CsPHYB–CsPIF3/4 Regulates Hypocotyl Elongation by Coordinating the Auxin and Gibberellin Biosynthetic Pathways in Cucumber (Cucumis sativus L.)
Abstract
1. Introduction
2. Results
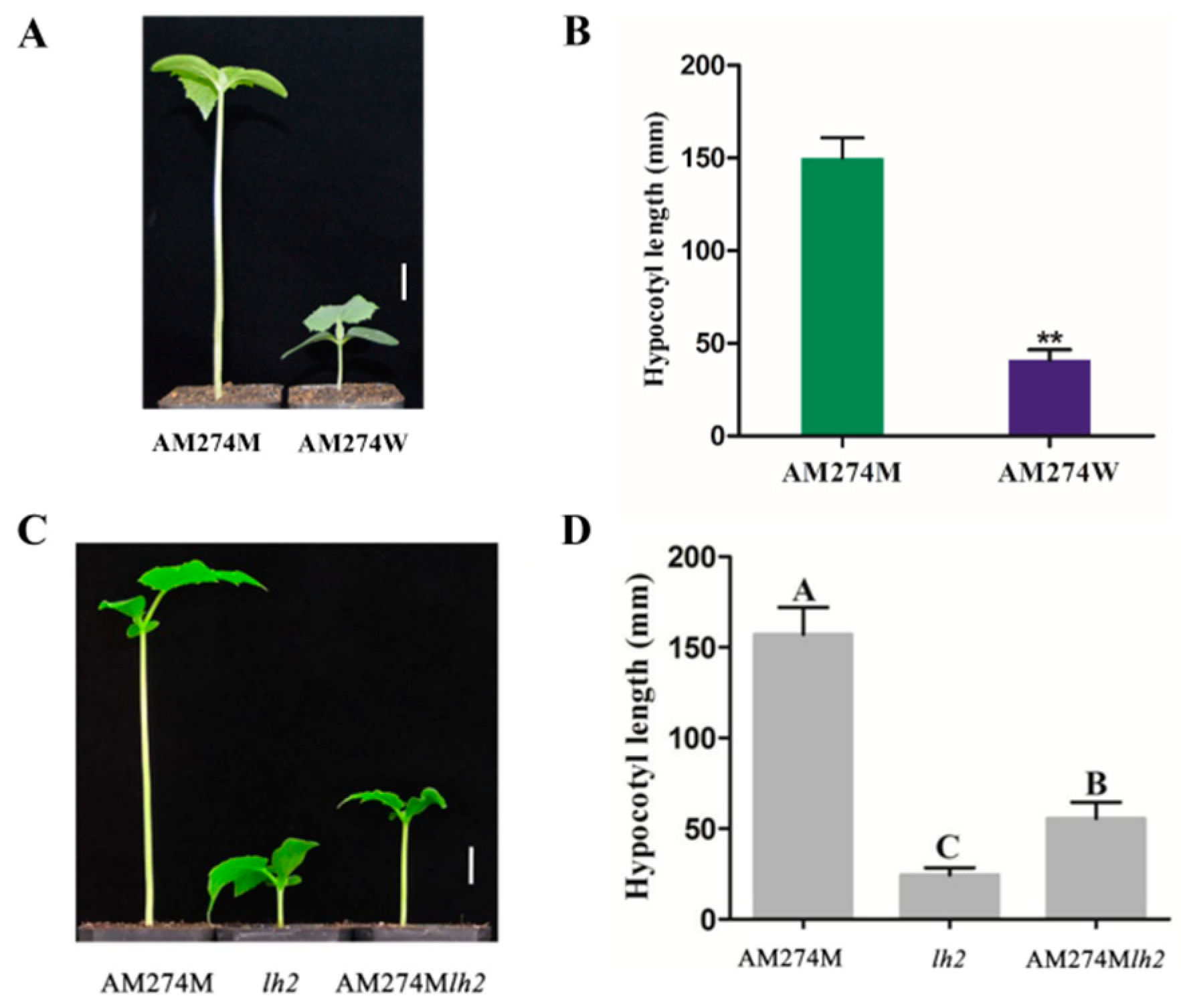
2.1. Phenotypic Characterization of Mutant AM274M and Double-Mutant AM274Mlh2
2.2. GA and IAA Content in Mutant AM274M
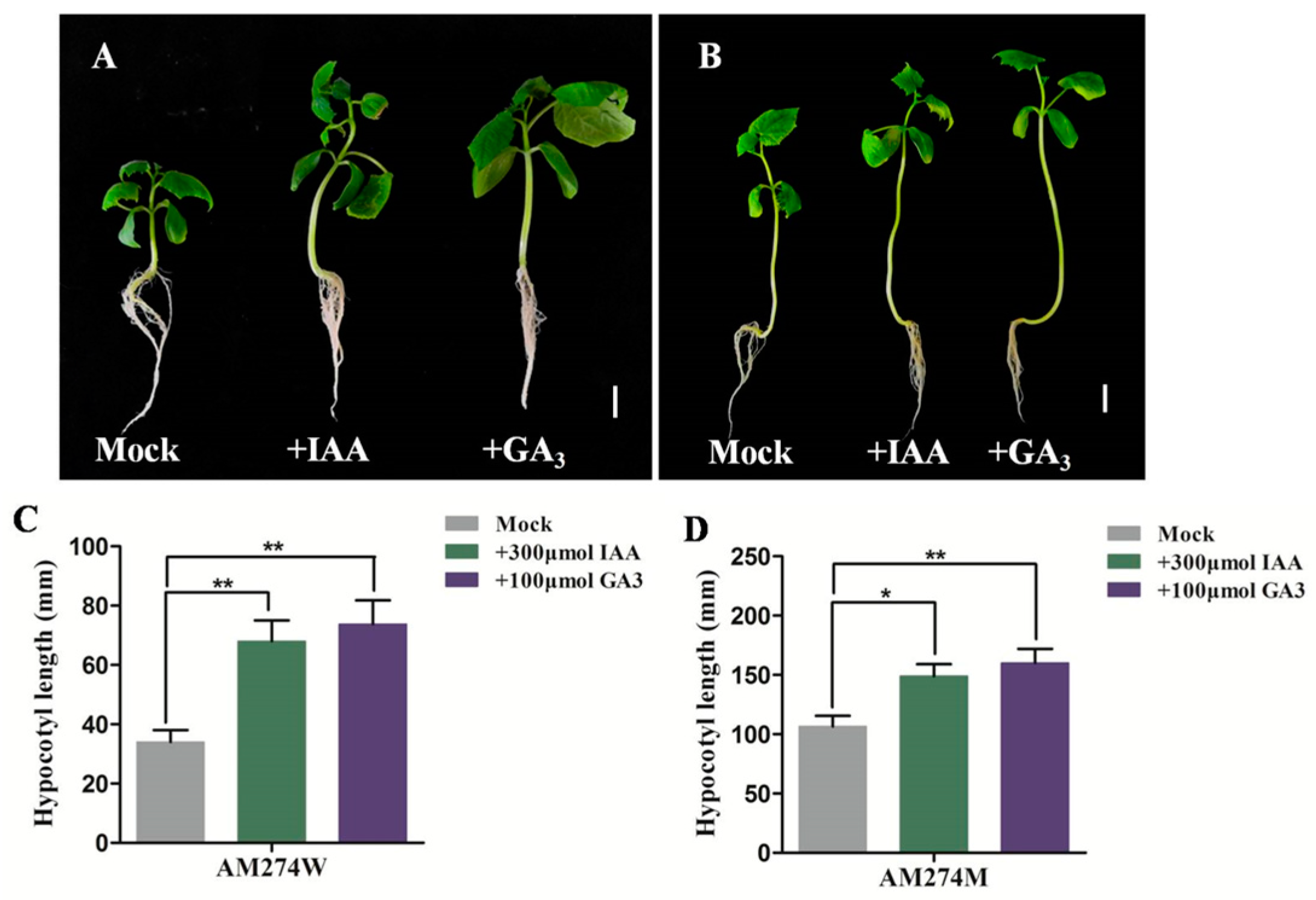
2.3. Exogenous GA and IAA Promote Hypocotyl Elongation
2.4. Comparative Transcriptome Profiling Analysis
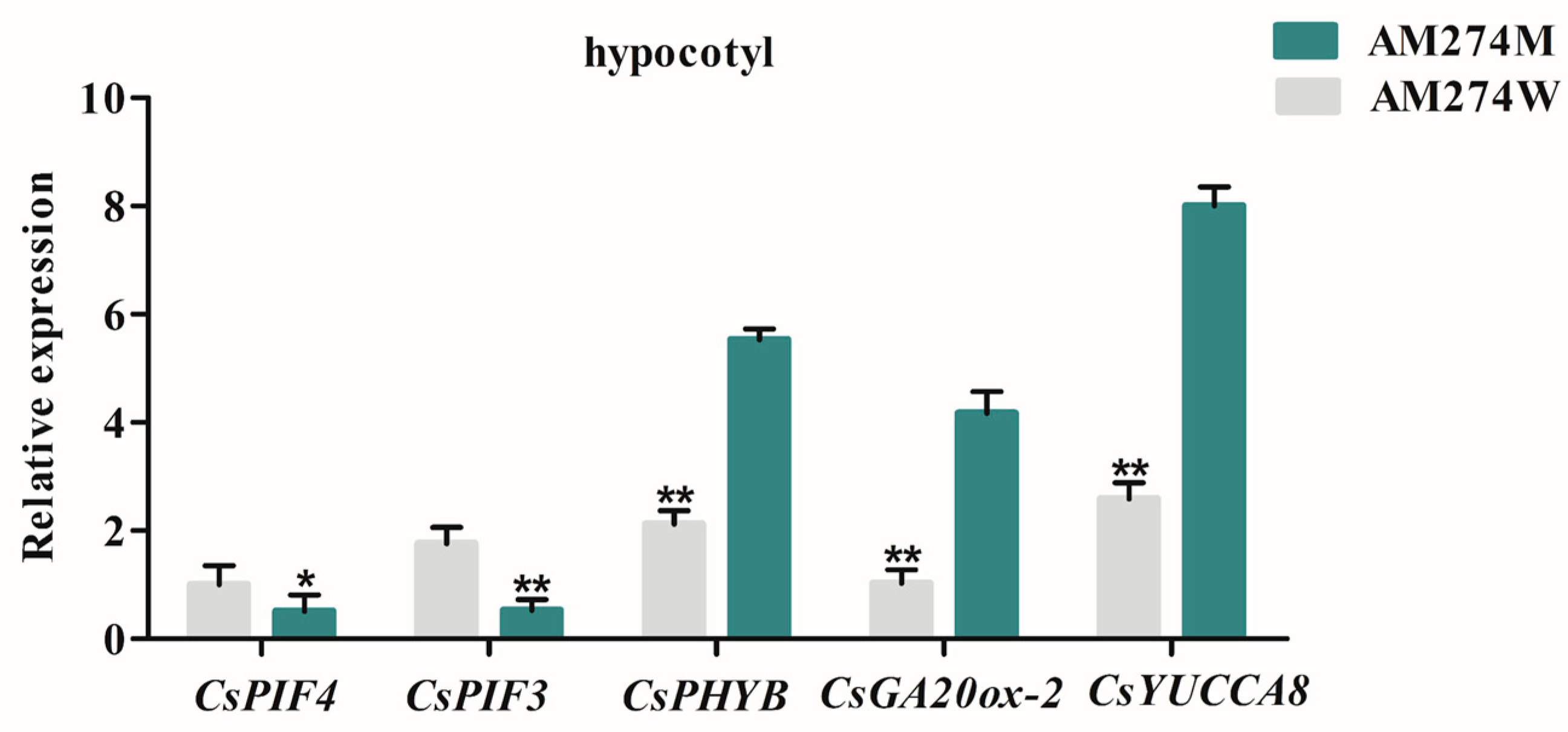
2.5. Expression Analysis of CsGA20ox-2 and CsYUCCA8 Genes
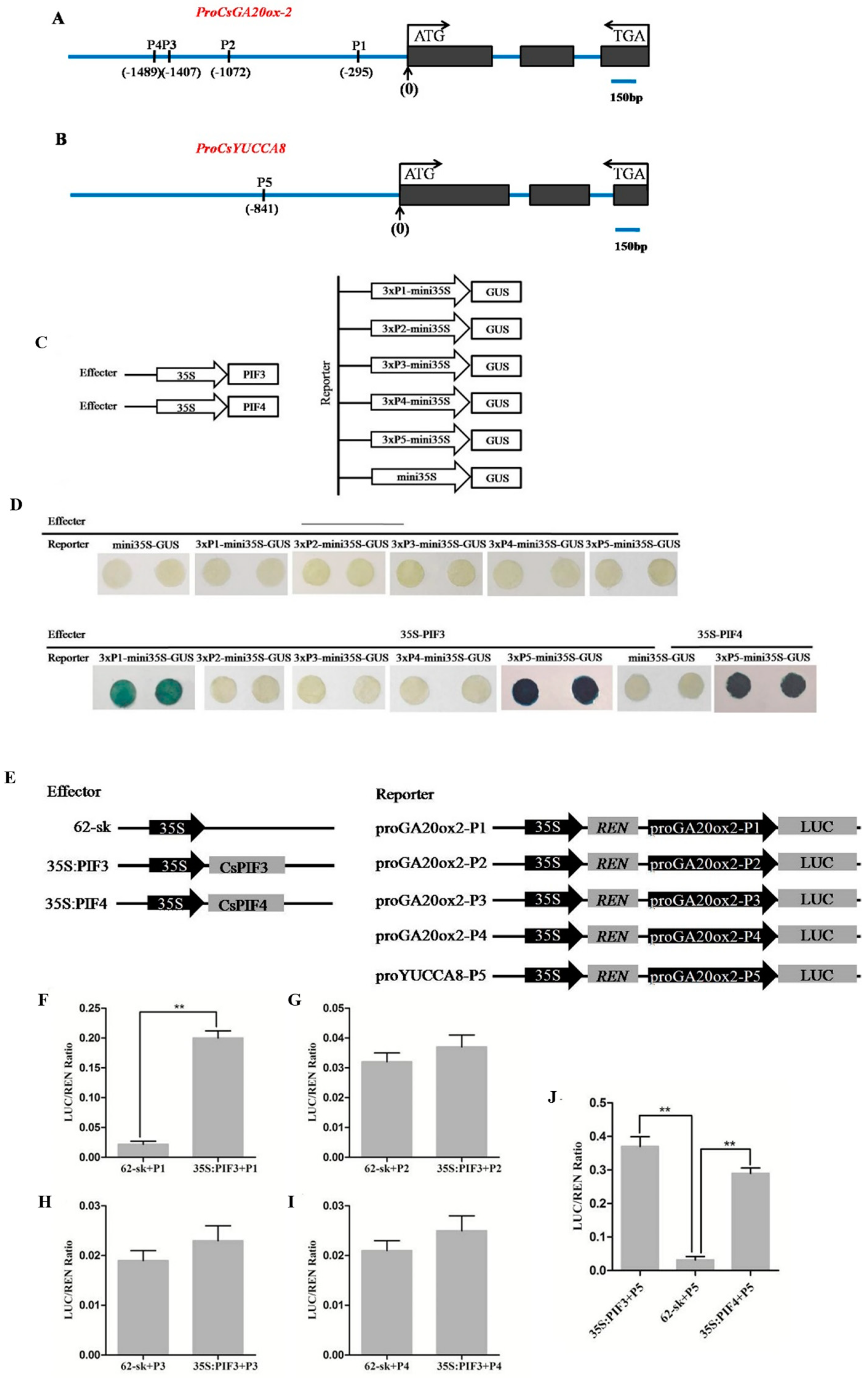
2.6. CsPIF3/4 Can Bind to the E(G)-Box of the CsGA20ox-2 and CsYUCCA8 Promoters
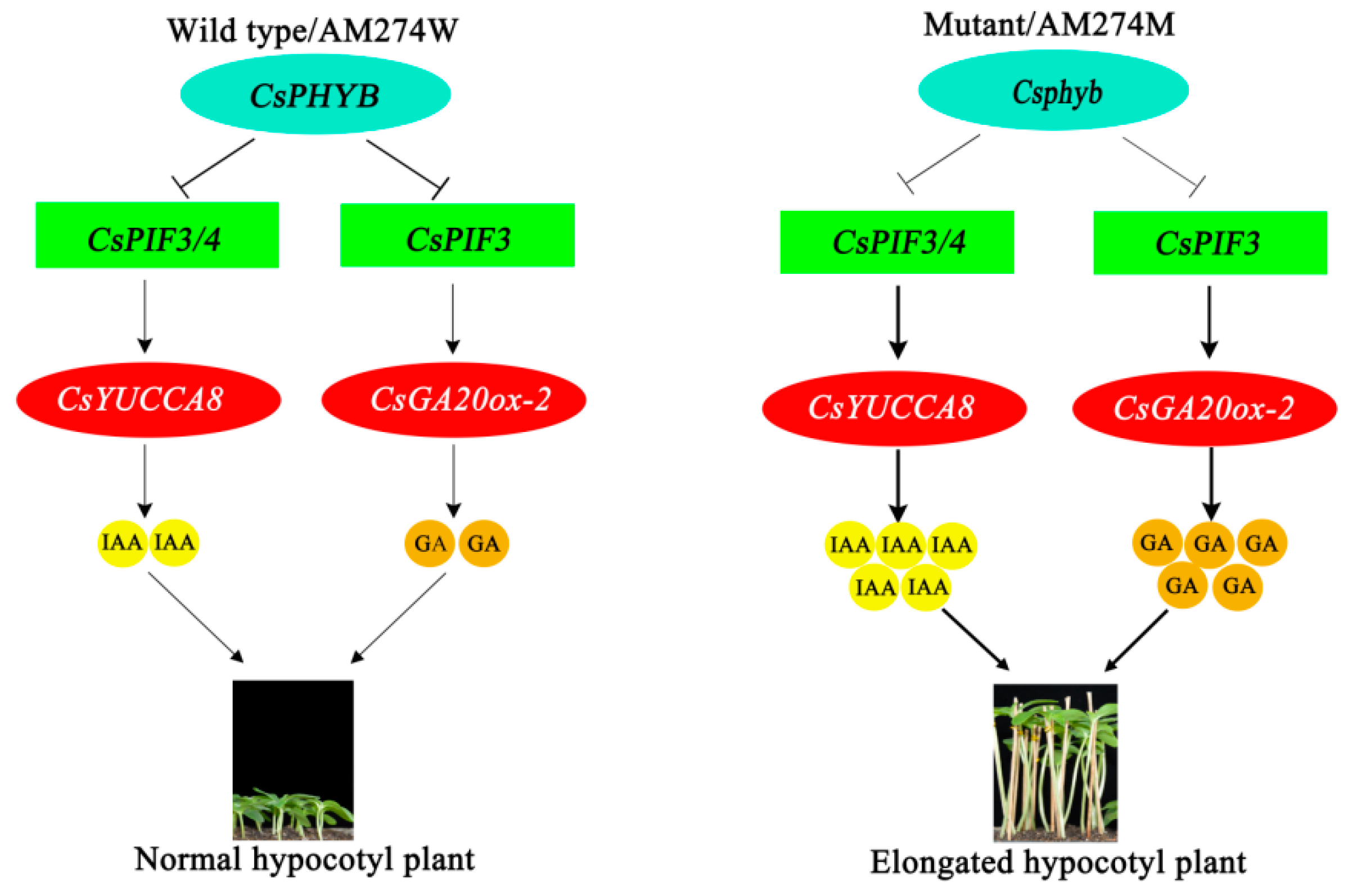
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Extraction and Quantification of Endogenous Hormones
4.3. Exogenous Hormone Treatment
4.4. RNA-Seq Analysis
4.5. qRT-PCR Analysis
4.6. Promoter Cis-Acting Element Prediction of CsGA20ox-2 and CsYUCCA8
4.7. GUS Staining Analysis and Dual-Luciferase Reporter Assay
4.8. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nakano, T. Hypocotyl elongation: A molecular mechanism for the first event in plant growth that infuences its physiology. Plant Cell Physiol. 2019, 60, 933–934. [Google Scholar] [CrossRef]
- Nie, J.; Jiang, Y.; Lv, L.; Shi, Y.; Chen, P.; Zhang, Q.; Sui, X. The bHLH transcription factor CsPIF4 positively regulates high temperature-induced hypocotyl elongation in cucumber. Hortic. Plant. J. 2024, 10, 1187–1197. [Google Scholar] [CrossRef]
