Overexpression of OsPIN5b Alters Plant Architecture and Impairs Cold Tolerance in Rice (Oryza sativa L.)
Abstract
1. Introduction
2. Results
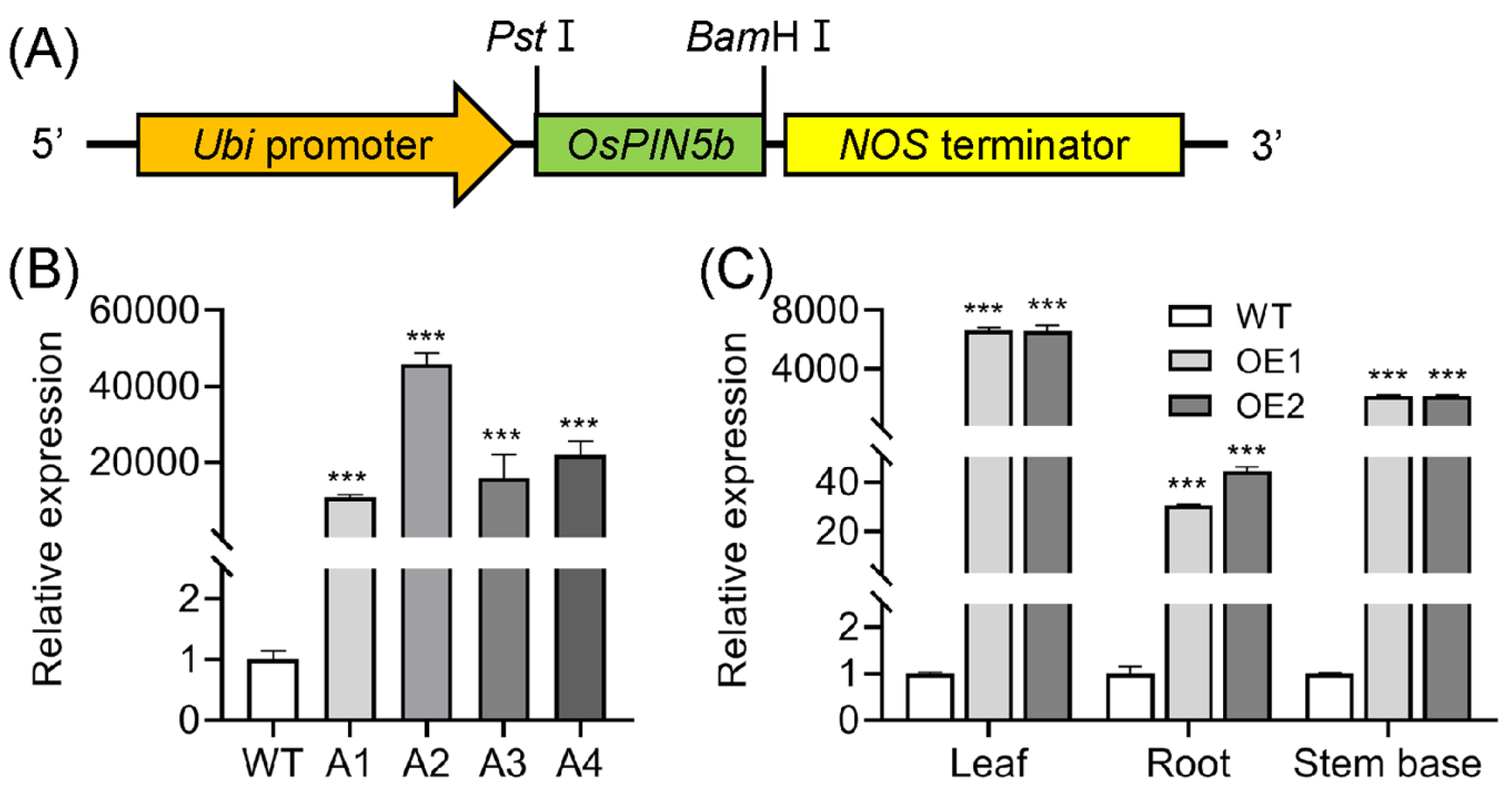
2.1. Generation of OsPIN5b-Overexpressing Lines and Phenotypes of Transformants
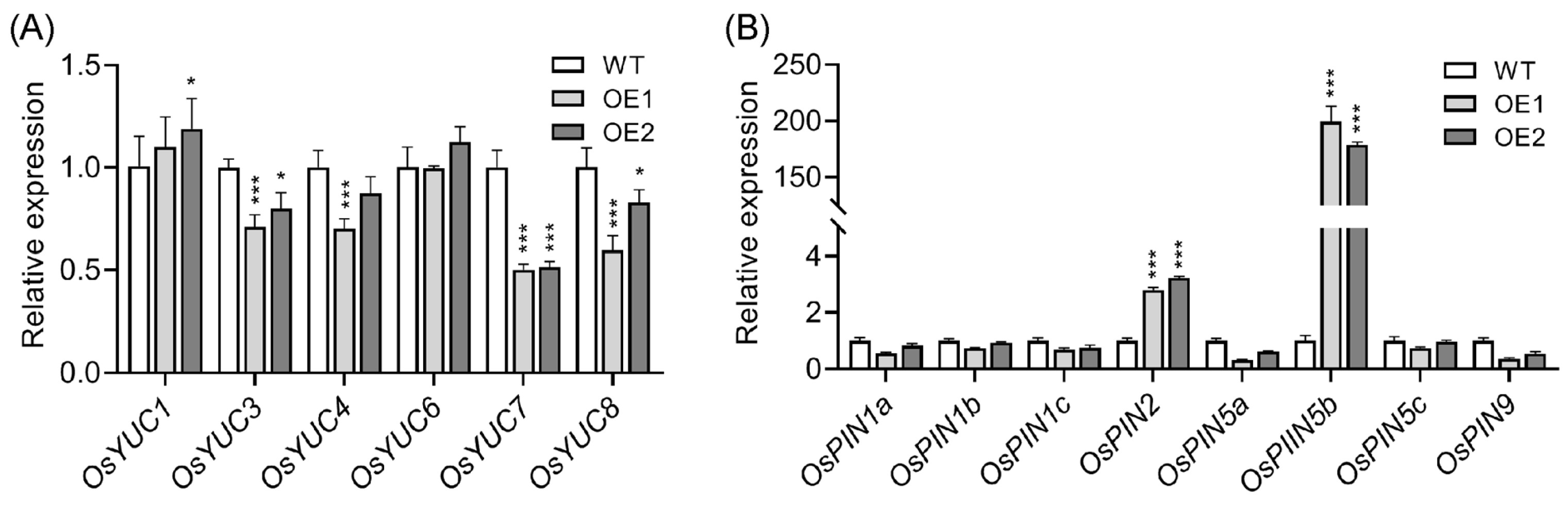
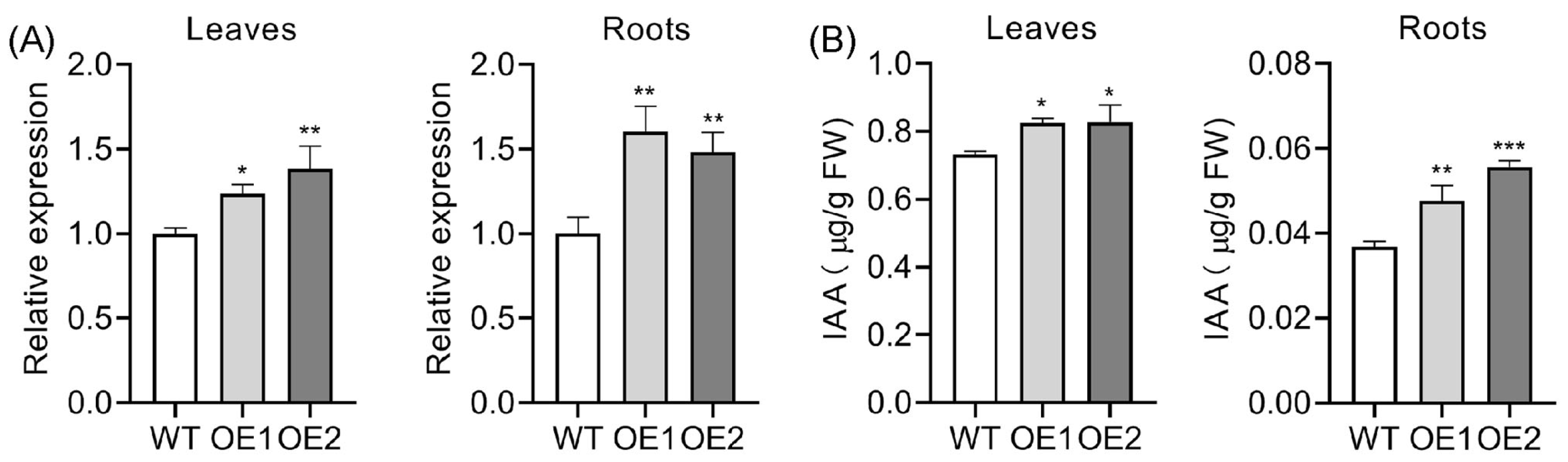
2.2. Upregulation of OsPIN5b Disrupts Auxin Homeostasis
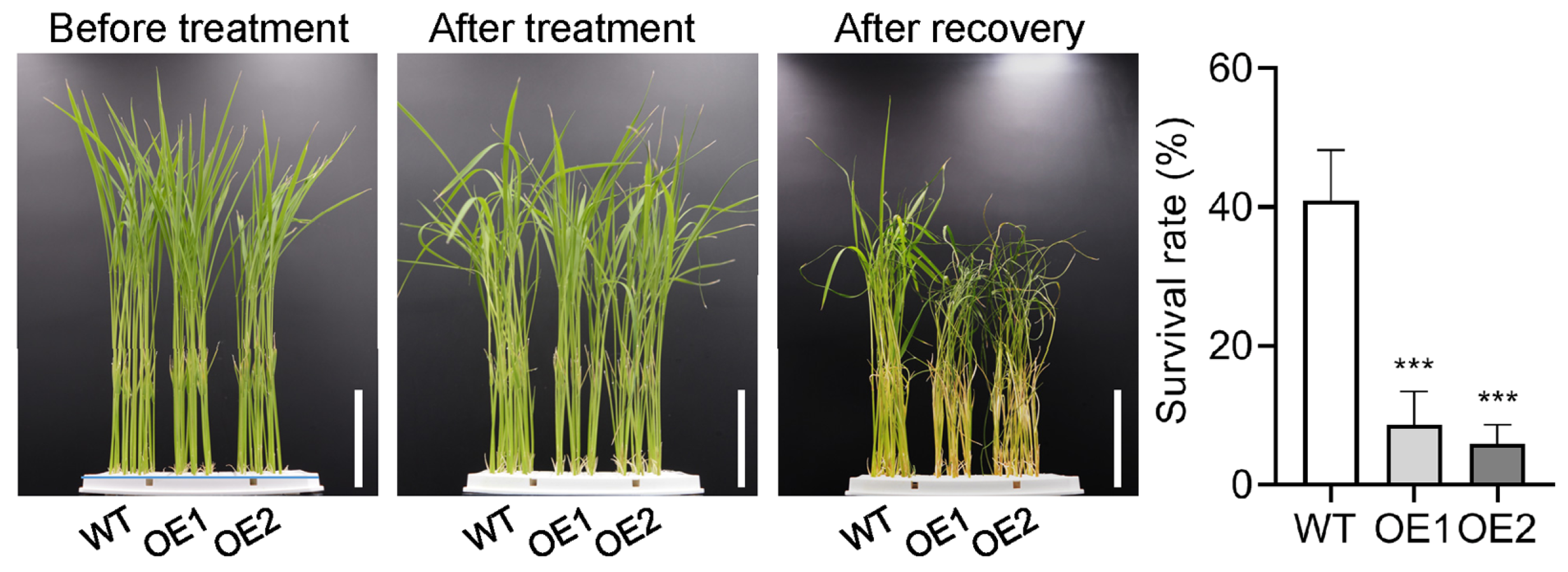
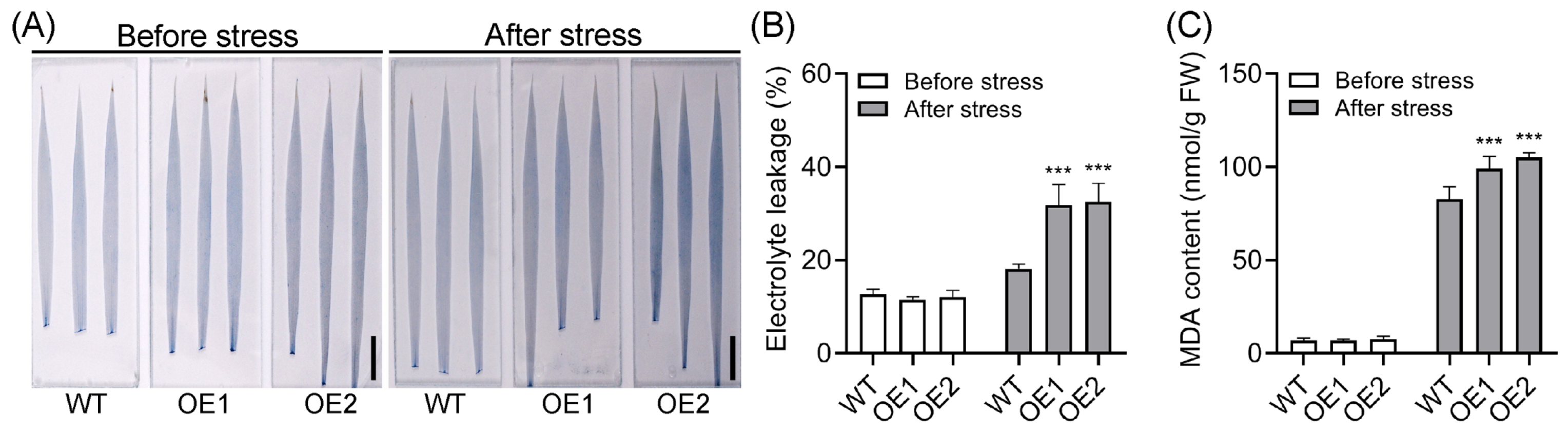
2.3. Overexpression of OsPIN5b Impairs Rice Cold Tolerance
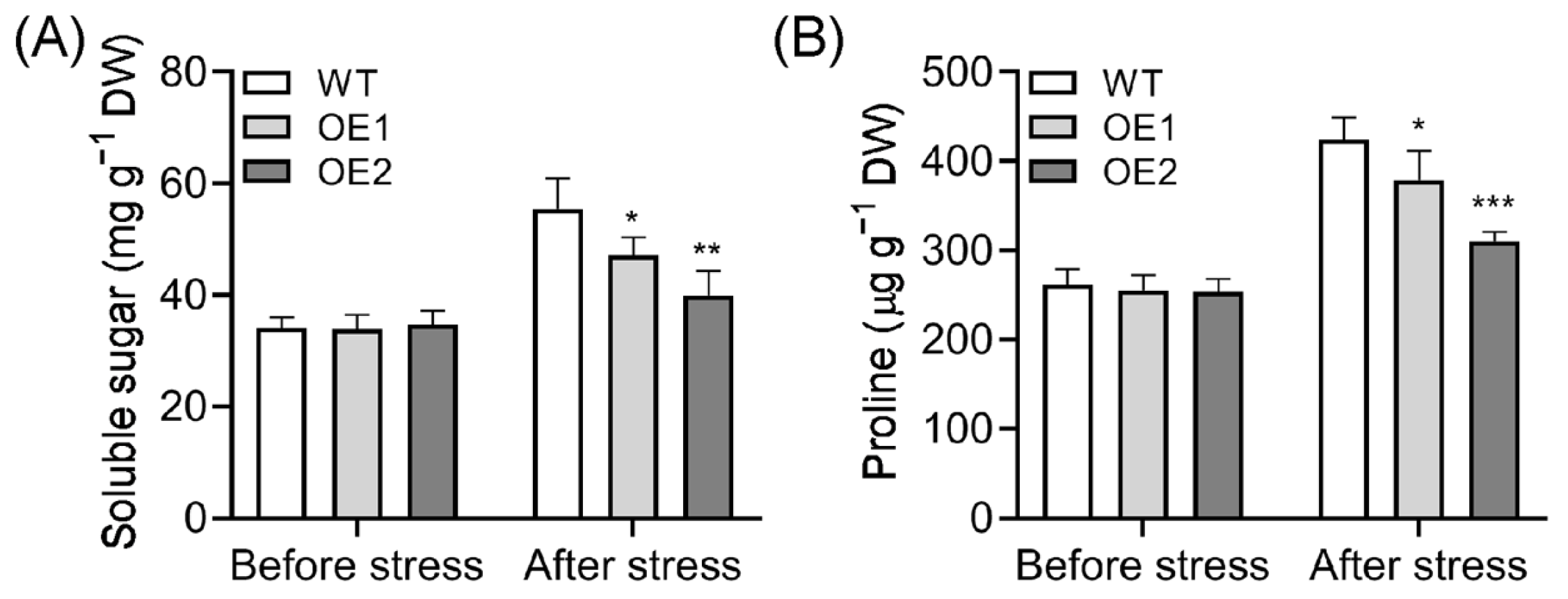
2.4. Soluble Sugar and Proline Content Decreased in OE Lines After Cold Treatment
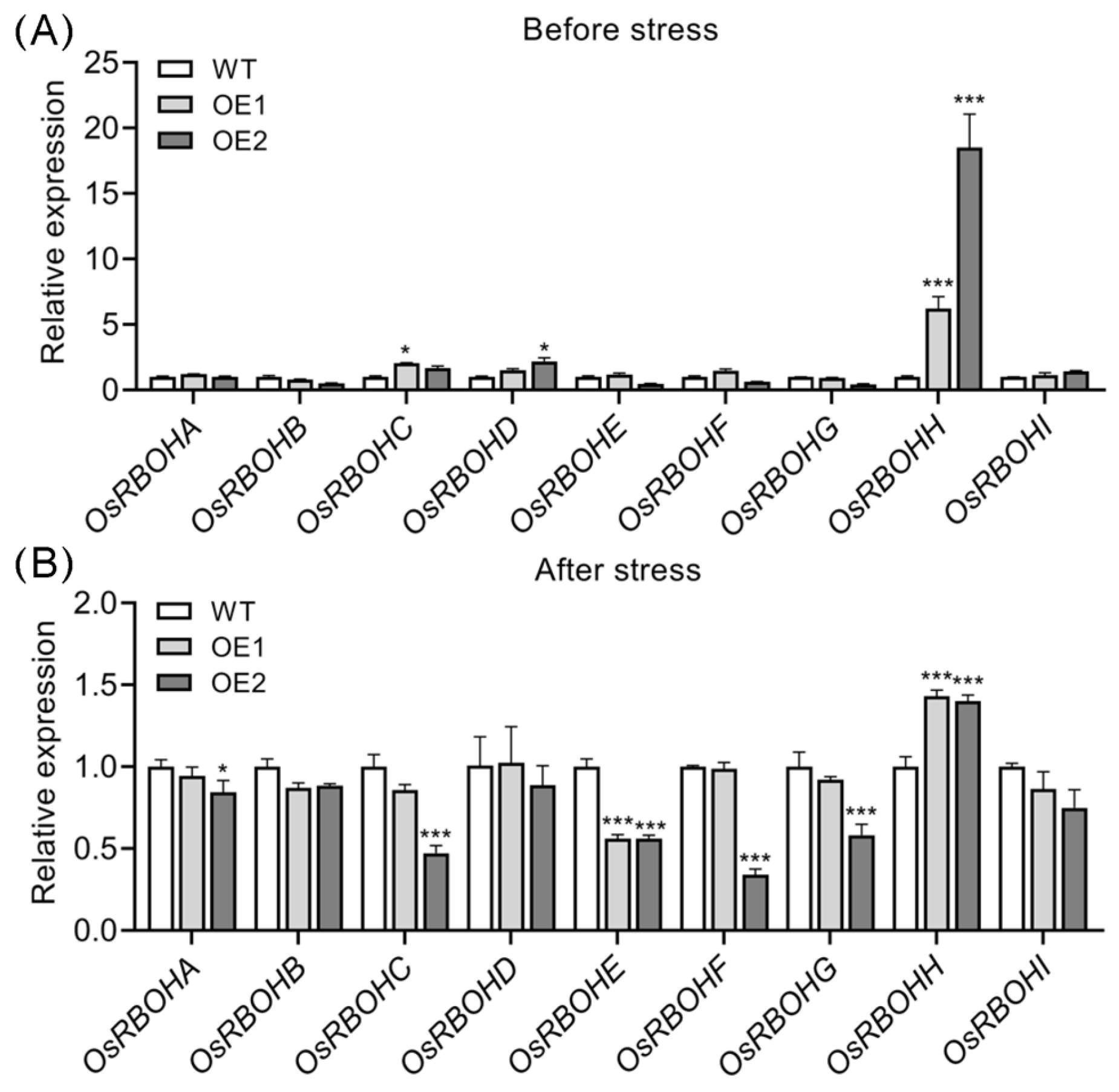
2.5. Overexpression of OsPIN5b Disturbs ROS Homeostasis
2.6. OsRBOHH Likely Plays a Role in ROS Production in OsPIN5-Overexpressing Lines
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Cold Treatment
4.2. Vector Construction and Generation of the Transgenic Plants
4.3. Measurement of Auxin Levels
4.4. Quantitative Real-Time PCR (qRT-PCR) Analysis
4.5. Physiological Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, J.L.; Li, C.G.; Kang, H.X.; Wang, Y.; Tan, X.Q.; Liu, M.H.; Deng, Y.F.; Wang, Z.L.; Liu, Y.; et al. Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 2016, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Xu, Y.; Wang, X.; Du, C.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ. 2011, 34, 52–64. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Molecular regulation of plant responses to environmental temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Cheng, C.; Yun, K.Y.; Ressom, H.W.; Mohanty, B.; Bajic, V.B.; Jia, Y.; Yun, S.J.; de los Reyes, B.G. An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genom. 2007, 8, 175. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, R.; Wang, Y.; Zhang, L.; Yao, S. A point mutation in LTT1 enhances cold tolerance at the booting stage in rice. Plant Cell Environ. 2020, 43, 992–1007. [Google Scholar] [CrossRef]
- Andaya, V.C.; Mackill, D.J. Mapping of QTLs associated with cold tolerance during the vegetative stage in rice. J. Exp. Bot. 2003, 54, 2579–2585. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, S. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.M.; Lin, H.X.; Chong, K. Crop improvement through temperature resilience. Annu. Rev. Plant Biol. 2019, 70, 753–780. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef]
