Genome-Wide Identification and Characterization of SNPs and InDels of Capsicum annuum var. glabriusculum from Mexico Based on Whole Genome Sequencing
Abstract
1. Introduction
2. Results
2.1. Preprocessing Reads and Mapping of CHMX_Ch1 and QO to the Reference Genome
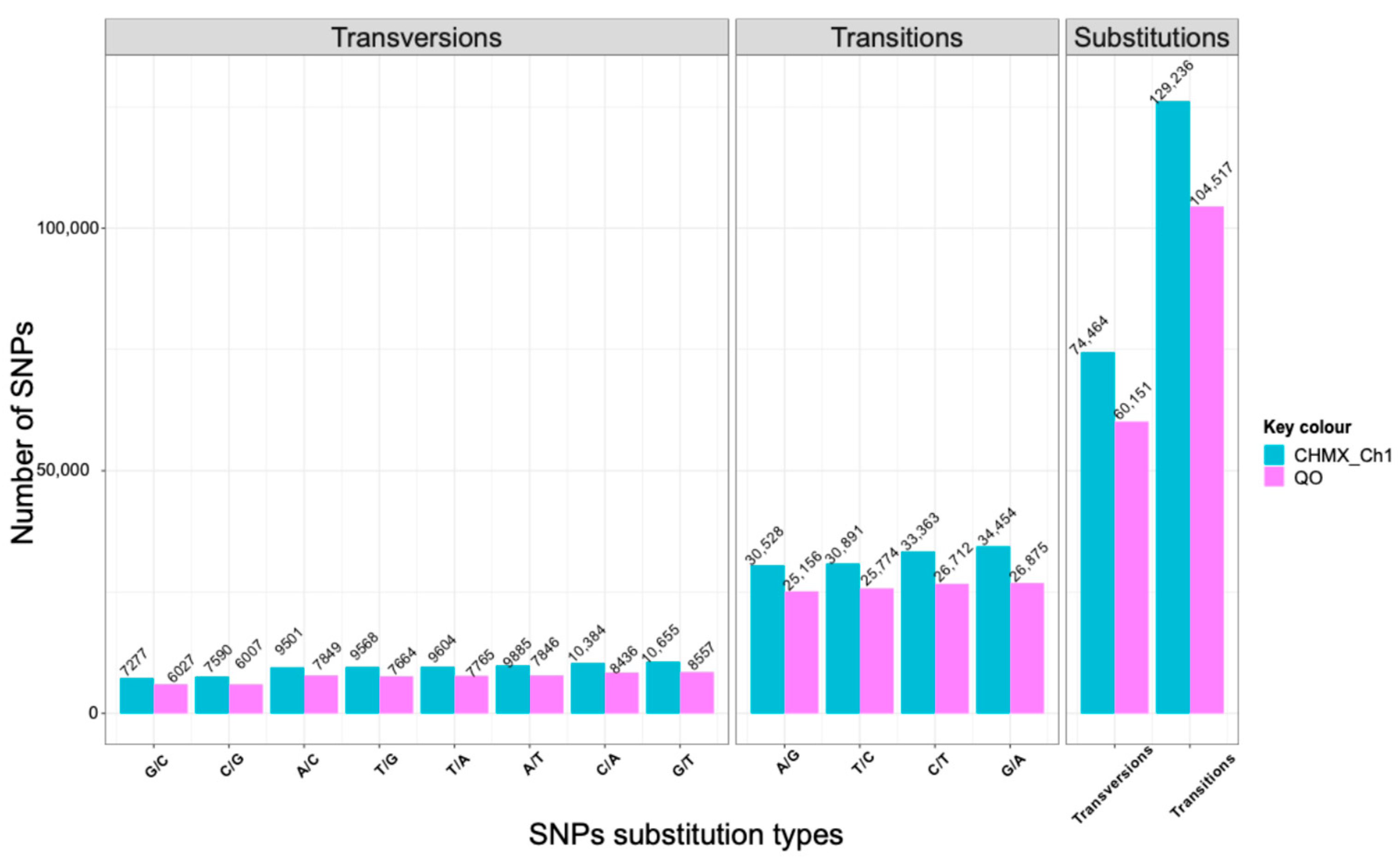
2.2. Genome-Wide Identification and Characterization of SNPs and InDels
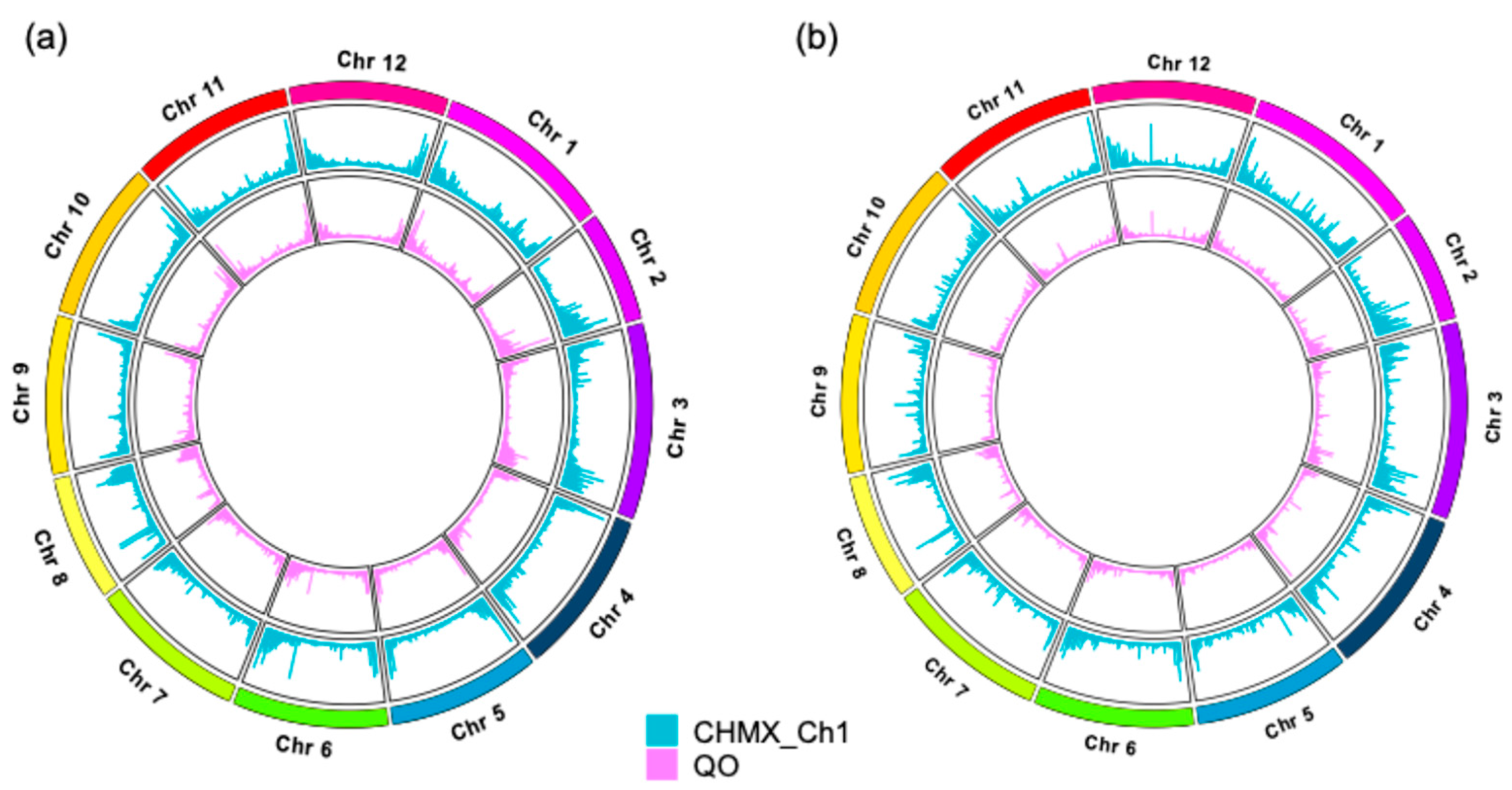
2.3. Distribution of SNPs and InDels
2.4. Effects of Mutations
2.5. Gene Ontology Analysis and Annotation of High- and Moderate-Impact Genetic Variants
3. Discussion
4. Materials and Methods
4.1. Plant Material and DNA Extraction
4.2. Library Preparation and Whole Genome Sequencing
4.3. SNP/InDel Calling and Variant Annotation
4.4. Gene Ontology Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bie, M.; Han, C.; Wang, X.; Xiao, W.; Song, K. Characterization and phylogenetic relationships analysis of the complete chloroplast genome of Capsicum annuum (Solanaceae). Mitochondrial DNA Part B 2020, 5, 570–571. [Google Scholar] [CrossRef] [PubMed]
- Acquadro, A.; Barchi, L.; Portis, E.; Nourdine, M.; Carli, C.; Monge, S.; Valentino, D.; Lanteri, S. Whole genome resequencing of four Italian sweet pepper landraces provides insights on sequence variation in genes of agronomic value. Sci. Rep. 2020, 10, 9189. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.K.; Tripathi, S.; Cho, Y., II; Kim, J.H.; Lee, H.E.; Kim, D.S.; Eta, L. Next generation transcriptomic sequencing and polymorphism detection in pepper varieties Saengryeg 211 and Saengryeg 213. Crop Sci. 2014, 54, 1690–1697. [Google Scholar] [CrossRef]
- Ahn, Y.K.; Manivannan, A.; Karna, S.; Jun, T.H.; Yang, E.Y.; Choi, S.; Kim, J.H.; Kim, D.S.; Lee, E.S. Whole Genome Resequencing of Capsicum baccatum and Capsicum annuum to Discover Single Nucleotide Polymorphism Related to Powdery Mildew Resistance. Sci. Rep. 2018, 8, 5188. [Google Scholar] [CrossRef] [PubMed]
- Barchi, L.; Pietrella, M.; Venturini, L.; Minio, A.; Toppino, L.; Acquadro, A.; Andolfo, G.; Aprea, G.; Avanzato, C.; Bassolino, L.; et al. A chromosome-anchored eggplant genome sequence reveals key events in Solanaceae evolution. Sci. Rep. 2019, 9, 11769. [Google Scholar] [CrossRef]
- Kim, S.; Park, M.; Yeom, S.I.; Kim, Y.M.; Lee, J.M.; Lee, H.A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef]
- Li, W.; Cheng, J.; Wu, Z.; Qin, C.; Tan, S.; Tang, X.; Cui, J.; Zhang, L.; Hu, K. An InDel-based linkage map of hot pepper (Capsicum annuum). Mol. Breed. 2015, 35, 32. [Google Scholar] [CrossRef]
