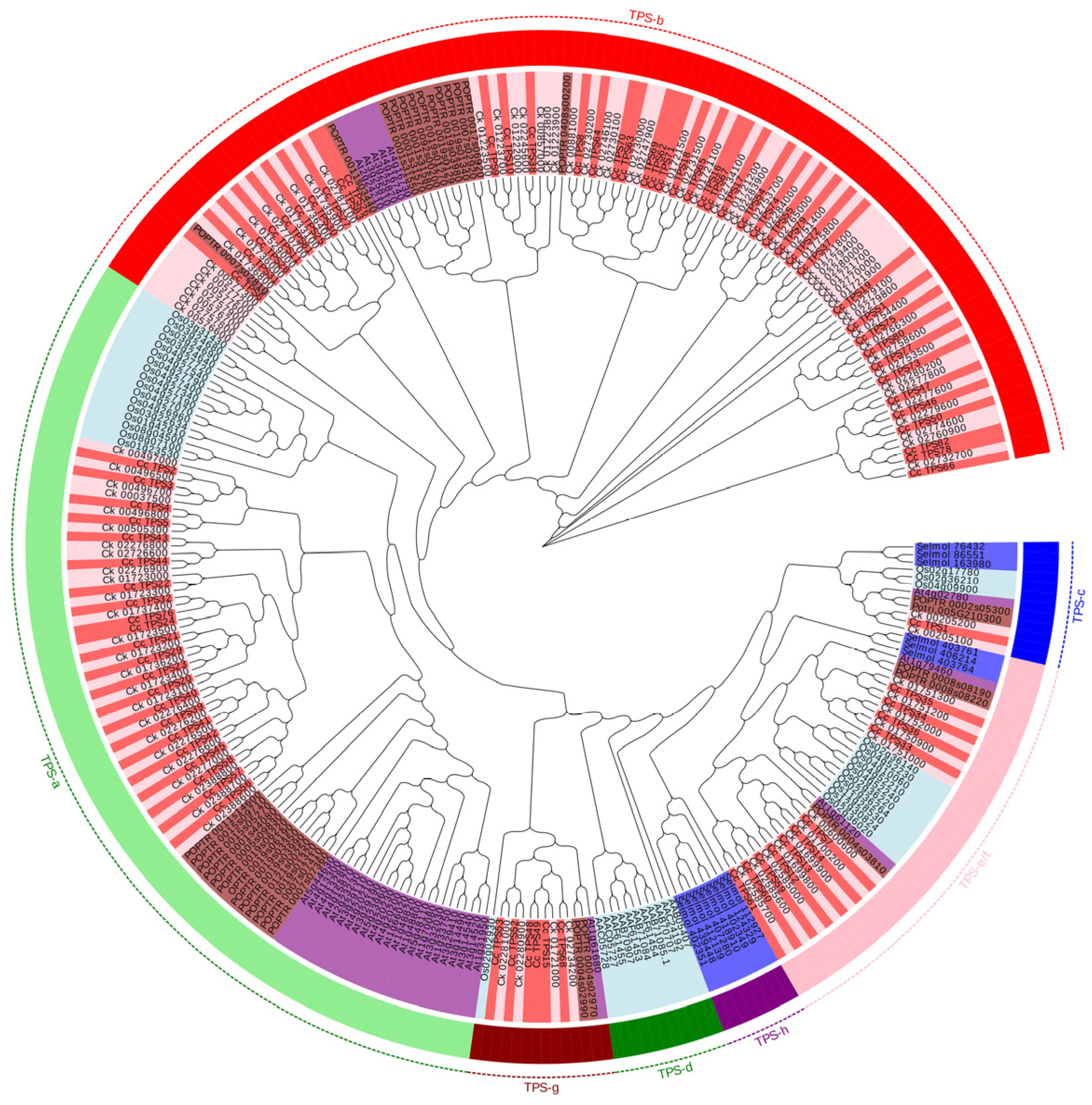
2.5. Analysis of TPS Families in C. camphora
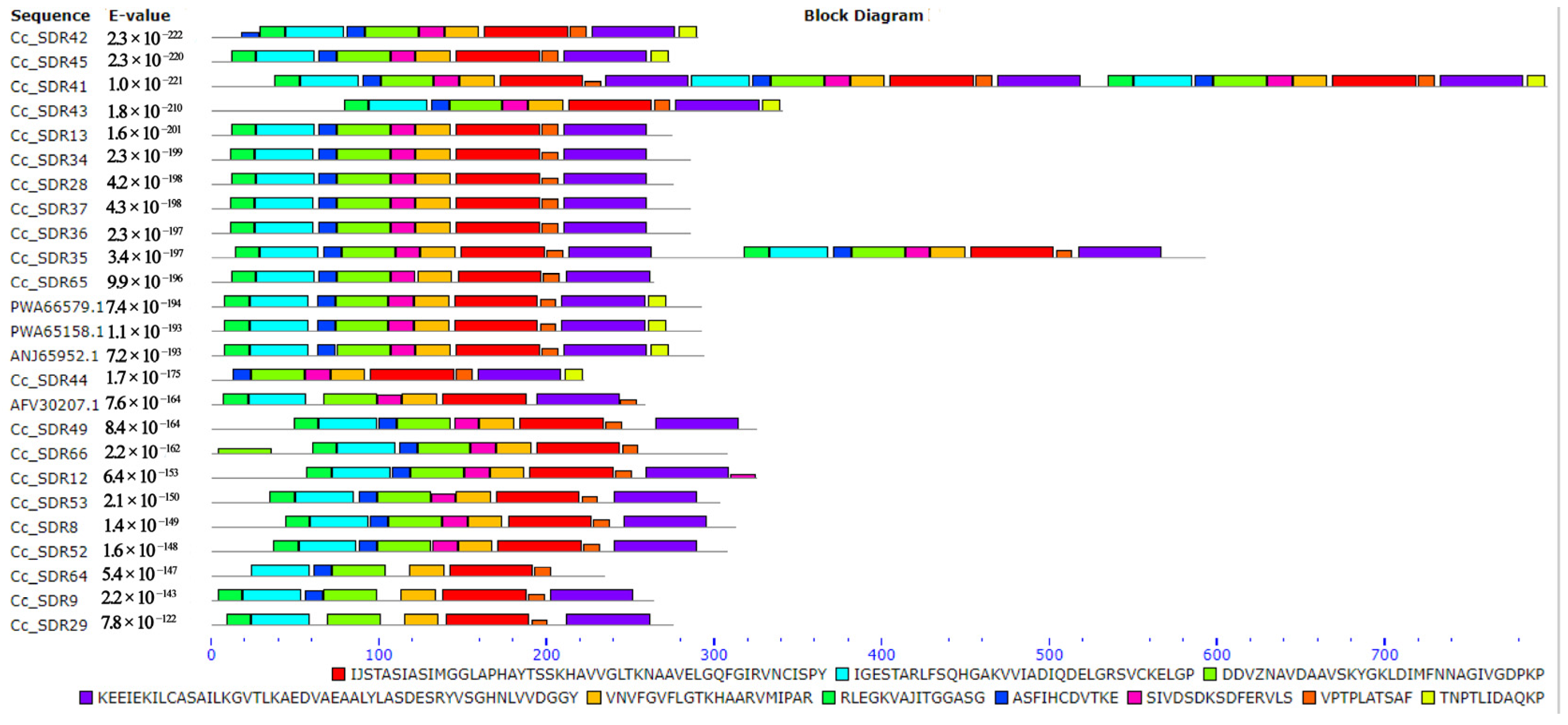
2.6. Analysis of SDR Families in C. camphora
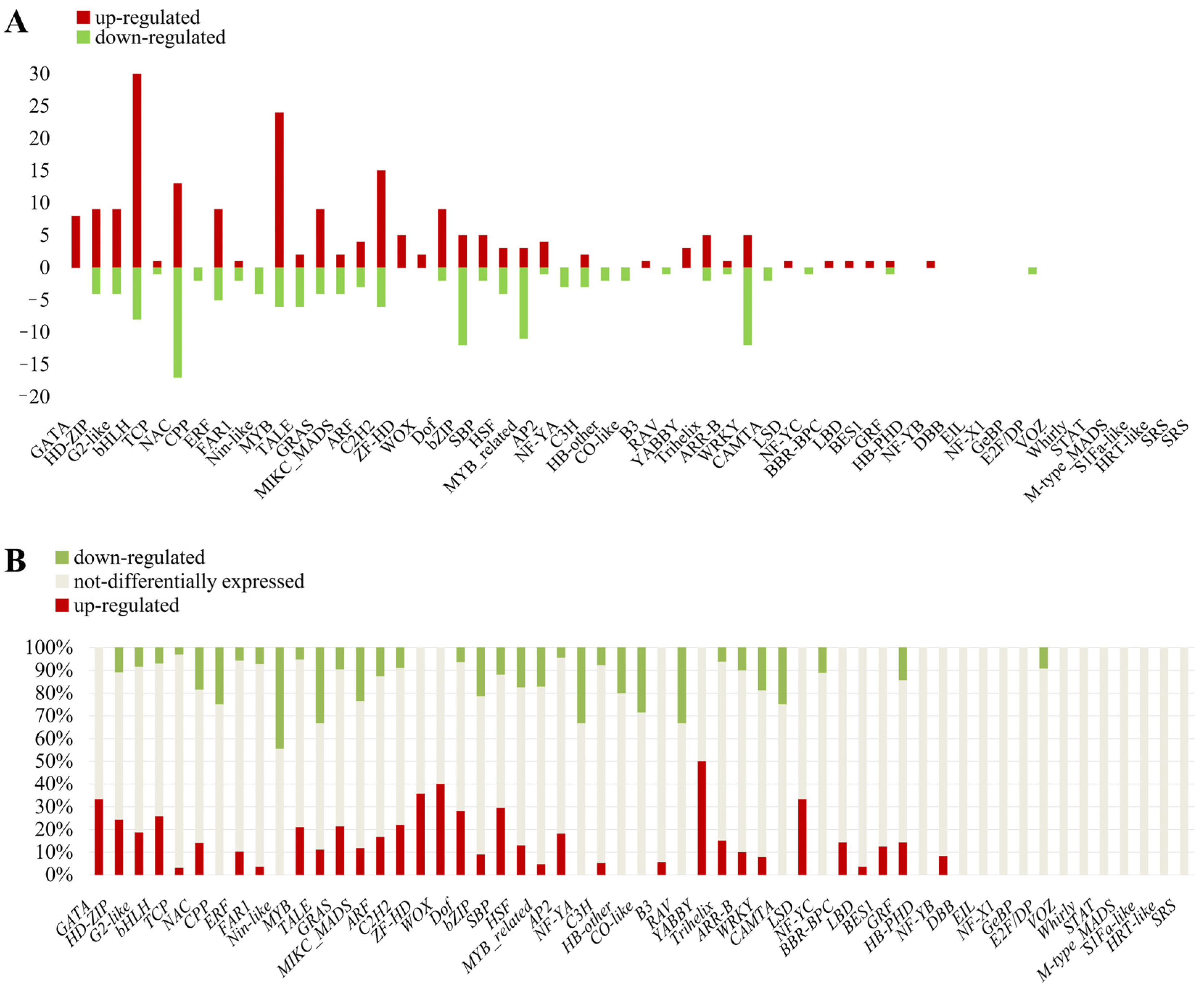
2.7. Analysis of Transcription Factor Genes Related to Terpenoid Biosynthesis
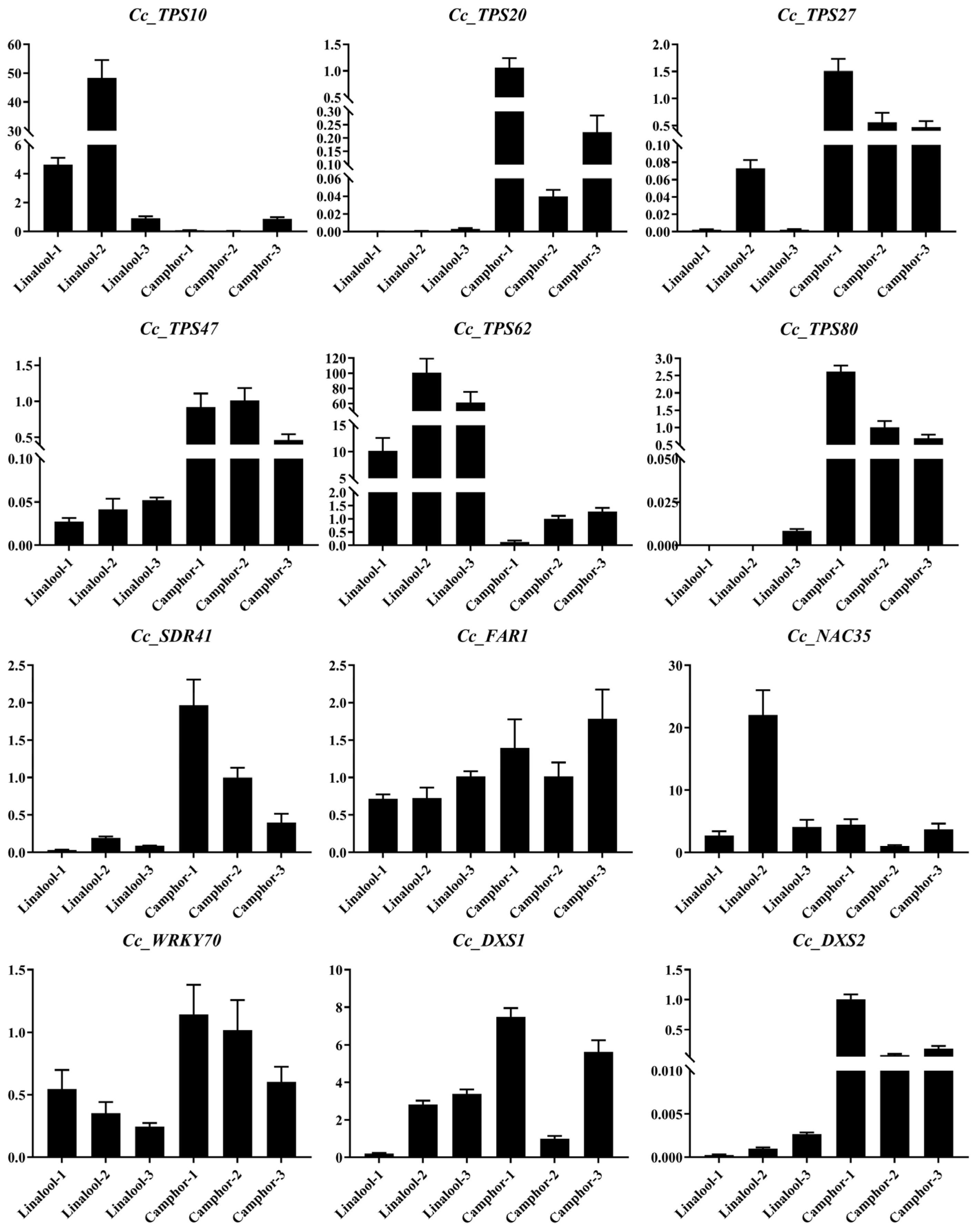
2.8. RT-qPCR Confirmation of RNA-Seq Data
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Phytochemical Analysis
4.3. RNA Sequencing and Transcriptome Analysis
4.4. Analysis of the Differentially Expressed Unigenes
4.5. RT-qPCR Analysis
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| EO(s) | Essential oil(s). |
| TPS(s) | Terpene synthase(s). |
| mTPS(s) | Monoterpene synthase(s). |
| sTPS(s) | Sesquiterpene synthase(s). |
| GPP | Geranyl diphosphate. |
| NPP | Neryl diphosphate. |
| LiLINS | L. × intermedia linalool synthase. |
| LiBDH | L. × intermedia borneol dehydrogenase. |
| SDR | Short-chain dehydrogenase/reductase. |
References
- Desjardins, A.E. Natural Product Chemistry Meets Genetics: When Is a Genotype a Chemotype? J. Agric. Food Chem. 2008, 56, 7587–7592. [Google Scholar] [CrossRef] [PubMed]
- Kamol, P.; Nukool, W.; Pumjaroen, S.; Inthima, P.; Kongbangkerd, A.; Suphrom, N.; Buddhachat, K. Assessing the genetic diversity of Centella asiatica (L.) Urb. and seasonal influence on chemotypes and morphotypes in Thailand. Ind. Crops Prod. 2024, 218, 118976. [Google Scholar] [CrossRef]
- Soltani Howyzeh, M.; Sadat Noori, S.A.; Shariati, J.V.; Niazian, M. Essential Oil Chemotype of Iranian Ajowan (Trachyspermum ammi L.). J. Essent. Oil Bear. Plants 2018, 21, 273–276. [Google Scholar] [CrossRef]