- Ito, Y.; Katsura, K.; Maruyama, K.; Taji, T.; Kobayashi, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006, 47, 141–153. [Google Scholar] [CrossRef]
- Wang, Q.; Guan, Y.; Wu, Y.; Chen, H.; Chen, F.; Chu, C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008, 67, 589–602. [Google Scholar] [CrossRef]
- Moon, S.J.; Min, M.K.; Kim, J.A.; Kim, D.Y.; Yoon, I.S.; Kwon, T.R.; Byun, M.O.; Kim, B.G. Ectopic expression of OsDREB1G, a member of the OsDREB1 subfamily, confers cold stress tolerance in rice. Front. Plant Sci. 2019, 10, 297. [Google Scholar] [CrossRef]
- Zhang, M.; Zhao, R.; Huang, K.; Huang, S.; Wang, H.; Wei, Z.; Li, Z.; Bian, M.; Jiang, W.; Wu, T.; et al. The OsWRKY63-OsWRKY76-OsDREB1B module regulates chilling tolerance in rice. Plant J. 2022, 112, 383–398. [Google Scholar] [CrossRef]
- Mao, D.; Chen, C. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold-responsive pathway. PLoS ONE 2012, 7, e47275. [Google Scholar] [CrossRef]
- Zhao, C.; Lang, Z.; Zhu, J.K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 2015, 20, 466–468. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Luo, W.; Zhao, Y.; Xu, Y.; Song, S.; Chong, K. Comparative metabolomic analysis reveals a reactive oxygen species-dominated dynamic model underlying chilling environment adaptation and tolerance in rice. New Phytol. 2016, 211, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Li, R.; Song, Y.; Wang, X.; Zheng, C.; Liu, B.; Zhang, H.; Ke, J.; Wu, X.; Wu, L.; Yang, R.; et al. OsNAC5 orchestrates OsABI5 to fine-tune cold tolerance in rice. J. Integr. Plant Biol. 2024, 66, 660–682. [Google Scholar] [CrossRef]
- Sun, Y.; Xie, Z.; Jin, L.; Qin, T.; Zhan, C.; Huang, J. Histone deacetylase OsHDA716 represses rice chilling tolerance by deacetylating OsbZIP46 to reduce its transactivation function and protein stability. Plant Cell 2024, 36, 1913–1936. [Google Scholar] [CrossRef]
- Liu, D.; Luo, S.; Li, Z.; Liang, G.; Guo, Y.; Xu, Y.; Chong, K. COG3 confers the chilling tolerance to mediate OsFtsH2-D1 module in rice. New Phytol. 2024, 241, 2143–2157. [Google Scholar] [CrossRef]
- Ji, L.; Zhang, Z.; Liu, S.; Zhao, L.; Li, Q.; Xiao, B.; Suzuki, N.; Burks, D.J.; Azad, R.K.; Xie, G. The OsTIL1 lipocalin protects cell membranes from reactive oxygen species damage and maintains the 18:3-containing glycerolipid biosynthesis under cold stress in rice. Plant J. 2024, 117, 72–91. [Google Scholar] [CrossRef]
- Liu, J.; Liu, J.; He, M.; Zhang, C.; Liu, Y.; Li, X.; Wang, Z.; Jin, X.; Sui, J.; Zhou, W.; et al. OsMAPK6 positively regulates rice cold tolerance at seedling stage via phosphorylating and stabilizing OsICE1 and OsIPA1. Theor. Appl. Genet. 2024, 137, 10. [Google Scholar] [CrossRef]