- Liu, S.; Li, W.; Wu, Y.; Chen, C.; Lei, J. De Novo Transcriptome Assembly in Chili Pepper (Capsicum frutescens) to Identify Genes Involved in the Biosynthesis of Capsaicinoids. PLoS ONE 2013, 8, e48156. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef]
- Ashrafi, H.; Hill, T.; Stoffel, K.; Kozik, A.; Yao, J.; Chin-Wo, S.R.; Van Deynze, A. De novo assembly of the pepper transcriptome (Capsicum annuum): A benchmark for in silico discovery of SNPs, SSRs and candidate genes. BMC Genom. 2012, 13, 571. [Google Scholar] [CrossRef]
- Ahn, Y.K.; Tripathi, S.; Cho, Y.I.; Kim, J.H.; Lee, H.E.; Kim, D.S.; Woo, J.G.; Cho, M.C. De novo transcriptome assembly and novel microsatellite marker information in Capsicum annuum varieties Saengryeg 211 and Saengryeg 213. Bot. Stud. 2013, 54, 58. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhao, Z.; Li, B.; Qin, C.; Wu, Z.; Trejo-Saavedra, D.L.; Luo, X.; Cui, J.; Rivera-Bustamante, R.F.; Li, S.; et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 2016, 6, 18919. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Natarajan, P.; Bhandari, M.; Abburi, V.; Dutta, S.K.; Yadav, L.; Stommel, J.; Nimmakayala, P.; Reddy, U.K. The ankyrin repeat gene family in Capsicum spp: Genome-wide survey, characterization and gene expression profile. Sci. Rep. 2020, 10, 4044. [Google Scholar] [CrossRef]
- Igwe, D.O.; Afiukwa, C.A.; Acquaah, G.; Ude, G.N. Genetic diversity and structure of Capsicum annuum as revealed by start codon targeted and directed amplified minisatellite DNA markers. Hereditas 2019, 156, 1–13. [Google Scholar] [CrossRef]
- Hill, T.A.; Chunthawodtiporn, J.; Ashrafi, H.; Stoffel, K.; Weir, A.; Van Deynze, A. Regions Underlying Population Structure and the Genomics of Organ Size Determination in Capsicum annuum. Plant Genome 2017, 10, plantgenome2017-03. [Google Scholar] [CrossRef]
- Esposito, S.; Aiese Cigliano, R.; Cardi, T.; Tripodi, P. Whole-genome resequencing reveals genomic footprints of Italian sweet and hot pepper heirlooms giving insight into genes underlying key agronomic and qualitative traits. BMC Genom. Data 2022, 23, 21. [Google Scholar] [CrossRef]
- Colonna, V.; D’Agostino, N.; Garrison, E.; Albrechtsen, A.; Meisner, J.; Facchiano, A.; Cardi, T.; Tripodi, P. Genomic diversity and novel genome-wide association with fruit morphology in Capsicum, from 746k polymorphic sites. Sci. Rep. 2019, 9, 10067. [Google Scholar] [CrossRef]
- Naegele, R.P.; Mitchell, J.; Hausbeck, M.K. Genetic diversity, population structure, and heritability of fruit traits in Capsicum annuum. PLoS ONE 2016, 11, e0156969. [Google Scholar] [CrossRef]
