Transcriptomics and Metabolomics Reveal Biosynthetic Pathways and Regulatory Mechanisms of Phenylpropanes in Different Ploidy of Capsicum frutescens
Abstract
1. Introduction
2. Results
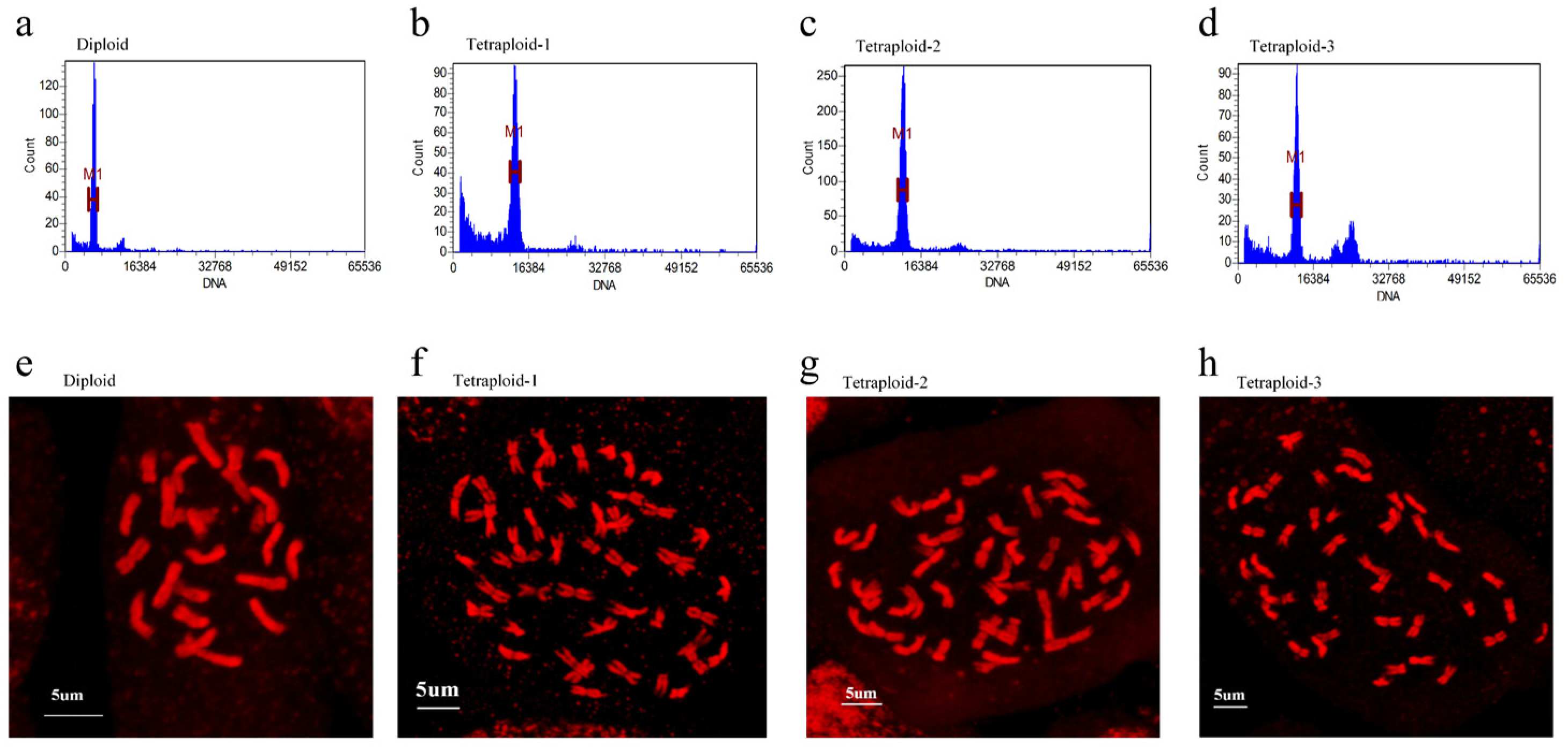
2.1. Identification of Different Ploidy of C. frutescens and Morphological Observations
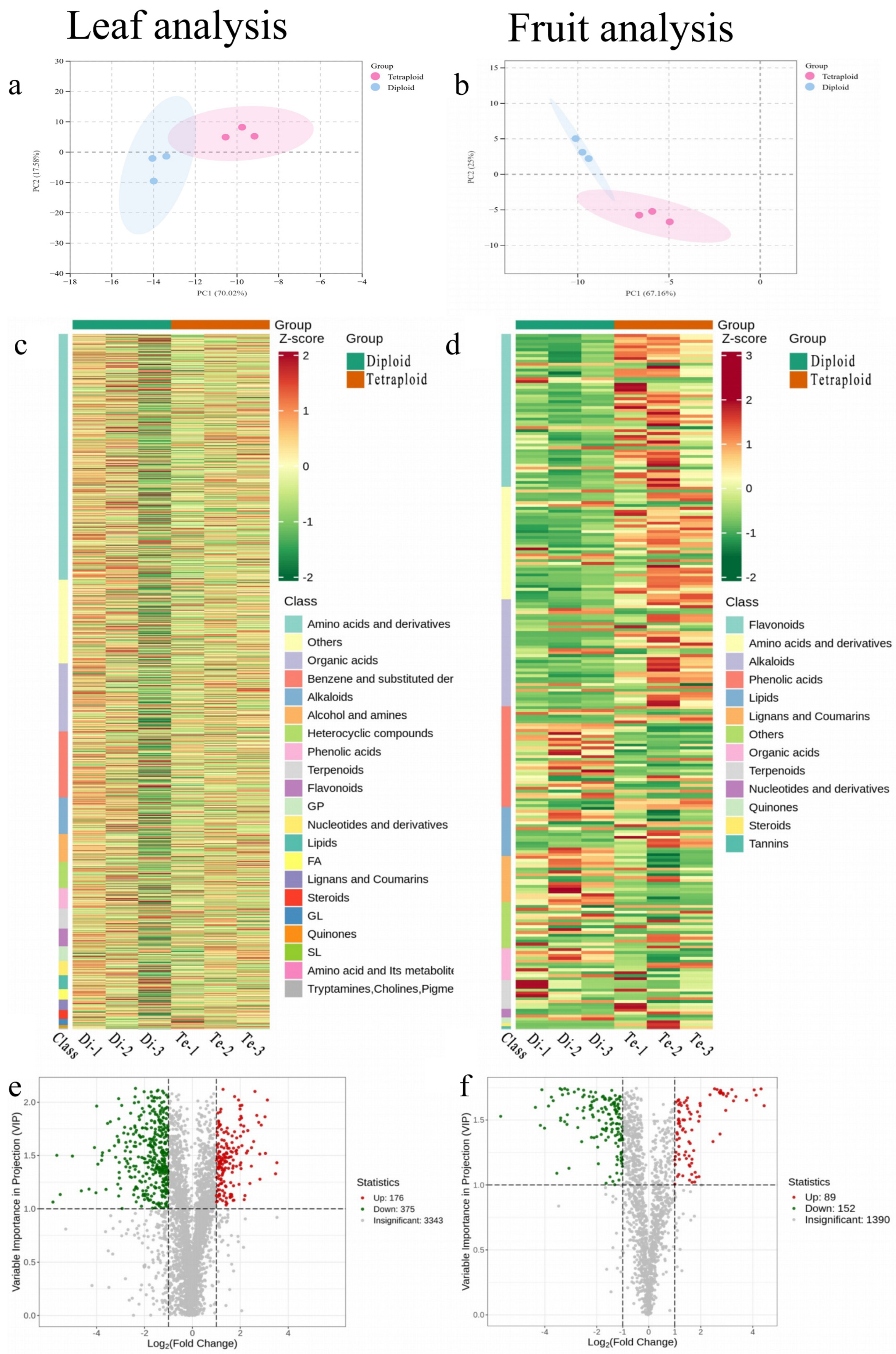
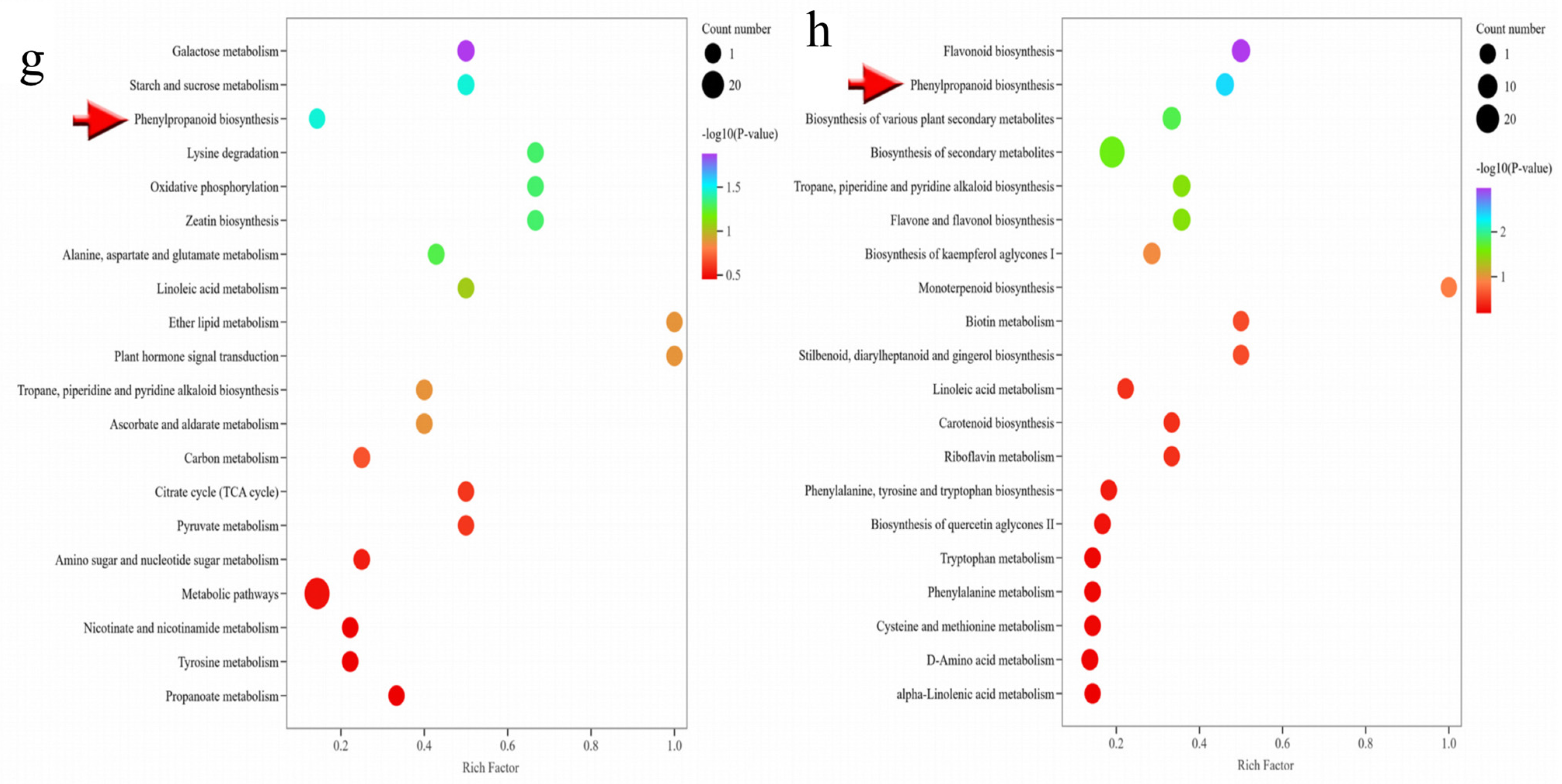
2.2. Metabolome Data Analysis of C. frutescens with Different Ploidy Levels
2.3. Transcriptome Data Analysis of C. frutescens
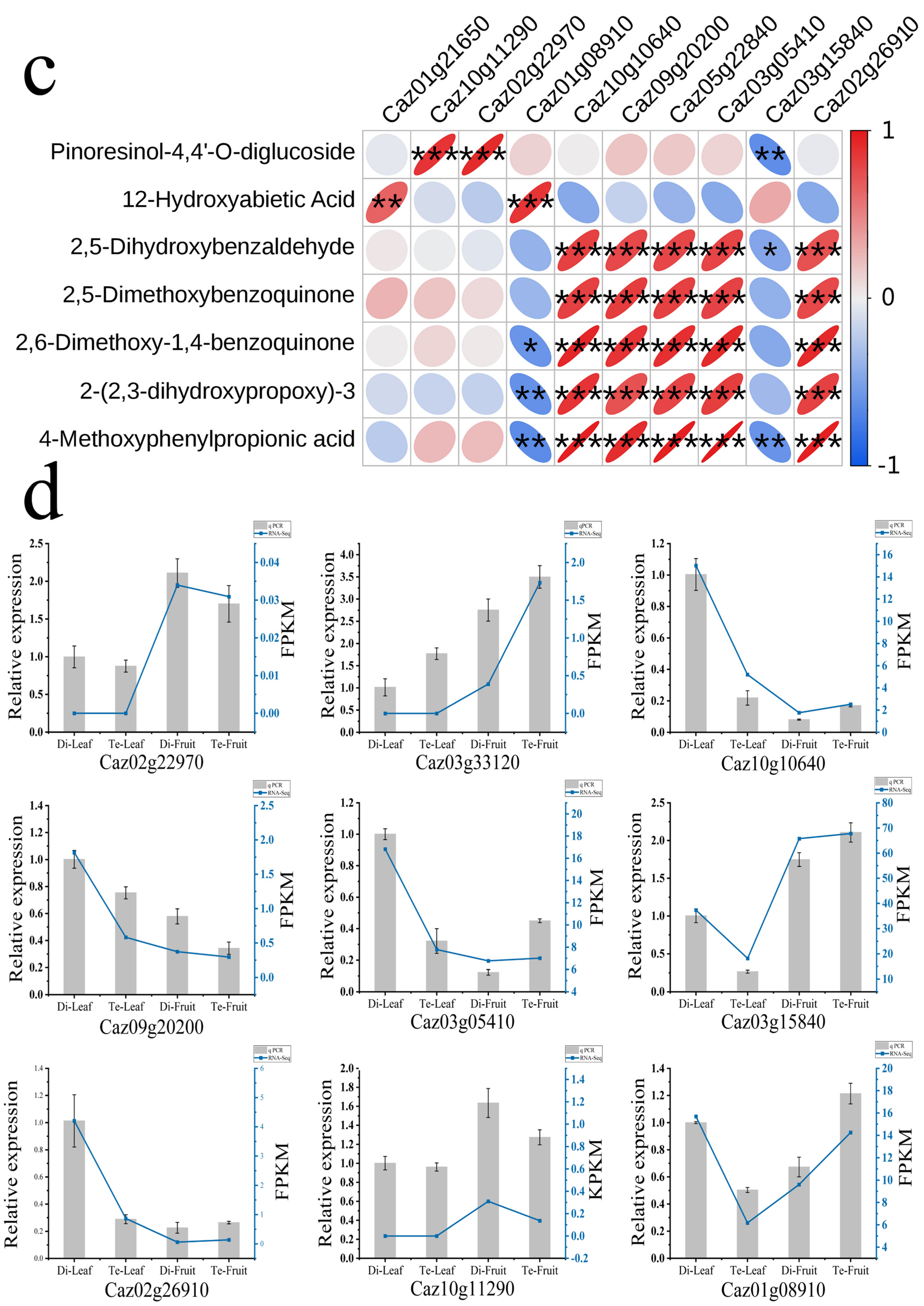
2.4. Conjoint Transcriptome–Metabolome Analysis
3. Discussion
4. Materials and Methods
4.1. C. frutescens Material, Growth Condition, and Treatment
4.2. Sample Preparation and Extraction
4.3. Differential Metabolites Selected
4.4. KEGG Annotation and Enrichment Analysis
4.5. Transcriptome Sequencing and Data Analysis
4.6. Quantitative Analysis of qRT-PCR
4.7. Scanning Electron Microscope Analysis
4.8. Paraffin Section Analysis
4.9. Chromosome Preparation Analysis
4.10. Flow Cytometer Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wood, T.E.; Takebayashi, N.; Barker, M.S.; Mayrose, I.; Greenspoon, P.B.; Rieseberg, L.H. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 2009, 106, 13875–13879. [Google Scholar] [CrossRef] [PubMed]
- Aïnouche, M.L.; Fortune, P.; Salmon, A.; Parisod, C.; Grandbastien, M.-A.; Fukunaga, K.; Ricou, M.; Misset, M.-T. Hybridization, polyploidy and invasion: Lessons from Spartina (Poaceae). Biol. Invasions 2009, 11, 1159–1173. [Google Scholar] [CrossRef]
- Fawcett, J.A.; Maere, S.; Van De Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. French-Italian Public Consortium for Grapevine Genome Characterization The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef] [PubMed]
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Fasano, C.; Diretto, G.; Aversano, R.; D’Agostino, N.; Di Matteo, A.; Frusciante, L.; Giuliano, G.; Carputo, D. Transcriptome and metabolome of synthetic Solanum autotetraploids reveal key genomic stress events following polyploidization. New Phytol. 2016, 210, 1382–1394. [Google Scholar] [CrossRef]