- Xia, C.; Liang, G.; Chong, K.; Xu, Y. The COG1-OsSERL2 complex senses cold to trigger signaling network for chilling tolerance in japonica rice. Nat. Commun. 2023, 14, 3104. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Zhang, Z.; Li, J.; Sun, J.; Cui, Z.; Li, F.; Zhuang, J.; Chen, W.; Su, C.; Wu, L.; et al. Natural variation in OsSEC13 HOMOLOG 1 modulates redox homeostasis to confer cold tolerance in rice. Plant Physiol. 2023, 193, 2180–2196. [Google Scholar] [CrossRef] [PubMed]
- Benkova, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertova, D.; Jurgens, G.; Friml, J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef]
- Lavy, M.; Estelle, M. Mechanisms of auxin signaling. Development 2016, 143, 3226–3229. [Google Scholar] [CrossRef]
- Mroue, S.; Simeunovic, A.; Robert, H.S. Auxin production as an integrator of environmental cues for developmental growth regulation. J. Exp. Bot. 2018, 69, 201–212. [Google Scholar] [CrossRef]
- Waadt, R.; Seller, C.A.; Hsu, P.K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Shi, Y.; Ding, Y.; Yang, S. Cold signal transduction and its interplay with phytohormones during cold acclimation. Plant Cell Physiol. 2015, 56, 7–15. [Google Scholar] [CrossRef]
- Gaveliene, V.; Novickiene, L.; Pakalniskyte, L. Effect of auxin physiological analogues on rapeseed (Brassica napus) cold hardening, seed yield and quality. J. Plant Res. 2013, 126, 283–292. [Google Scholar] [CrossRef]
- Du, H.; Liu, H.; Xiong, L. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Zhang, J.; Peer, W.A. Auxin homeostasis: The DAO of catabolism. J. Exp. Bot. 2017, 68, 3145–3154. [Google Scholar] [CrossRef] [PubMed]
- Kramer, E.M.; Ackelsberg, E.M. Auxin metabolism rates and implications for plant development. Front. Plant Sci. 2015, 6, 150. [Google Scholar] [CrossRef] [PubMed]
- Shibasaki, K.; Uemura, M.; Tsurumi, S.; Rahman, A. Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. Plant Cell 2009, 21, 3823–3838. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Peret, B. AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef]
- Konstantinova, N.; Korbei, B.; Luschnig, C. Auxin and root gravitropism: Addressing basic cellular processes by exploiting a defined growth response. Int. J. Mol. Sci. 2021, 22, 2749. [Google Scholar] [CrossRef]
- Petrasek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertova, D.; Wisniewska, J.; Tadele, Z.; Kubes, M.; Covanova, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef]