- Peñuela, M.; Arias, L.L.; Viáfara-Vega, R.; Rivera Franco, N.; Cárdenas, H. Morphological and molecular description of three commercial Capsicum varieties: A look at the correlation of traits and genetic distancing. Genet. Resour. Crop Evol. 2021, 68, 261–277. [Google Scholar] [CrossRef]
- Khoury, C.K.; Carver, D.; Barchenger, D.W.; Barboza, G.E.; van Zonneveld, M.; Jarret, R.; Bohs, L.; Kantar, M.; Uchanski, M.; Mercer, K.; et al. Modelled distributions and conservation status of the wild relatives of chile peppers (Capsicum L.). Divers. Distrib. 2020, 26, 209–225. [Google Scholar] [CrossRef]
- Jiménez-Leyva, A.; Orozco-Avitia, J.; Gutiérrez, A.; Vargas, G.; Sánchez, E.; Muñoz, E.; Esqueda, M. Functional plasticity of Capsicum annuum var. glabriusculum through multiple traits. AoB Plants. 2022, 14, plac017. [Google Scholar] [PubMed]
- Pang, X.; Chen, J.; Xu, Y.; Liu, J.; Zhong, Y.; Wang, L.; Zheng, J.; Wan, H. Genome-wide characterization of ascorbate peroxidase gene family in pepper (Capsicum annuum L.) in response to multiple abiotic stresses. Front. Plant Sci. 2023, 14, 1189020. [Google Scholar] [CrossRef] [PubMed]
- Lozada, D.N.; Bhatta, M.; Coon, D.; Bosland, P.W. Single nucleotide polymorphisms reveal genetic diversity in New Mexican chile peppers (Capsicum spp.). BMC Genom. 2021, 22, 356. [Google Scholar] [CrossRef] [PubMed]
- Villalon-Mendoza, H.; Medina-Martinez, T.; Ramirez-Meraz, M.; Urbina, S.E.S.; Maiti, R. Factors influencing the price of Chile piquin wild chili (Capsicum annuum L. var. glabriusculum) of North-East Mexico. Int. J. Bio-Resour. Stress. Manag. 2014, 5, 128. [Google Scholar] [CrossRef]
- Murillo-Amador, B.; Rueda-Puente, E.O.; Troyo-Diéguez, E.; Córdoba-Matson, M.V.; Hernández-Montiel, L.G.; Nieto-Garibay, A. Baseline study of morphometric traits of wild Capsicum annuum growing near two biosphere reserves in the Peninsula of Baja California for future conservation management. BMC Plant Biol. 2015, 15, 118. [Google Scholar] [CrossRef]
- Magdy, M.; Ouyang, B. The complete mitochondrial genome of the chiltepin pepper (Capsicum annuum var. glabriusculum), the wild progenitor of Capsicum annuum L. Mitochondrial DNA B Resour. 2020, 5, 683–684. [Google Scholar] [CrossRef]
- Cervantes-Hernández, F.; Ochoa-Alejo, N.; Martínez, O.; Ordaz-Ortiz, J.J. Metabolomic Analysis Identifies Differences Between Wild and Domesticated Chili Pepper Fruits During Development (Capsicum annuum L.). Front. Plant Sci. 2022, 13, 893055. [Google Scholar] [CrossRef]