- De Souza, T.D.S.; da Silva Ferreira, M.F.; Menini, L.; de Lima Souza, J.R.C.; de Oliveira Bernardes, C.; Ferreira, A. Chemotype Diversity of Psidium guajava L. Phytochemistry 2018, 153, 129–137. [Google Scholar] [CrossRef]
- Xie, P.; Yang, Q.; Chen, J.; Tu, T.; Lian, H.; He, B.; Cai, Y. Unpredictable Chemical Diversity of Essential Oils in Cinnamomum burmanni (Lauraceae) Living Collections: Beyond Maternally Inherited Phylogenetic Relationships. Molecules 2024, 29, 1206. [Google Scholar] [CrossRef]
- Guo, X.; Cui, M.; Deng, M.; Liu, X.; Huang, X.; Zhang, X.; Luo, L. Molecular Differentiation of Five Cinnamomum camphora Chemotypes Using Desorption Atmospheric Pressure Chemical Ionization Mass Spectrometry of Raw Leaves. Sci. Rep. 2017, 7, 46579. [Google Scholar] [CrossRef]
- Marcial, G.; De Lampasona, M.P.; Vega, M.I.; Lizarraga, E.; Viturro, C.I.; Slanis, A.; Juárez, M.A.; Elechosa, M.A.; Catalán, C.A. Intraspecific Variation in Essential Oil Composition of the Medicinal Plant Lippia integrifolia (Verbenaceae). Evidence for Five Chemotypes. Phytochemistry 2016, 122, 203–212. [Google Scholar] [CrossRef]
- Lee, S.-H.; Kim, D.-S.; Park, S.-H.; Park, H. Phytochemistry and Applications of Cinnamomum camphora Essential Oils. Molecules 2022, 27, 2695. [Google Scholar] [CrossRef]
- IUCN. Iucn Red List of Threatened Species; The International Union for Conservation of Nature: Gland, Switzerland, 2012. [Google Scholar]
- Shi, S.; Wu, Q.; Su, J.; Li, C.; Zhao, X.; Xie, J.; Gui, S.; Su, Z.; Zeng, H. Composition Analysis of Volatile Oils from Flowers, Leaves and Branches of Cinnamomum camphora Chvar. Borneol in China. J. Essent. Oil Res. 2013, 25, 7. [Google Scholar] [CrossRef]