- Morris, D.A. The effect of temperature on the velocity of exogenous auxin transport in intact chilling-sensitive and chilling-resistant plants. Planta 1979, 146, 603–605. [Google Scholar] [CrossRef]
- Nadella, V.; Shipp, M.J.; Muday, G.K.; Wyatt, S.E. Evidence for altered polar and lateral auxin transport in the gravity persistent signal (gps) mutants of Arabidopsis. Plant Cell Environ. 2006, 29, 682–690. [Google Scholar] [CrossRef]
- Steinmann, T.; Geldner, N.; Grebe, M.; Mangold, S.; Jackson, C.L.; Paris, S.; Galweiler, L.; Palme, K.; Jurgens, G. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 1999, 286, 316–318. [Google Scholar] [CrossRef]
- Geldner, N.; Anders, N.; Wolters, H.; Keicher, J.; Kornberger, W.; Muller, P.; Delbarre, A.; Ueda, T.; Nakano, A.; Jurgens, G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 2003, 112, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, J.; Wang, L.; Wang, X.; Xue, Y.; Wu, P.; Shou, H. Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 2009, 19, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.A.; Rahman, A. Cold stress response in Arabidopsis thaliana is mediated by GNOM ARF-GEF. Plant J. 2019, 97, 500–516. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yang, X.; Zhang, Y.; Wang, H.; Wu, S.; Zhang, Z.; Ahammed, G.J.; Zhao, C.; Liu, H. CRISPR/Cas9-mediated mutation in auxin efflux carrier OsPIN9 confers chilling tolerance by modulating reactive oxygen species homeostasis in rice. Front. Plant Sci. 2022, 13, 967031. [Google Scholar] [CrossRef]
- Ouyang, Q.; Zhang, Y.; Yang, X.; Yang, C.; Hou, D.; Liu, H.; Xu, H. Overexpression of OsPIN9 impairs chilling tolerance via disturbing ROS homeostasis in rice. Plants 2023, 12, 2809. [Google Scholar] [CrossRef]
- Yang, C.; Wang, H.; Ouyang, Q.; Chen, G.; Fu, X.; Hou, D.; Xu, H. Deficiency of auxin efflux carrier OsPIN1b impairs chilling and drought tolerance in rice. Plants 2023, 12, 4058. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, Y.; Yang, X.; Wang, H.; Hou, D. Tissue specificity and responses to abiotic stresses and hormones of PIN genes in rice. Biologia 2022, 77, 1459–1470. [Google Scholar] [CrossRef]
- Wang, J.R.; Hu, H.; Wang, G.H.; Li, J.; Chen, J.Y.; Wu, P. Expression of PIN genes in rice (Oryza sativa L.): Tissue specificity and regulation by hormones. Mol. Plant 2009, 2, 823–831. [Google Scholar] [CrossRef]