- Rivera, A.; Monteagudo, A.B.; Igartua, E.; Taboada, A.; García-Ulloa, A.; Pomar, F.; Riveiro-Leira, M.; Silvar, C. Assessing genetic and phenotypic diversity in pepper (Capsicum annuum L.) landraces from North-West Spain. Sci. Hortic. 2016, 203, 1–11. [Google Scholar] [CrossRef]
- Razo-Mendivili, F.G.; Hernandez-Godínez, F.; Kanashiro, C.H.; Martínez, O. Transcriptomic analysis of a wild and a cultivated varieties of Capsicum annuum over fruit development and ripening. PLoS ONE 2021, 16, e0256319. [Google Scholar] [CrossRef]
- Lopez-Moreno, H.; Basurto-Garduño, A.C.; Torres-Meraz, M.A.; Diaz-Valenzuela, E.; Arellano-Arciniega, S.; Zalapa, J.; Sawers, R.J.; Cibrián-Jaramillo, A.; Diaz-Garcia, L. Genetic analysis and QTL mapping of domestication-related traits in chili pepper (Capsicum annuum L.). Front. Genet. 2023, 14, 1101401. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Mejía, C.; Bello-Bedoy, R.; Arteaga, M.C.; Castillo, G.R. Does Domestication Affect Structural and Functional Leaf Epidermal Traits? A Comparison between Wild and Cultivated Mexican Chili Peppers (Capsicum annuum). Plants 2022, 11, 3062. [Google Scholar] [CrossRef] [PubMed]
- Mares-Quiñones, M.D.; Valiente-Banuet, J.I. Horticultural aspects for the cultivated production of piquin peppers (Capsicum annuum L. var. glabriusculum)—A review. HortScience 2019, 54, 70–75. [Google Scholar] [CrossRef]
- Zhang, B.; Hu, F.; Cai, X.; Cheng, J.; Zhang, Y.; Lin, H.; Hu, K.; Wu, Z. Integrative Analysis of the Metabolome and Transcriptome of a Cultivated Pepper and Its Wild Progenitor Chiltepin (Capsicum annuum L. var. glabriusculum) Revealed the Loss of Pungency During Capsicum Domestication. Front. Plant Sci. 2022, 12, 783496. [Google Scholar] [PubMed]
- Hayano-Kanashiro, C.; Gámez-Meza, N.; Medina-Juárez, L.Á. Wild Pepper Capsicum annuum L. var. glabriusculum: Taxonomy, plant morphology, distribution, genetic diversity, genome sequencing, and phytochemical compounds. Crop Sci. 2016, 56, 1–11. [Google Scholar]
- Quintero, C.M.F.; Castillo, O.G.; Sánchez, P.D.; Marín-Sánchez, J.; Guzmán, A.I.; Sánchez, A.; Guzmán, J.M. Relieving dormancy and improving germination of Piquín chili pepper (Capsicum annuum var. glabriusculum) by priming techniques. Cogent Food Agric. 2018, 4, 1550275. [Google Scholar] [CrossRef]
- Song, X.; Liu, Z.; Wan, H.; Chen, W.; Zhou, R.; Duan, W. Editorial: Comparative Genomics and Functional Genomics Analyses in Plants. Front. Genet. 2021, 12, 12–14. [Google Scholar] [CrossRef]
- Kang, Y.J.; Ahn, Y.K.; Kim, K.T.; Jun, T.H. Resequencing of Capsicum annuum parental lines (YCM334 and Taean) for the genetic analysis of bacterial wilt resistance. BMC Plant Biol. 2016, 16, 235. [Google Scholar] [CrossRef]