- Tate, J.A.; Joshi, P.; Soltis, K.A.; Soltis, P.S.; Soltis, D.E. On the road to diploidization? Homoeolog loss in independently formed populations of the allopolyploid Tragopogon miscellus (Asteraceae). BMC Plant Biol. 2009, 9, 80. [Google Scholar] [CrossRef]
- Tate, J.A.; Ni, Z.; Scheen, A.-C.; Koh, J.; Gilbert, C.A.; Lefkowitz, D.; Chen, Z.J.; Soltis, P.S.; Soltis, D.E. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 2006, 173, 1599–1611. [Google Scholar] [CrossRef]
- Lim, K.Y.; Soltis, D.E.; Soltis, P.S.; Tate, J.; Matyasek, R.; Srubarova, H.; Kovarik, A.; Pires, J.C.; Xiong, Z.; Leitch, A.R. Rapid chromosome evolution in recently formed polyploids in Tragopogon (Asteraceae). PLoS ONE 2008, 3, e3353. [Google Scholar] [CrossRef]
- Buggs, R.; Doust, A.; Tate, J.; Koh, J.; Soltis, K.; Feltus, F.; Paterson, A.; Soltis, P.; Soltis, D. Gene loss and silencing in Tragopogon miscellus (Asteraceae): Comparison of natural and synthetic allotetraploids. Heredity 2009, 103, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Koh, J.; Chen, S.; Zhu, N.; Yu, F.; Soltis, P.S.; Soltis, D.E. Comparative proteomics of the recently and recurrently formed natural allopolyploid Triticum aestivum (Asteraceae) and its parents. New Phytol. 2012, 196, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Sehrish, T.; Symonds, V.V.; Soltis, D.E.; Soltis, P.S.; Tate, J.A. Gene silencing via DNA methylation in naturally occurring Tragopogon miscellus (Asteraceae) allopolyploids. BMC Genom. 2014, 15, 701. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dobešová, E.; Malinská, H.; Matyášek, R.; Leitch, A.; Soltis, D.; Soltis, P.; Kovařík, A. Silenced rRNA genes are activated and substitute for partially eliminated active homeologs in the recently formed allotetraploid, Triticum aestivum (Asteraceae). Heredity 2015, 114, 356–365. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pumphrey, M.; Bai, J.; Laudencia-Chingcuanco, D.; Anderson, O.; Gill, B.S. Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics 2009, 181, 1147–1157. [Google Scholar] [CrossRef]
- Akhunova, A.R.; Matniyazov, R.T.; Liang, H.; Akhunov, E.D. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genom. 2010, 11, 505. [Google Scholar] [CrossRef]