- Xu, F.; He, S.; Zhang, J.; Mao, Z.; Wang, W.; Li, T.; Hua, J.; Du, S.; Xu, P.; Li, L.; et al. Photoactivated CRY1 and phyB Interact Directly with AUX/IAA Proteins to Inhibit Auxin Signaling in Arabidopsis. Mol. Plant 2018, 11, 523–541. [Google Scholar] [CrossRef]
- Paik, I.; Chen, F.; Ngoc, V.; Zhu, L.; Kim, J.I.; Huq, E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat. Commun. 2019, 10, 4216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, R.; Zeng, X.Y.; Lee, S.; Ye, L.H.; Tian, S.L.; Zhang, Y.J.; Busch, W.; Zhou, W.B.; Zhu, X.G.; et al. GLK transcription factors accompany ELONGATED HYPOCOTYL5 to orchestrate light-induced seedling development in Arabidopsis. Plant Physiol. 2024, 194, 2400–2421. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, W.; Zhao, D.; Song, Y.; Lin, X.; Shen, M.; Chi, C.; Xu, B.; Zhao, J.; Deng, X.W.; et al. Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor. Cell 2024, 187, 6235–6250. [Google Scholar] [CrossRef]
- Park, E.; Park, J.; Kim, J.; Nagatani, A.; Lagarias, J.C.; Choi, G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012, 72, 537–546. [Google Scholar] [CrossRef]
- Sadanandom, A.; Ádám, É.; Orosa, B.; Viczián, A.; Klose, C.; Zhang, C.; Josse, E.M.; Kozma-Bognár, L.; Nagy, F. SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 11108–11113. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell. Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef] [PubMed]
- Job, N.; Datta, S. PIF3/HY5 module regulates BBX11 to suppress protochlorophyllide levels in dark and promote photomorphogenesis in light. New Phytol. 2021, 230, 190–204. [Google Scholar] [CrossRef]