- Riaz, R.; Younis, W.; Uttra, A.M.; Malik, M.N.H.; Manzoor, W.; Qasim, S.; Hasan, U.H.; Mushtaq, M.N.; Kluck, A.J.; Lívero, F.A.d.R.; et al. Anti-arthritic and anti-inflammatory activity of linalool against formaldehyde and complete Freund’s adjuvant induced arthritis in rats. Biochem. Biophys. Res. 2025, 752, 151462. [Google Scholar] [CrossRef]
- Mo, J.; Yang, Y.; Huang, Z.; Xie, Y.; Lu, A.; Lao, Y.; Zhou, X.; Wu, Y.; Pan, Y.; Qin, Z. Study on Variations of Content and Main Constituent of Essential Oil from Stem and Leaf of Linalool-Type Cinnamomum camphora (Linn.) Presl in Guangxi Province. Chem. Ind. For. Prod. 2010, 30, 72–76. [Google Scholar]
- Tsang, H.L.; Huang, J.L.; Lin, Y.H.; Huang, K.F.; Lu, P.L.; Lin, G.H.; Khine, A.A.; Hu, A.; Chen, H.P. Borneol Dehydrogenase from Pseudomonas sp. Strain TCU-HL1 Catalyzes the Oxidation of (+)-Borneol and Its Isomers to Camphor. Appl. Environ. Microbiol. 2016, 82, 6378–6385. [Google Scholar] [CrossRef]
- Cardoso, J.M.; Correia, I.; Galvão, A.M.; Marques, F.; Carvalho, M.F.N. Synthesis of Ag(I) Camphor Sulphonylimine Complexes and Assessment of Their Cytotoxic Properties against Cisplatin -Resistant A2780cisr and A2780 Cell Lines. J. Inorg. Biochem. 2017, 166, 55–63. [Google Scholar] [CrossRef]
- Koksal, M.; Jin, Y.; Coates, R.M.; Croteau, R.; Christianson, D.W. Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 2011, 469, 116–120. [Google Scholar] [CrossRef]
- Masyita, A.; Mustika Sari, R.; Dwi Astuti, A.; Yasir, B.; Rahma Rumata, N.; Emran, T.B.; Nainu, F.; Simal-Gandara, J. Terpenes and terpenoids as main bioactive compounds of essential oils, their roles in human health and potential application as natural food preservatives. Food Chem. X 2022, 13, 100217. [Google Scholar] [CrossRef]
- Gershenzon, J.; McConkey, M.E.; Croteau, R.B. Regulation of Monoterpene Accumulation in Leaves of Peppermint. Plant Physiol. 2000, 122, 205–214. [Google Scholar] [CrossRef]