- Jindal, S.K.; Dhaliwal, M.S.; Meena, O.P. Molecular advancements in male sterility systems of Capsicum: A review. Plant Breed. 2020, 139, 42–64. [Google Scholar] [CrossRef]
- Nimmakayala, P.; Abburi, V.L.; Saminathan, T.; Alaparthi, S.B.; Almeida, A.; Davenport, B.; Nadimi, M.; Davidson, J.; Tonapi, K.; Yadav, L.; et al. Genome-wide Diversity and Association Mapping for Capsaicinoids and Fruit Weight in Capsicum annuum L. Sci. Rep. 2016, 6, 38081. [Google Scholar] [CrossRef]
- Hulse-Kemp, A.M.; Maheshwari, S.; Stoffel, K.; Hill, T.A.; Jaffe, D.; Williams, S.R.; Weisenfeld, N.; Ramakrishnan, S.; Kumar, V.; Shah, P.; et al. Reference quality assembly of the 3.5-Gb genome of Capsicum annuum from a single linked-read library. Hortic. Res. 2018, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Tripodi, P.; Acquadro, A.; Lanteri, S.; D’Agostino, N. Genome Sequencing of Capsicum Species: Strategies, Assembly, and Annotation of Genes. In The Capsicum Genome. Compendium of Plant Genomes; Springer: Berlin/Heidelberg, Germany, 2019; pp. 139–152. [Google Scholar]
- INEGI Uso del suelo y vegetación, escala 1, 250000, serie VII (continuo nacional). 2021. Available online: https://www.inegi.org.mx/contenidos/temas/usosuelo/doc/USV_250K_SVII.pdf (accessed on 1 November 2024).
- Secretaría General de Gobierno. Plan Municipal de Dsarrollo 2021–2024 Municipio de Chinipas. 2021. Available online: https://chihuahua.gob.mx/atach2/anexo/anexo_03-2022_pmd_chinipas_2021-2024.pdf (accessed on 1 November 2024).
- Herrera Aguilar, A.; Cervantes Ortiz, F.; Grijalva, O.A.; Guadalupe, J.; Rodríguez, G.; Mercado, D.R.; Rodrííguez Herrera, S.A.; Andrío Enríquez, E.; Mendoza Elos, M. Deterioration of the Quality of the Piquín Pepper Seed from Four Collections in Querétaro and Guanajuato. 2018. Available online: https://www.scielo.org.mx/scielo.php?pid=S2007-09342018001001627&script=sci_arttext&tlng=en (accessed on 1 November 2024).
- Alcalá-Rico, J.S.G.J.; López-Benítez, A.; Vázquez-Badillo, M.E.; Sánchez-Aspeytia, D.; Rodríguez-Herrera, S.A.; Pérez-Rodríguez, M.Á.; Eta, L. Seed physiological potential of Capsicum annuum var. glabriusculum genotypes and their answers to pre-germination treatments. Agronomy 2019, 9, 325. [Google Scholar] [CrossRef]
- Ahn, Y.K.; Karna, S.; Jun, T.H.; Yang, E.Y.; Lee, H.E.; Kim, J.H.; Kim, J.H. Complete genome sequencing and analysis of Capsicum annuum varieties. Mol. Breed. 2016, 36, 140. [Google Scholar] [CrossRef]