- Wei, X.; Wang, W.; Xu, P.; Wang, W.; Guo, T.; Kou, S.; Liu, M.; Niu, Y.; Yang, H.Q.; Mao, Z. Phytochrome B interacts with SWC6 and ARP6 to regulate H2A.Z deposition and photomorphogensis in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 1133–1146. [Google Scholar] [CrossRef]
- Won, J.H.; Park, J.; Lee, H.G.; Shim, S.; Lee, H.; Oh, E.; Seo, P.J. The PRR-EC complex and SWR1 chromatin remodeling complex function cooperatively to repress nighttime hypocotyl elongation by modulating PIF4 expression in Arabidopsis. Plant Commun. 2024, 5, 100981. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Jiang, Y.; Li, J.; Yan, T.; Fan, L.; Liang, J.; Chen, Z.J.; Xu, D.; Deng, X.W. B-BOX DOMAIN PROTEIN28 Negatively Regulates Photomorphogenesis by Repressing the Activity of Transcription Factor HY5 and Undergoes COP1-Mediated Degradation. Plant Cell 2018, 30, 2006–2019. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Hayashi, K.; Kinoshita, T. Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiol. 2012, 159, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhou, X.; Tang, W.; Takahashi, K.; Pan, X.; Dai, J.; Ren, H.; Zhu, X.; Pan, S.; Zheng, H.; et al. TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature 2021, 599, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Che, G.; Ding, L.; Chen, Z.; Liu, X.; Wang, H.; Zhao, W.; Ning, K.; Zhao, J.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef]
- Effendi, Y.; Jones, A.M.; Scherer, G.F. AUXIN-BINDING-PROTEIN1 (ABP1) in phytochrome-B-controlled responses. J. Exp. Bot. 2013, 64, 5065–5074. [Google Scholar] [CrossRef] [PubMed]