- Tian, N.; Tang, Y.; Xiong, S.; Tian, D.; Chen, Y.; Wu, D.; Liu, Z.; Liu, S. Molecular Cloning and Functional Identification of a Novel Borneol Dehydrogenase from Artemisia annua L. Ind. Crops Prod. 2015, 77, 190–195. [Google Scholar] [CrossRef]
- Kavanagh, K.L.; Jörnvall, H.; Persson, B.; Oppermann, U. Medium- and Short-Chain Dehydrogenase/Reductase Gene and Protein Families: The Sdr Superfamily: Functional and Structural Diversity within a Family of Metabolic and Regulatory Enzymes. Cell. Mol. Life Sci. 2008, 65, 3895–3906. [Google Scholar] [CrossRef]
- Persson, B.; Kallberg, Y.; Bray, J.E.; Bruford, E.; Dellaporta, S.L.; Favia, A.D.; Duarte, R.G.; Jörnvall, H.; Kavanagh, K.L.; Kedishvili, N.; et al. The Sdr (Short-Chain Dehydrogenase/Reductase and Related Enzymes) Nomenclature Initiative. Chem. Biol. Interact. 2009, 178, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Persson, B.; Kallberg, Y. Classification and Nomenclature of the Superfamily of Short-Chain Dehydrogenases/Reductases (Sdrs). Chem. Biol. Interact. 2013, 202, 111–115. [Google Scholar] [CrossRef]
- Wei, J.; Yang, Y.; Peng, Y.; Wang, S.; Zhang, J.; Liu, X.; Liu, J.; Wen, B.; Li, M. Biosynthesis and the Transcriptional Regulation of Terpenoids in Tea Plants (Camellia sinensis). Int. J. Mol. Sci. 2023, 24, 6937. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The Family of Terpene Synthases in Plants: A Mid-Size Family of Genes for Specialized Metabolism That Is Highly Diversified Throughout the Kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Martin, D.M.; Faldt, J.; Bohlmann, J. Functional Characterization of Nine Norway Spruce Tps Genes and Evolution of Gymnosperm Terpene Synthases of the Tps-D Subfamily. Plant Physiol. 2004, 135, 1908–1927. [Google Scholar] [CrossRef]