- Guo, G.; Zhang, G.; Pan, B.; Diao, W.; Liu, J.; Ge, W.; Gao, C.; Zhang, Y.; Jiang, C.; Wang, S. Development and Application of InDel Markers for Capsicum spp. Based on Whole-Genome Re-Sequencing. Sci. Rep. 2019, 9, 3691. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Q.; Xu, Z.; Yang, W.; Guo, Y.; Lu, J.; Pan, H.; Cheng, T.; Cai, M. Genome-wide DNA polymorphisms in two cultivars of mei (Prunus mume sieb. et zucc.). BMC Genet. 2013, 14, 98. [Google Scholar] [CrossRef]
- Ritter, E.J.; Niederhuth, C.E. Intertwined evolution of plant epigenomes and genomes. Curr. Opin. Plant Biol. 2021, 61, 101990. [Google Scholar] [CrossRef]
- Dong, S.; Zhang, L.; Pang, W.; Zhang, Y.; Wang, C.; Li, Z.; Ma, L.; Tang, W.; Yang, G.; Song, H. Comprehensive analysis of coding sequence architecture features and gene expression in Arachis duranensis. Physiol. Mol. Biol. Plants 2021, 27, 213–222. [Google Scholar] [CrossRef]
- Yadav, C.B.; Bhareti, P.; Muthamilarasan, M.; Mukherjee, M.; Khan, Y.; Rathi, P.; Prasad, M. Genome-wide SNP identification and characterization in two soybean cultivars with contrasting mungbean yellow mosaic india virus disease resistance traits. PLoS ONE 2015, 10, e0123897. [Google Scholar] [CrossRef]
- Cortaga, C.Q.; Lachica, J.A.P.; Lantican, D.V.; Ocampo, E.T.M. Genome-wide SNP and InDel analysis of three Philippine mango species inferred from whole-genome sequencing. J. Genet. Eng. Biotechnol. 2022, 20, 46. [Google Scholar] [CrossRef]
- Stafuzza, N.B.; Zerlotini, A.; Lobo, F.P.; Yamagishi, M.E.B.; Chud, T.C.S.; Caetano, A.R.; Munari, D.P.; Garrick, D.J.; Machado, M.A.; Martins, M.F.; et al. Single nucleotide variants and InDels identified from whole-genome re-sequencing of Guzerat, Gyr, Girolando and Holstein cattle breeds. PLoS ONE 2017, 12, e0173954. [Google Scholar] [CrossRef]
- Zou, Z.; Zhang, J. Are Nonsynonymous Transversions Generally More Deleterious than Nonsynonymous Transitions? Mol. Biol. Evol. 2021, 38, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Singhabahu, S.; Wijesinghe, C.; Gunawardana, D.; Senarath-Yapa, M.D.; Kannangara, M.; Edirisinghe, R.; Dissanayake, V.H.W. Whole Genome Sequencing and Analysis of Godawee, a Salt Tolerant Indica Rice Variety. Rice Res. Open Access 2017, 5, 2. [Google Scholar] [CrossRef]
- Chen, J.; Guo, J.T. Comparative assessments of indel annotations in healthy and cancer genomes with next-generation sequencing data. BMC Med. Genom. 2020, 13, 170. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.; Iqbal, Z.; Vauterin, P.; Pearson, R.; Campino, S.; Theron, M.; Gould, K.; Mead, D.; Drury, E.; O’Brien, J.; et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016, 26, 1288–1299. [Google Scholar] [CrossRef]