- De-Wit, M.; Keuskamp, D.H.; Bongers, F.J.; Hornitschek, P.; Gommers, C.M.M.; Reinen, E.; Martínez-Cerón, C.; Fankhauser, C.; Pierik, R. Integration of Phytochrome and Cryptochrome Signals Determines Plant Growth during Competition for Light. Curr. Biol. 2016, 26, 3320–3326. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liu, K.; Ma, S.; Su, H.; Yang, J.; Sun, L.; Liu, Z.; Qin, Y. Gibberellin and cytokinin signaling antagonistically control female-germline cell specification in Arabidopsis. Dev. Cell 2024. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Xu, R.R.; Wang, X.N.; Zhang, X.W.; You, C.X.; Han, Y. MdbHLH162 connects the gibberellin and jasmonic acid signals to regulate anthocyanin biosynthesis in apple. J. Integr. Plant Biol. 2024, 66, 265–284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, L.; Wu, Y.; Wang, D.; He, X.Q. Gibberellin promotes cambium reestablishment during secondary vascular tissue regeneration after girdling in an auxin-dependent manner in Populus. J. Integr. Plant Biol. 2024, 66, 86–102. [Google Scholar] [CrossRef]
- Ramon, U.; Weiss, D.; Illouz-Eliaz, N. Underground gibberellin activity: Differential gibberellin response in tomato shoots and roots. New Phytol. 2021, 229, 1196–1200. [Google Scholar] [CrossRef]
- Sun, J.; Zheng, Y.; Guo, J.; Zhang, Y.; Liu, Y.; Tao, Y.; Wang, M.; Liu, T.; Liu, Y.; Li, X.; et al. GmGAMYB-BINDING PROTEIN 1 promotes small auxin-up RNA gene transcription to modulate soybean maturity and height. Plant Physiol. 2023, 193, 775–791. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Zhang, M.; Shang, J.; Liu, Z.; Weng, Y.; Yue, H.; Li, Y.; Chen, P. A 5.5-kb LTR-retrotransposon insertion Inside phytochrome B gene (CsPHYB) results in long hypocotyl and early fowering in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2023, 136, 68. [Google Scholar] [CrossRef]