- Li, M.; Xu, J.; Algarra Alarcon, A.; Carlin, S.; Barbaro, E.; Cappellin, L.; Velikova, V.; Vrhovsek, U.; Loreto, F.; Varotto, C. In Planta Recapitulation of Isoprene Synthase Evolution from Ocimene Synthases. Mol. Biol. Evol. 2017, 34, 2583–2599. [Google Scholar] [CrossRef]
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Faldt, J.; Miller, B.; Bohlmann, J. (E)-Β-Ocimene and Myrcene Synthase Genes of Floral Scent Biosynthesis in Snapdragon: Function and Expression of Three Terpene Synthase Genes of a New Terpene Synthase Subfamily. Plant Cell 2003, 15, 1227–1241. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Song, A.; Guan, Z.; Zhang, X.; Sun, H.; Wang, Y.; Yu, Q.; Fu, X.; Fang, W.; Chen, F. CmWRKY41 activates CmHMGR2 and CmFPPS2 to positively regulate sesquiterpenes synthesis in Chrysanthemum morifolium. Plant Physiol. Biochem. 2023, 196, 821–829. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis Myc2 Interacts with Della Proteins in Regulating Sesquiterpene Synthase Gene Expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuizen, N.J.; Chen, X.; Wang, M.Y.; Matich, A.J.; Perez, R.L.; Allan, A.C.; Green, S.A.; Atkinson, R.G. Natural variation in monoterpene synthesis in kiwifruit: Transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 2015, 167, 1243–1258. [Google Scholar] [CrossRef]
- Pathak, S.; Lakhwani, D.; Gupta, P.; Mishra, B.K.; Shukla, S.; Asif, M.H.; Trivedi, P.K. Comparative Transcriptome Analysis Using High Papaverine Mutant of Papaver somniferum Reveals Pathway and Uncharacterized Steps of Papaverine Biosynthesis. PLoS ONE 2013, 8, e65622. [Google Scholar] [CrossRef]