- Jilani, M.; Turcan, A.; Haspel, N.; Jagodzinski, F. Elucidating the Structural Impacts of Protein InDels. Biomolecules 2022, 12, 1435. [Google Scholar] [CrossRef]
- Ajawatanawong, P.; Baldauf, S.L. Evolution of protein indels in plants, animals and fungi. BMC Evol. Biol. 2013, 13, 140. [Google Scholar] [CrossRef]
- Scaldaferro, M.A.; Grabiele, M.; Moscone, E.A. Heterochromatin type, amount and distribution in wild species of chili peppers (Capsicum, Solanaceae). Genet. Resour. Crop Evol. 2013, 60, 693–709. [Google Scholar] [CrossRef]
- Shirasawa, K.; Hosokawa, M.; Yasui, Y.; Toyoda, A.; Isobe, S. Chromosome-scale genome assembly of a Japanese chili pepper landrace, Capsicum annuum “Takanotsume”. DNA Res. 2022, 30, 1–9. [Google Scholar] [CrossRef]
- Bekele, W.A.; Wieckhorst, S.; Friedt, W.; Snowdon, R.J. High-throughput genomics in sorghum: From whole-genome resequencing to a SNP screening array. Plant Biotechnol. J. 2013, 11, 1112–1125. [Google Scholar] [CrossRef]
- Arai-Kichise, Y.; Shiwa, Y.; Nagasaki, H.; Ebana, K.; Yoshikawa, H.; Yano, M.; Wakasa, K. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of japonica rice by whole-genome sequencing. Plant Cell Physiol. 2011, 52, 274–282. [Google Scholar] [CrossRef]
- Bohry, D.; Ramos, H.C.C.; Dos Santos, P.H.D.; Boechat, M.S.B.; Arêdes, F.A.S.; Pirovani, A.A.V.; Pereira, M.G. Discovery of SNPs and InDels in papaya genotypes and its potential for marker assisted selection of fruit quality traits. Sci. Rep. 2021, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Liu, X.; Yang, T.; Huang, Z.; Hanif, Q.; Asif, M.; Khan, Q.M.; Mansoor, S. Genomic variants identified from whole genome resequencing of indicine cattle breeds from Pakistan. PLoS ONE 2019, 14, e0215065. [Google Scholar] [CrossRef] [PubMed]
- Raherison, E.; Majidi, M.M.; Goessen, R.; Hughes, N.; Cuthbert, R.; Knox, R.; Lukens, L. Evidence for the accumulation of nonsynonymous mutations and favorable pleiotropic alleles during wheat breeding. G3 Genes Genomes Genet. 2020, 10, 4001–4011. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Rau, D.; Benazzo, A.; Bellucci, E.; Goretti, D.; Biagetti, E.; Panziera, A.; Laido, G.; Rodriguez, M.; Gioia, T.; et al. High level of nonsynonymous changes in common bean suggests that selection under domestication increased functional diversity at target traits. Front. Plant Sci. 2017, 7, 2005. [Google Scholar] [CrossRef] [PubMed]