- Chagué, V.; Just, J.; Mestiri, I.; Balzergue, S.; Tanguy, A.M.; Huneau, C.; Huteau, V.; Belcram, H.; Coriton, O.; Jahier, J. Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 2010, 187, 1181–1194. [Google Scholar] [CrossRef]
- Qi, B.; Huang, W.; Zhu, B.; Zhong, X.; Guo, J.; Zhao, N.; Xu, C.; Zhang, H.; Pang, J.; Han, F. Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol. 2012, 10, 3. [Google Scholar] [CrossRef]
- Li, A.; Liu, D.; Wu, J.; Zhao, X.; Hao, M.; Geng, S.; Yan, J.; Jiang, X.; Zhang, L.; Wu, J. mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 2014, 26, 1878–1900. [Google Scholar] [CrossRef]
- Pignatta, D.; Dilkes, B.P.; Yoo, S.Y.; Henry, I.M.; Madlung, A.; Doerge, R.W.; Jeffrey Chen, Z.; Comai, L. Differential sensitivity of the Arabidopsis thaliana transcriptome and enhancers to the effects of genome doubling. New Phytol. 2010, 186, 194–206. [Google Scholar] [CrossRef]
- Riddle, N.C.; Jiang, H.; An, L.; Doerge, R.; Birchler, J.A. Gene expression analysis at the intersection of ploidy and hybridity in maize. Theor. Appl. Genet. 2010, 120, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Tadeo, F.; Froelicher, Y.; Talon, M.; Navarro, L.; Ollitrault, P.; Morillon, R. Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J. Exp. Bot. 2011, 62, 2507–2519. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.-Q.; Tu, H.; Liang, W.-J.; Long, J.-M.; Wu, X.-M.; Zhang, H.-Y.; Guo, W.-W. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 2015, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Stupar, R.M.; Bhaskar, P.B.; Yandell, B.S.; Rensink, W.A.; Hart, A.L.; Ouyang, S.; Veilleux, R.E.; Busse, J.S.; Erhardt, R.J.; Buell, C.R.; et al. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics 2007, 176, 2055–2067. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Haberer, G.; Matthes, M.; Rattei, T.; Mayer, K.F.; Gierl, A.; Torres-Ruiz, R.A. Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 17809–17814. [Google Scholar] [CrossRef]
- Tossi, V.E.; Martínez Tosar, L.J.; Laino, L.E.; Iannicelli, J.; Regalado, J.J.; Escandón, A.S.; Baroli, I.; Causin, H.F.; Pitta-Álvarez, S.I. Impact of polyploidy on plant tolerance to abiotic and biotic stresses. Front. Plant Sci. 2022, 13, 869423. [Google Scholar] [CrossRef]