- Soltani Howyzeh, M.; Sadat Noori, S.A.; Shariati, J.V.; Amiripour, M. Comparative Transcriptome Analysis to Identify Putative Genes Involved in Thymol Biosynthesis Pathway in Medicinal Plant Trachyspermum ammi L. Sci. Rep. 2018, 8, 13405. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Zhong, Y.; Wu, Y.; Li, Z.; Xu, L.A.; Xu, M. Transcriptome Analysis and Identification of Genes Related to Terpenoid Biosynthesis in Cinnamomum camphora. BMC Genom. 2018, 19, 550. [Google Scholar] [CrossRef] [PubMed]
- Friesen, J.A.; Rodwell, V.W. The 3-Hydroxy-3-Methylglutaryl Coenzyme-A (HMG-CoA) Reductases. Genome Biol. 2004, 5, 248. [Google Scholar] [CrossRef]
- He, S.; Bekhof, A.S.M.; Popova, E.Z.; van Merkerk, R.; Quax, W.J. Improved taxadiene production by optimizing DXS expression and fusing short-chain prenyltransferases. New Biotechnol. 2024, 83, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y.; Mann, F.; Huang, Y.; Saunders, R.M.K.; Chappell, J. Diterpene Cyclases and the Nature of the Isoprene Unit. Proc. Natl. Acad. Sci. USA 2010, 107, 9127–9132. [Google Scholar]
- Taibi, M.; Elbouzidi, A.; Haddou, M.; Baraich, A.; Gharsallaoui, A.; Mothana, R.A.; Alqahtani, A.M.; Asehraou, A.; Bellaouchi, R.; Addi, M.; et al. Evaluation of the Interaction Between Menthol and Camphor, Major Compounds of Clinopodium nepeta Essential Oil: Antioxidant, Anti-inflammatory and Anticancer Activities Against Breast Cancer Cell Lines. Chem. Biodivers. 2025, e202403098. [Google Scholar] [CrossRef]
- Zulak, K.G.; Liscombe, M.A.; Liscombe, D.K.; Facchini, P.J. Molecular Characterization of Terpenoid Indole Alkaloid Biosynthesis in Catharanthus roseus. Phytochem. Rev. 2009, 8, 321–337. [Google Scholar]