- Miyashita, Y.; Takasugi, T.; Ito, Y. Identification and expression analysis of PIN genes in rice. Plant Sci. 2010, 178, 424–428. [Google Scholar] [CrossRef]
- Lu, G.; Coneva, V.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Nambara, E.; Bi, Y.M.; Rothstein, S.J. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015, 83, 913–925. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, T.T.; Xiao, D.W.; Zhao, S.M.; Lin, J.S.; Wang, T.; Li, Y.J.; Hou, B.K. OsIAGT1 is a glucosyltransferase gene involved in the glucose conjugation of auxins in rice. Rice 2019, 12, 92. [Google Scholar] [CrossRef] [PubMed]
- Sang, D.; Chen, D.; Liu, G.; Liang, Y.; Huang, L.; Meng, X.; Chu, J.; Sun, X.; Dong, G.; Yuan, Y.; et al. Strigolactones regulate rice tiller angle by attenuating shoot gravitropism through inhibiting auxin biosynthesis. Proc. Natl. Acad. Sci. USA 2014, 111, 11199–11204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Yu, H.; Yu, H.; Cai, Y.; Huang, L.; Xu, C.; Xiong, G.; Meng, X.; Wang, J.; Chen, H.; et al. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell 2018, 30, 1461–1475. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liang, Y.; Yuan, Y.D.; Wang, L.; Meng, X.B.; Xiong, G.S.; Zhou, J.; Cai, Y.Y.; Han, N.P.; Hua, L.K.; et al. OsBRXL4 regulates shoot gravitropism and rice tiller angle through affecting LAZY1 nuclear localization. Mol. Plant 2019, 12, 1143–1156. [Google Scholar] [CrossRef]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Fu, J.; Yu, H.; Li, X.; Xiao, J.; Wang, S. Rice GH3 gene family: Regulators of growth and development. Plant Signal Behav. 2011, 6, 570–574. [Google Scholar] [CrossRef]
- Du, H.; Wu, N.; Fu, J.; Wang, S.; Li, X.; Xiao, J.; Xiong, L. A GH3 family member, OsGH3-2, modulates auxin and abscisic acid levels and differentially affects drought and cold tolerance in rice. J. Exp. Bot. 2012, 63, 6467–6480. [Google Scholar] [CrossRef]
- Abraham, E.; Rigo, G.; Szekely, G.; Nagy, R.; Koncz, C.; Szabados, L. Light-dependent induction of proline biosynthesis by abscisic acid and salt stress is inhibited by brassinosteroid in Arabidopsis. Plant Mol. Biol. 2003, 51, 363–372. [Google Scholar] [CrossRef]
- Sun, S.J.; Guo, S.Q.; Yang, X.; Bao, Y.M.; Tang, H.J.; Sun, H.; Huang, J.; Zhang, H.S. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 2010, 61, 2807–2818. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, M.; Wu, C.; Chen, C.; Meng, L.; Zhang, G.; Zhuang, K.; Shi, Q. SlWRKY51 regulates proline content to enhance chilling tolerance in tomato. Plant Cell Environ. 2024, 47, 5104–5114. [Google Scholar] [CrossRef]