- Mishra, B.; Pathak, S.; Sharma, A.; Trivedi, P.; Shukla, S. Modulated gene expression in newly synthesized auto-tetraploid of Papaver somniferum L. S. Afr. J. Bot. 2010, 76, 447–452. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant–environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef]
- Koeduka, T.; Sugimoto, K.; Watanabe, B.; Someya, N.; Kawanishi, D.; Gotoh, T.; Ozawa, R.; Takabayashi, J.; Matsui, K.; Hiratake, J. Bioactivity of natural O-prenylated phenylpropenes from I llicium anisatum leaves and their derivatives against spider mites and fungal pathogens. Plant Biol. 2014, 16, 451–456. [Google Scholar] [CrossRef]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Zheng, K.; Cai, Y.; Qu, Y.; Teng, L.; Wang, C.; Gao, J.; Chen, Q. Effect of the HCT gene on lignin synthesis and fiber development in Gossypium barbadense. Plant Sci. 2024, 338, 111914. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.G.; Muchero, W. Regulation of Lignin Biosynthesis and Its Role in Growth-Defense Tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef] [PubMed]
- Cesarino, I. Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr. Opin. Biotechnol. 2019, 56, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; He, Y.; Kabahuma, M.; Chaya, T.; Kelly, A.; Borrego, E.; Bian, Y.; El Kasmi, F.; Yang, L.; Teixeira, P.; et al. A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat. Genet. 2017, 49, 1364–1372. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; He, Q.; Zhang, F.; Yu, J.; Li, C.; Zhao, T.; Zhang, Y.; Xie, Q.; Su, B.; Mei, L.; et al. Melatonin enhances cotton immunity to Verticillium wilt via manipulating lignin and gossypol biosynthesis. Plant J. 2019, 100, 784–800. [Google Scholar] [CrossRef]
- Silva, F.L.B.; Vieira, L.G.E.; Ribas, A.F.; Moro, A.L.; Neris, D.M.; Pacheco, A.C. Proline accumulation induces the production of total phenolics in transgenic tobacco plants under water deficit without increasing the G6PDH activity. Theor. Exp. Plant Physiol. 2018, 30, 251–260. [Google Scholar] [CrossRef]