- Degenhardt, J.; Kollner, T.G.; Gershenzon, J. Monoterpene and Sesquiterpene Synthases and the Origin of Terpene Skeletal Diversity in Plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef]
- Liu, J.; Xu, Y.; Liang, J.; Chen, X. Alternative Splicing of Terpene Synthase Genes in Artemisia annua and Its Implications for Terpenoid Biosynthesis. Plant Physiol. Biochem. 2018, 132, 160–168. [Google Scholar]
- Suo, J.; Tong, K.; Wu, J.; Ding, M.; Chen, W.; Yang, Y.; Lou, H.; Hu, Y.; Yu, W.; Song, L. Comparative Transcriptome Analysis Reveals Key Genes in the Regulation of Squalene and β-Sitosterol Biosynthesis in Torreya grandis. Ind. Crops Prod. 2019, 131, 182–193. [Google Scholar] [CrossRef]
- Sarker, L.S.; Galata, M.; Demissie, Z.A.; Mahmoud, S.S. Molecular cloning and functional characterization of borneol dehydrogenase from the glandular trichomes of Lavandula x intermedia. Arch. Biochem. Biophys. 2012, 528, 163–170. [Google Scholar] [CrossRef]
- He, X.; Wang, H.; Yang, J.; Deng, K.; Wang, T. RNA sequencing on Amomum villosum Lour. induced by MeJA identifies the genes of WRKY and terpene synthases involved in terpene biosynthesis. Genome 2018, 61, 91–102. [Google Scholar]
- Kumar, A.; Sharma, P.; Srivastava, R.; Verma, P.C. Chapter 11—Demystifying the role of transcription factors in plant terpenoid biosynthesis. In Plant Transcription Factors; Srivastava, V., Mishra, S., Mehrotra, S., Upadhyay, S.K., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 233–249. [Google Scholar]
- Li, M.; Shao, Y.; Pan, B.; Liu, C.; Tan, H. Regulation of important natural products biosynthesis by WRKY transcription factors in plants. J. Adv. Res. 2025. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.Y.; Chen, J.; Xu, W.X.; Qiu, J.R.; Song, L.; Wang, J.T.; Tang, R.; Chen, D.; Jiang, C.Z.; Huang, Z. Dehydration-Induced WRKY Transcriptional Factor MfWRKY70 of Myrothamnus flabellifolia Enhanced Drought and Salinity Tolerance in Arabidopsis. Biomolecules 2021, 11, 327. [Google Scholar] [CrossRef] [PubMed]