- Jiang, H.; Shui, Z.; Xu, L.; Yang, Y.; Li, Y.; Yuan, X.; Shang, J.; Asghar, M.A.; Wu, X.; Yu, L.; et al. Gibberellins modulate shade-induced soybean hypocotyl elongation downstream of the mutual promotion of auxin and brassinosteroids. Plant Physiol. Biochem. 2020, 150, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Liu, J.; He, G.; Liu, P.; Sun, J. Photoexcited phytochrome B interacts with brassinazole resistant 1 to repress brassinosteroid signaling in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 652–667. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhao, S.; Li, P.; Yin, Y.; Niu, Q.; Yan, J.; Huang, D. Plant buffering against the high-light stress-induced accumulation of CsGA2ox8 transcripts via alternative splicing to finely tune gibberellin levels and maintain hypocotyl elongation. Hortic. Res. 2021, 8, 2. [Google Scholar] [CrossRef]
- Seo, D.H.; Yoon, G.M. Light-induced stabilization of ACS contributes to hypocotyl elongation during the dark-to-light transition in Arabidopsis seedlings. Plant J. 2019, 98, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Lorrai, R.; Boccaccini, A.; Ruta, V.; Possenti, M.; Costantino, P.; Vittorioso, P. Abscisic acid inhibits hypocotyl elongation acting on gibberellins, DELLA proteins and auxin. AoB Plants 2018, 10, 61. [Google Scholar]