- Liu, W.; Jiang, Y.; Wang, C.; Zhao, L.; Jin, Y.; Xing, Q.; Li, M.; Lv, T.; Qi, H. Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance. Plant Mol. Biol. 2020, 103, 689–704. [Google Scholar] [CrossRef]
- Domon, J.M.; Baldwin, L.; Acket, S.; Caudeville, E.; Arnoult, S.; Zub, H.; Gillet, F.; Lejeune-Hénaut, I.; Brancourt-Hulmel, M.; Pelloux, J.; et al. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry 2013, 85, 51–61. [Google Scholar] [CrossRef]
- Zhao, H.; Liu, H.; Jin, J.; Ma, X.; Li, K. Physiological and Transcriptome Analysis on Diploid and Polyploid Populus ussuriensis Kom. under Salt Stress. Int. J. Mol. Sci. 2022, 23, 7529. [Google Scholar] [CrossRef]
- Gray, J.; Caparrós-Ruiz, D.; Grotewold, E. Grass phenylpropanoids: Regulate before using! Plant Sci. 2012, 184, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q. Lignification: Flexibility, biosynthesis and regulation. Trends Plant Sci. 2016, 21, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Kraft, K.H.; Brown, C.H.; Nabhan, G.P.; Luedeling, E.; Luna Ruiz, J.d.J.; Coppens d’Eeckenbrugge, G.; Hijmans, R.J.; Gepts, P. Multiple lines of evidence for the origin of domesticated chili pepper, Capsicum annuum, in Mexico. Proc. Natl. Acad. Sci. USA 2014, 111, 6165–6170. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef] [PubMed]
- Feldman, M.; Levy, A. Allopolyploidy—A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- Osborn, T.C.; Pires, J.C.; Birchler, J.A.; Auger, D.L.; Chen, Z.J.; Lee, H.-S.; Comai, L.; Madlung, A.; Doerge, R.; Colot, V. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef]
- Xu, C.; Bai, Y.; Lin, X.; Zhao, N.; Hu, L.; Gong, Z.; Wendel, J.F.; Liu, B. Genome-wide disruption of gene expression in allopolyploids but not hybrids of rice subspecies. Mol. Biol. Evol. 2014, 31, 1066–1076. [Google Scholar] [CrossRef]
- Bertrand, B.; Bardil, A.; Baraille, H.; Dussert, S.; Doulbeau, S.; Dubois, E.; Severac, D.; Dereeper, A.; Etienne, H. The greater phenotypic homeostasis of the allopolyploid Coffea arabica improved the transcriptional homeostasis over that of both diploid parents. Plant Cell Physiol. 2015, 56, 2035–2051. [Google Scholar] [CrossRef]