- Hu, C.H.; Wang, P.Q.; Zhang, P.P.; Nie, X.M.; Li, B.B.; Tai, L.; Liu, W.T.; Li, W.Q.; Chen, K.M. NADPH oxidases: The vital performers and center hubs during plant growth and signaling. Cells 2020, 9, 437. [Google Scholar] [CrossRef] [PubMed]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.Y.; Wang, H.Q.; Shi, W.; Zhang, W.W.; Zhao, F.J. The respiratory burst oxidase homologue OsRBOHE is crucial for root hair formation, drought resistance and tillering in rice. Plant Cell Environ. 2025, 48, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T.; et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 2007, 19, 4022–4034. [Google Scholar] [CrossRef]
- Keller, T.; Damude, H.G.; Werner, D.; Doerner, P.; Dixon, R.A.; Lamb, C. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell 1998, 10, 255–266. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yoshioka, M.; Fukazawa, A.; Mori, H.; Nishizawa, N.K.; Tsutsumi, N.; Yoshioka, H.; Nakazono, M. An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 2017, 29, 775–790. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Regulatory networks underlying plant responses and adaptation to cold stress. Annu. Rev. Genet. 2024, 58, 43–65. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Ashraf, M.A.; Cheng, Y.; Wang, B.R.; Qin, Y. Plant low-temperature stress: Signaling and response. Agronomy 2022, 12, 702. [Google Scholar] [CrossRef]
- Xu, M.; Zhu, L.; Shou, H.; Wu, P. A PIN1 family gene, OsPIN1, involved in auxin-dependent adventitious root emergence and tillering in rice. Plant Cell Physiol. 2005, 46, 1674–1681. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Wu, L.; Shao, Y.; Wu, Y.; Mao, C. Functional divergence of PIN1 paralogous genes in rice. Plant Cell Physiol. 2019, 60, 2720–2732. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guo, M.; Li, Y.; Ruan, W.; Mo, X.; Wu, Z.; Sturrock, C.J.; Yu, H.; Lu, C.; Peng, J.; et al. LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J. Exp. Bot. 2018, 69, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Inahashi, H.; Shelley, I.J.; Yamauchi, T.; Nishiuchi, S.; Takahashi-Nosaka, M.; Matsunami, M.; Ogawa, A.; Noda, Y.; Inukai, Y. OsPIN2, which encodes a member of the auxin efflux carrier proteins, is involved in root elongation growth and lateral root formation patterns via the regulation of auxin distribution in rice. Physiol. Plant 2018, 164, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Q.; Zhang, M.J.; Qiao, L.; Chen, Y.B.; Zhang, D.P.; Jing, X.Q.; Gan, P.F.; Huang, Y.B.; Gao, J.R.; Liu, W.T.; et al. Characterization of wavy root 1, an agravitropism allele, reveals the functions of OsPIN2 in fine regulation of auxin transport and distribution and in ABA biosynthesis and response in rice (Oryza sativa L.). Crop J. 2022, 10, 980–992. [Google Scholar] [CrossRef]