- Chaw, S.M.; Liu, Y.C.; Wu, Y.W.; Wang, H.Y.; Lin, C.Y.I.; Wu, C.S.; Ke, H.M.; Chang, L.Y.; Hsu, C.Y.; Yang, H.T.; et al. Stout Camphor Tree Genome Fills Gaps in Understanding of Flowering Plant Genome Evolution. Nat. Plants 2019, 5, 63–73. [Google Scholar] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. Tophat: Discovering Splice Junctions with Rna-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching During Cell Differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA Homology Search with Profile Hmms. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Zhou, S.; Ni, M.; Zhang, Y.; Lin, S.; Zhang, J.; Tong, Z. The Mining of Candidate Genes Involved in the Camphor Biosynthesis Pathway of Cinnamomum camphora. Plants 2025, 14, 991. https://doi.org/10.3390/plants14070991
Yang Y, Zhou S, Ni M, Zhang Y, Lin S, Zhang J, Tong Z. The Mining of Candidate Genes Involved in the Camphor Biosynthesis Pathway of Cinnamomum camphora. Plants. 2025; 14(7):991. https://doi.org/10.3390/plants14070991
Chicago/Turabian StyleYang, Yan, Shengcai Zhou, Mingyang Ni, Yuting Zhang, Shixiong Lin, Junhong Zhang, and Zaikang Tong. 2025. "The Mining of Candidate Genes Involved in the Camphor Biosynthesis Pathway of Cinnamomum camphora" Plants 14, no. 7: 991. https://doi.org/10.3390/plants14070991
APA StyleYang, Y., Zhou, S., Ni, M., Zhang, Y., Lin, S., Zhang, J., & Tong, Z. (2025). The Mining of Candidate Genes Involved in the Camphor Biosynthesis Pathway of Cinnamomum camphora. Plants, 14(7), 991. https://doi.org/10.3390/plants14070991