- Tamayo-Ordóñez, M.; Rodriguez-Zapata, L.; Narváez-Zapata, J.; Tamayo-Ordóñez, Y.; Ayil-Gutiérrez, B.; Barredo-Pool, F.; Sánchez-Teyer, L. Morphological features of different polyploids for adaptation and molecular characterization of CC-NBS-LRR and LEA gene families in Agave L. J. Plant Physiol. 2016, 195, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.J.; Liu, W.G. Progress on salt resistance in autopolyploid plants. Yi Chuan= Hered. 2018, 40, 315–326. [Google Scholar]
- Tsukaya, H. Does ploidy level directly control cell size? Counterevidence from Arabidopsis genetics. PLoS ONE 2013, 8, e83729. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.; Kareem, A.; Deng, Z. In vivo induction and characterization of polyploids in gerbera daisy. Sci. Hortic. 2021, 282, 110054. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, L.; Yang, B.; Cui, Q.; Dai, Y.; Li, X.; Chen, Y.; Dai, X.; Zou, X.; Ou, L.; et al. A multi-omics approach identifies bHLH71-like as a positive regulator of yellowing leaf pepper mutants exposed to high-intensity light. Hortic. Res. 2023, 10, uhad098. [Google Scholar] [CrossRef]
- Rawoof, A.; Ahmad, I.; Islam, K.; Momo, J.; Kumar, A.; Jaiswal, V.; Ramchiary, N. Integrated omics analysis identified genes and their splice variants involved in fruit development and metabolites production in Capsicum species. Funct. Integr. Genom. 2022, 22, 1189–1209. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, S.T.; Nemati, F. Identification of GUCA2A and COL3A1 as prognostic biomarkers in colorectal cancer by integrating analysis of RNA-Seq data and qRT-PCR validation. Sci. Rep. 2023, 13, 17086. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Zhu, Y.; Wang, Y.; Wang, T.; Zhao, S.; Feng, K.; Li, L.; Wu, P. Molecular identification of phenylalanine ammonia lyase-encoding genes EfPALs and EfPAL2-interacting transcription factors in Euryale ferox. Front. Plant Sci. 2023, 14, 1114345. [Google Scholar] [CrossRef]
- Balao, F.; Herrera, J.; Talavera, S. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: A multivariate morphological approach. New Phytol. 2011, 192, 256–265. [Google Scholar] [CrossRef]
- Dai, F.; Wang, Z.; Luo, G.; Tang, C. Phenotypic and Transcriptomic Analyses of Autotetraploid and Diploid Mulberry (Morus alba L.). Int. J. Mol. Sci. 2015, 16, 22938–22956. [Google Scholar] [CrossRef]