- Loewe, L.; Charlesworth, B.; Bartolomé, C.; Nöel, V. Estimating selection on nonsynonymous mutations. Genetics 2006, 172, 1079–1092. [Google Scholar] [CrossRef][Green Version]
- D’Agostino, N.; Tripodi, P. NGS-based genotyping, high-throughput phenotyping and genome-wide association studies laid the foundations for next-generation breeding in horticultural crops. Diversity 2017, 9, 38. [Google Scholar] [CrossRef]
- Zhang, F.; Qu, K.; Chen, N.; Hanif, Q.; Jia, Y.; Huang, Y.; Dang, R.; Zhang, J.; Lan, X.; Chen, H.; et al. Genome-wide SNPs and indels characteristics of three Chinese cattle breeds. Animals 2019, 9, 596. [Google Scholar] [CrossRef]
- Ramakrishna, G.; Kaur, P.; Nigam, D.; Chaduvula, P.K.; Yadav, S.; Talukdar, A.; Singh, N.K.; Gaikwad, K. Genome-wide identification and characterization of InDels and SNPs in Glycine max and Glycine soja for contrasting seed permeability traits. BMC Plant Biol. 2018, 18, 141. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118, iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreno-Contreras, V.I.; Delgado-Gardea, M.C.E.; Ramos-Hernández, J.A.; Mendez-Tenorio, A.; Varela-Rodríguez, H.; Sánchez-Ramírez, B.; Muñoz-Ramírez, Z.Y.; Infante-Ramírez, R. Genome-Wide Identification and Characterization of SNPs and InDels of Capsicum annuum var. glabriusculum from Mexico Based on Whole Genome Sequencing. Plants 2024, 13, 3248. https://doi.org/10.3390/plants13223248
Moreno-Contreras VI, Delgado-Gardea MCE, Ramos-Hernández JA, Mendez-Tenorio A, Varela-Rodríguez H, Sánchez-Ramírez B, Muñoz-Ramírez ZY, Infante-Ramírez R. Genome-Wide Identification and Characterization of SNPs and InDels of Capsicum annuum var. glabriusculum from Mexico Based on Whole Genome Sequencing. Plants. 2024; 13(22):3248. https://doi.org/10.3390/plants13223248
Chicago/Turabian StyleMoreno-Contreras, Valeria Itzel, Ma. Carmen E. Delgado-Gardea, Jesús A. Ramos-Hernández, Alfonso Mendez-Tenorio, Hugo Varela-Rodríguez, Blanca Sánchez-Ramírez, Zilia Y. Muñoz-Ramírez, and Rocío Infante-Ramírez. 2024. "Genome-Wide Identification and Characterization of SNPs and InDels of Capsicum annuum var. glabriusculum from Mexico Based on Whole Genome Sequencing" Plants 13, no. 22: 3248. https://doi.org/10.3390/plants13223248
APA StyleMoreno-Contreras, V. I., Delgado-Gardea, M. C. E., Ramos-Hernández, J. A., Mendez-Tenorio, A., Varela-Rodríguez, H., Sánchez-Ramírez, B., Muñoz-Ramírez, Z. Y., & Infante-Ramírez, R. (2024). Genome-Wide Identification and Characterization of SNPs and InDels of Capsicum annuum var. glabriusculum from Mexico Based on Whole Genome Sequencing. Plants, 13(22), 3248. https://doi.org/10.3390/plants13223248