- Hao, B.; Zhang, R.; Zhang, C.; Wen, N.; Xia, Y.; Zhao, Y.; Li, Q.; Qiao, L.; Li, W. Characterization of OsPIN2 mutants reveal novel roles for reactive oxygen species in modulating not only root gravitropism but also hypoxia tolerance in rice seedlings. Plants 2024, 13, 476. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 with the CRISPR-Cas9 System. Front. Plant Sci. 2019, 10, 1663. [Google Scholar] [CrossRef]
- Han, X.L.; Zhao, F.Y. OsYUCCA2 deficiency in rice growth and development. Cienc. Rural. 2022, 52, e20210327. [Google Scholar] [CrossRef]
- Xu, X.Y.; Zhiguo, E.; Zhang, D.P.; Yun, Q.B.; Zhou, Y.; Niu, B.X.; Chen, C. OsYUC11-mediated auxin biosynthesis is essential for endosperm development of rice. Plant Physiol. 2021, 185, 934–950. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef]
- Hou, M.; Luo, F.; Wu, D.; Zhang, X.; Lou, M.; Shen, D.; Yan, M.; Mao, C.; Fan, X.; Xu, G.; et al. OsPIN9, an auxin efflux carrier, is required for the regulation of rice tiller bud outgrowth by ammonium. New Phytol. 2021, 229, 935–949. [Google Scholar] [CrossRef]
- Suzuki, N.; Koussevitzky, S.; Mittler, R.; Miller, G. ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 2012, 35, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Song, S.; Zhang, H.; Li, Y.; Niu, L.; Zhang, J.; Wang, W. Signaling transduction of ABA, ROS, and Ca2+ in plant stomatal closure in response to drought. Int. J. Mol. Sci. 2022, 23, 14824. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ullah, F.; Zhou, D.X.; Yi, M.; Zhao, Y. Mechanisms of ROS regulation of plant development and stress responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef]
- Sun, M.; Shen, Y.; Chen, Y.; Wang, Y.; Cai, X.; Yang, J.; Jia, B.; Dong, W.; Chen, X.; Sun, X. Osa-miR1320 targets the ERF transcription factor OsERF096 to regulate cold tolerance via JA-mediated signaling. Plant Physiol. 2022, 189, 2500–2516. [Google Scholar] [CrossRef]
- Ben Rejeb, K.; Abdelly, C.; Savouré, A. How reactive oxygen species and proline face stress together. Plant Physiol. Biochem. 2014, 80, 278–284. [Google Scholar] [CrossRef]