- Shi, X.; Qi, Y.; Lin, L.; Wang, J.; Qin, X.; Niu, B. Comparative Analysis of the Transcriptome and Metabolome in Leaves of Diploid and Tetraploid Fagopyrum tataricum. Phyton 2023, 92, 3149–3162. [Google Scholar] [CrossRef]
- Liao, T.; Cheng, S.; Zhu, X.; Min, Y.; Kang, X. Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees 2016, 30, 1137–1147. [Google Scholar] [CrossRef]
- Allario, T.; Brumos, J.; COLMENERO-FLORES, J.M.; Iglesias, D.J.; Pina, J.A.; Navarro, L.; Talon, M.; Ollitrault, P.; Morillon, R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 2013, 36, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.C.; RAMIREZ-PARRA, E. Deciphering the molecular bases for drought tolerance in A rabidopsis autotetraploids. Plant Cell Environ. 2014, 37, 2722–2737. [Google Scholar] [CrossRef]
- You, Q.; Yi, X.; Zhang, K.; Wang, C.; Ma, X.; Zhang, X.; Xu, W.; Li, F.; Su, Z. Genome-wide comparative analysis of H3K4me3 profiles between diploid and allotetraploid cotton to refine genome annotation. Sci. Rep. 2017, 7, 9098. [Google Scholar] [CrossRef]
- Zhu, W.; Hu, B.; Becker, C.; Doğan, E.S.; Berendzen, K.W.; Weigel, D.; Liu, C. Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 2017, 18, 157. [Google Scholar] [CrossRef]
- Ding, M.; Chen, Z.J. Epigenetic perspectives on the evolution and domestication of polyploid plant and crops. Curr. Opin. Plant Biol. 2018, 42, 37–48. [Google Scholar] [CrossRef]
- Zou, L.; Liu, W.; Zhang, Z.; Edwards, E.J.; Gathunga, E.K.; Fan, P.; Duan, W.; Li, S.; Liang, Z. Gene body demethylation increases expression and is associated with self-pruning during grape genome duplication. Hortic. Res. 2020, 7, 84. [Google Scholar] [CrossRef]
- Dolezel, J.; Bartos, J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]



| Sampling Place | Sampling Time | Clean Reads | Q30 (%) | Reads Mapped |
|---|---|---|---|---|
| The third pair of true leaves of diploid | 50 d after seeding | 7.25–8.04 G | ≥95 | 95.37–95.74% |
| The third pair of true leaves of tetraploid | 50 d after seeding | 7.27–7.98 G | ≥95 | 94.85–95.88% |