- Zhao, J.; Bo, K.; Pan, Y.; Li, Y.; Yu, D.; Li, C.; Chang, J.; Wu, S.; Wang, Z.; Zhang, X.; et al. Phytochrome-interacting factor PIF3 integrates phytochrome B and UVB signaling pathways to regulate gibberellin- and auxin-dependent growth in cucumber hypocotyls. J. Exp. Bot. 2023, 74, 4520–4539. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Rolff, E.; Spruit, C.J.P. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 1980, 100, 147–160. [Google Scholar] [CrossRef]
- Reed, J.W.; Nagpal, P.; Poole, D.S.; Furuya, M.; Chory, J. Mutations in the gene for red/far-red light receptor phytochrome B alter cell elongtion and physiological responses throught Arabidopsis development. Plant Cell 1993, 5, 147–157. [Google Scholar] [PubMed]
- Franklin, K.A.; Quail, P.H. Phytochrome functions in Arabidopsis development. J. Exp. Bot. 2010, 61, 11–24. [Google Scholar] [CrossRef]
- Yoshida, Y.; Sarmiento, M.R.; Yamori, W.; Ponce, M.R.; Micol, J.L.; Tsukaya, H. The Arabidopsis phyB-9 mutant has a second-site mutation in the VENOSA4 gene that alters chloroplast size, photosynthetic traits, and leaf growth. Plant Physiol. 2018, 178, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Sheerin, D.J.; Menon, C.; Oven-Krockhaus, S.Z.; Enderle, B.; Zhu, L.; Johnen, P.; Schleifenbaum, F.; Stierhof, Y.D.; Huq, E.; Hiltbrunner, A. Light-activated phytochrome A and B interact with memebers of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell 2015, 27, 189–201. [Google Scholar] [CrossRef]
- Xu, P.B.; Lian, H.L.; Xu, F.; Zhang, T.; Wang, S.; Wang, W.X.; Du, S.S.; Huang, J.R.; Yang, H.Q. Phytochrome B and AGB1 coordinately regulate photomorphogenesis by antagonistically modulating PIF3 stability in Arabidopsis. Mol. Plant 2019, 12, 229–247. [Google Scholar] [CrossRef] [PubMed]