- Zhao, Z.F.; Sun, A.Q.; Shan, W.F.; Zheng, X.H.; Wang, Y.; Bai, L.; Xu, Y.C.; An, Z.; Wang, X.Y.; Wang, Y.M.; et al. OsRbohI is the indispensable NADPH oxidase for molecular-patterns-induced reactive oxygen species production in rice. Plant Commun. 2024, 5, 101129. [Google Scholar] [CrossRef]
- Xu, H.W.; Ji, X.M.; He, Z.H.; Shi, W.P.; Zhu, G.H.; Niu, J.K.; Li, B.S.; Peng, X.X. Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves. J. Exp. Bot. 2006, 57, 1899–1908. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Manna, M.; Rengasamy, B.; Sinha, A.K. A rapid and robust colorimetric method for measuring relative abundance of auxins in plant tissues. Phytochem. Anal. 2024, 35, 1052–1062. [Google Scholar] [CrossRef]
- Choi, H.W.; Kim, Y.J.; Lee, S.C.; Hong, J.K.; Hwang, B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef]
- Hu, Z.; Huang, X.; Amombo, E.; Liu, A.; Fan, J.; Bi, A.; Ji, K.; Xin, H.; Chen, L.; Fu, J. The ethylene responsive factor CdERF1 from bermudagrass (Cynodon dactylon) positively regulates cold tolerance. Plant Sci. 2020, 294, 110432. [Google Scholar] [CrossRef] [PubMed]
- Morris, D.L. Quantitative determination of carbohydrates with dreywood’s anthrone reagent. Science 1948, 107, 254–255. [Google Scholar] [CrossRef] [PubMed]
- Troll, W.; Lindsley, J. A photometric method for the determination of proline. J. Biol. Chem. 1955, 215, 655–660. [Google Scholar] [PubMed]
- Wu, F.B.; Zhang, G.P.; Dominy, P. Four barley genotypes respond differently to cadmium: Lipid peroxidation and activities of antioxidant capacity. Environ. Exp. Bot. 2003, 50, 67–78. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Chen, G.; Ruan, X.; Kang, G.; Hou, D.; Xu, H. Overexpression of OsPIN5b Alters Plant Architecture and Impairs Cold Tolerance in Rice (Oryza sativa L.). Plants 2025, 14, 1026. https://doi.org/10.3390/plants14071026
Fu X, Chen G, Ruan X, Kang G, Hou D, Xu H. Overexpression of OsPIN5b Alters Plant Architecture and Impairs Cold Tolerance in Rice (Oryza sativa L.). Plants. 2025; 14(7):1026. https://doi.org/10.3390/plants14071026
Chicago/Turabian StyleFu, Xiaoyu, Guo Chen, Xinya Ruan, Guozhang Kang, Dianyun Hou, and Huawei Xu. 2025. "Overexpression of OsPIN5b Alters Plant Architecture and Impairs Cold Tolerance in Rice (Oryza sativa L.)" Plants 14, no. 7: 1026. https://doi.org/10.3390/plants14071026
APA StyleFu, X., Chen, G., Ruan, X., Kang, G., Hou, D., & Xu, H. (2025). Overexpression of OsPIN5b Alters Plant Architecture and Impairs Cold Tolerance in Rice (Oryza sativa L.). Plants, 14(7), 1026. https://doi.org/10.3390/plants14071026