| Diploid mature fruit | 80 d after flowering | 8.38–10.3 G | ≥95 | 96.75–96.91% |
| Tetraploid mature fruit | 80 d after flowering | 7.38–10.23 G | ≥95 | 96.71–96.88% |
| Gene ID | Gene Name | Forward Primer (5′->3′) | Reverse Primer (5′->3′) |
|---|---|---|---|
| Caz10g11290 | COMT 1 | AAGCCCCACAAATTCCTCGTAT | TAGGTACAACTGGCTCGCAA |
| Caz02g22970 | E1.11.1.7 1 | GCATCTTTGCTTCTGGATAATAGCA | TTGGTCCACCGGACAGAACA |
| Caz03g33120 | E2.1.1.104 2 | GAAAGGAGGCTCTGCGGTT | CCGCTGAGCAGAATCCATACA |
| Caz01g08910 | CCR 2 | CCTTCTTCACCTCAAAGTTTGGAA | AGGTCATTGATCGGTGGCTG |
| Caz10g10640 | PAL 1 | CTGAGGATGCAAGAGCTGGT | AAACTCCTGCATTCAAGAATCTAAT |
| Caz09g20200 | PAL 2 | TGCAGCTTTCGAGGACGAAT | AGTTCCAAGTTCCTTCCTCACA |
| Caz05g22840 | PAL 3 | AGGAAGACAGAGGAGGCACT | GGCTGCTAAACTCACTGCAC |
| Caz03g05410 | 4CL 1 | CACACTGGCGATATGGGGTT | TGCTCGTCTTTCATTGGGAC |
| Caz03g15840 | 4CL 2 | TCAAAGGTTTCCAGGTGCCA | ACTTCCCCTGCAGCATCATC |
| Caz06g27840 | UBI3 | GTCCATCTGCTCTCTGTTG | CACCCCAAGCACAATAAGAC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Cai, Q.; Yang, Y.; Wang, X.; Li, L.; Sun, Z.; Li, W. Transcriptomics and Metabolomics Reveal Biosynthetic Pathways and Regulatory Mechanisms of Phenylpropanes in Different Ploidy of Capsicum frutescens. Plants 2024, 13, 3393. https://doi.org/10.3390/plants13233393
Yang Y, Cai Q, Yang Y, Wang X, Li L, Sun Z, Li W. Transcriptomics and Metabolomics Reveal Biosynthetic Pathways and Regulatory Mechanisms of Phenylpropanes in Different Ploidy of Capsicum frutescens. Plants. 2024; 13(23):3393. https://doi.org/10.3390/plants13233393
Chicago/Turabian StyleYang, Yinxin, Qihang Cai, Yanbo Yang, Xuan Wang, Liping Li, Zhenghai Sun, and Weiwei Li. 2024. "Transcriptomics and Metabolomics Reveal Biosynthetic Pathways and Regulatory Mechanisms of Phenylpropanes in Different Ploidy of Capsicum frutescens" Plants 13, no. 23: 3393. https://doi.org/10.3390/plants13233393
APA StyleYang, Y., Cai, Q., Yang, Y., Wang, X., Li, L., Sun, Z., & Li, W. (2024). Transcriptomics and Metabolomics Reveal Biosynthetic Pathways and Regulatory Mechanisms of Phenylpropanes in Different Ploidy of Capsicum frutescens. Plants, 13(23), 3393. https://doi.org/10.3390/plants13233393