- Heng, Y.Q.; Jiang, Y.; Zhao, X.H.; Zhou, H.; Wang, X.C.; Deng, X.W.; Xu, D.Q. BBX4, a phyB- interacting and modulated regulator, directly interacts with PIF3 to fine tune red light-mediated photomorphogenesis. Proc. Natl. Acad. Sci. USA 2019, 116, 26049–26056. [Google Scholar] [CrossRef]
- Mao, Z.L.; He, S.B.; Xu, F.; Wei, X.X.; Jiang, L.; Wang, W.X.; Li, T.; Xu, P.B.; Du, S.S.; Li, L.; et al. Photoexcited CRY1 and phyB interact directly with ARF6 and ARF8 to regulate their DNA-binding activity and auxin-induced hypocotyl elongation in Arabidopsis. New Phytol. 2020, 225, 848–865. [Google Scholar] [CrossRef]
- Yan, Y.; Li, C.; Dong, X.J.; Li, H.; Zhang, D.; Zhou, Y.Y.; Jiang, B.C.; Peng, J.; Qin, X.Y.; Cheng, J.K. MYB30 is a key negative regulator of Arabidopsis photomorphogenic development that promotes PIF4 and PIF5 protein accumulation in the light. Plant Cell 2020, 32, 2196–2215. [Google Scholar] [CrossRef]
- Cowling, R.J.; Harberd, N.P. Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J. Exp. Bot. 1999, 50, 1351–1357. [Google Scholar] [CrossRef]
- Fernández, M.G.L.; Ballaré, C.L. Shade avoidance: Expanding the color and hormone palette. Trends Plant Sci. 2021, 26, 509–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Liu, Z.; Wang, Y.; Mu, S.; Yue, H.; Luo, Y.; Zhang, Z.; Li, Y.; Chen, P. A mutation in CsDWF7 gene encoding a delta7 sterol C-5(6) desaturase leads to the phenotype of super compact in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2024, 137, 20. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Hu, L.; Liu, P.; Jin, Z.; Sun, J.; Weng, Y.; Chen, P.; Du, S.; Wei, A.; Li, Y. A mutation in CsHY2 encoding a phytochromobilin (PΦB) synthase leads to an elongated hypocotyl 1(elh1) phenotype in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2021, 134, 2639–2652. [Google Scholar] [CrossRef]
- Li, X.Y. Infltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-Protocol 2011, 1, e95. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Qiu, Z.; Dong, J.; Bu, R.; Zhou, Y.; Wang, H.; Hu, L. CsPHYB–CsPIF3/4 Regulates Hypocotyl Elongation by Coordinating the Auxin and Gibberellin Biosynthetic Pathways in Cucumber (Cucumis sativus L.). Plants 2025, 14, 371. https://doi.org/10.3390/plants14030371
Chen L, Qiu Z, Dong J, Bu R, Zhou Y, Wang H, Hu L. CsPHYB–CsPIF3/4 Regulates Hypocotyl Elongation by Coordinating the Auxin and Gibberellin Biosynthetic Pathways in Cucumber (Cucumis sativus L.). Plants. 2025; 14(3):371. https://doi.org/10.3390/plants14030371
Chicago/Turabian StyleChen, Liqin, Zongqing Qiu, Jing Dong, Runhua Bu, Yu Zhou, Huilin Wang, and Liangliang Hu. 2025. "CsPHYB–CsPIF3/4 Regulates Hypocotyl Elongation by Coordinating the Auxin and Gibberellin Biosynthetic Pathways in Cucumber (Cucumis sativus L.)" Plants 14, no. 3: 371. https://doi.org/10.3390/plants14030371
APA StyleChen, L., Qiu, Z., Dong, J., Bu, R., Zhou, Y., Wang, H., & Hu, L. (2025). CsPHYB–CsPIF3/4 Regulates Hypocotyl Elongation by Coordinating the Auxin and Gibberellin Biosynthetic Pathways in Cucumber (Cucumis sativus L.). Plants, 14(3), 371. https://doi.org/10.3390/plants14030371





