Identification of RppSLN from an Elite Landrace: A Major Locus Conferring Resistance to Southern Corn Rust in Maize (Zea mays L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Phenotyping and Field Trials
2.3. Genotyping
2.4. Linkage Map Construction and QTL Mapping
2.5. Genome Assembly and Annotation
2.6. Artificial Inoculation Traits
2.7. Real-Time Quantitative Analysis
2.8. Transcriptome Analysis
2.9. Physiological and Biochemical Characteristics
2.10. Evaluation of Yield Breeding Value of RppSLN
3. Results
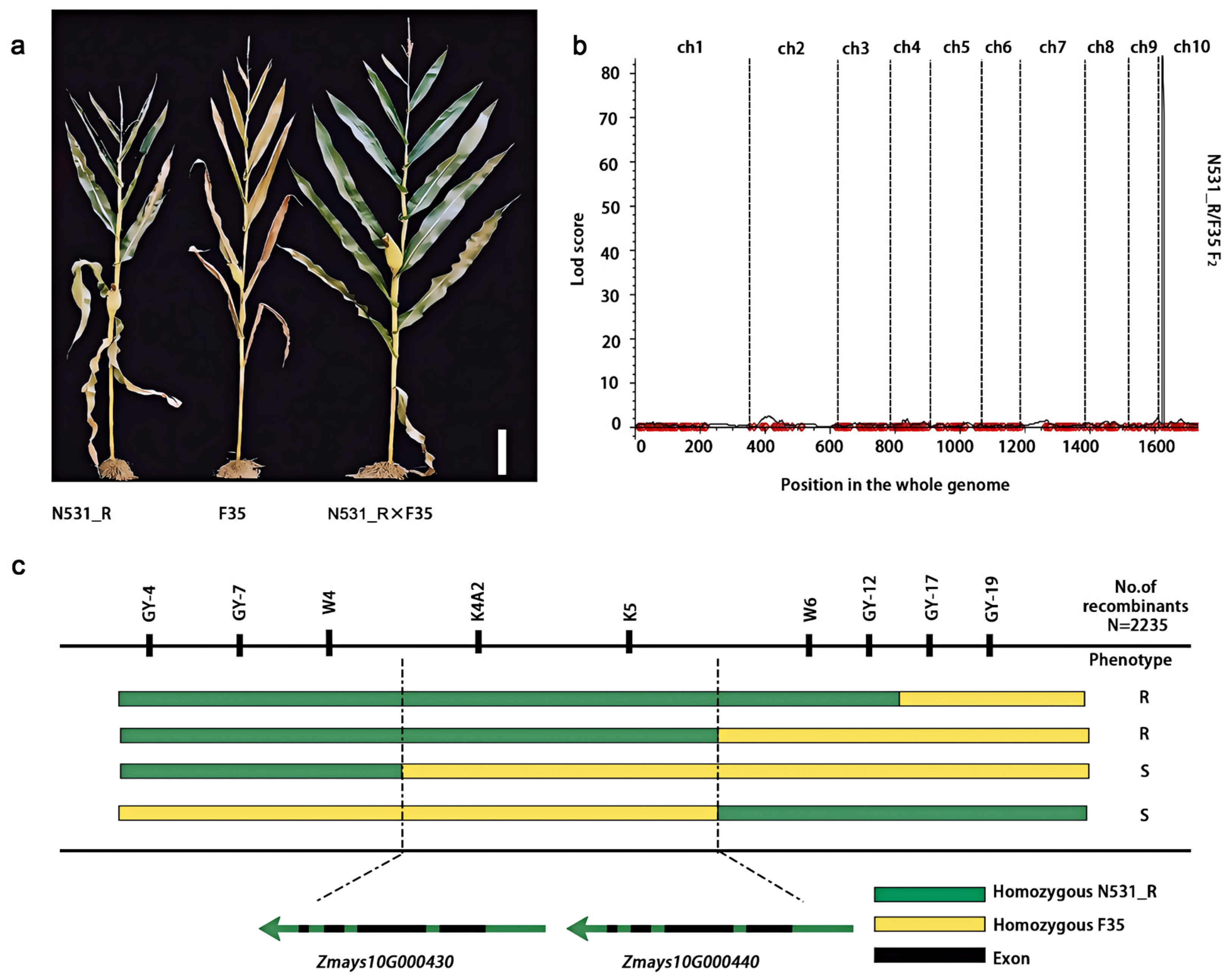
3.1. Identification of SCR-Resistant Locus RppSLN Isolated from Landrace
3.2. Fine Mapping of RppSLN
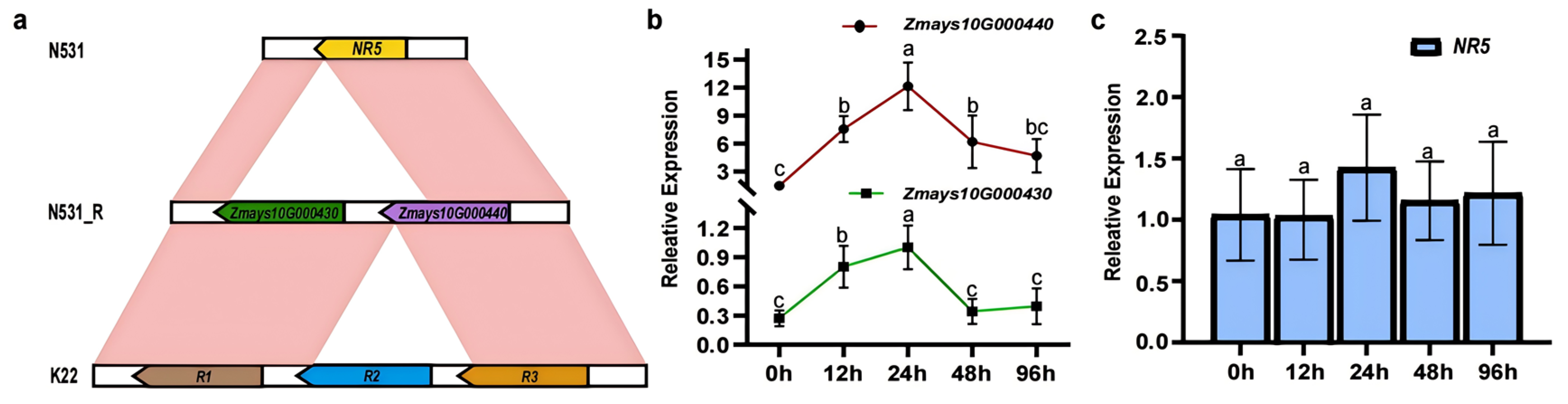
3.3. Identification of Candidate Genes for RppSLN
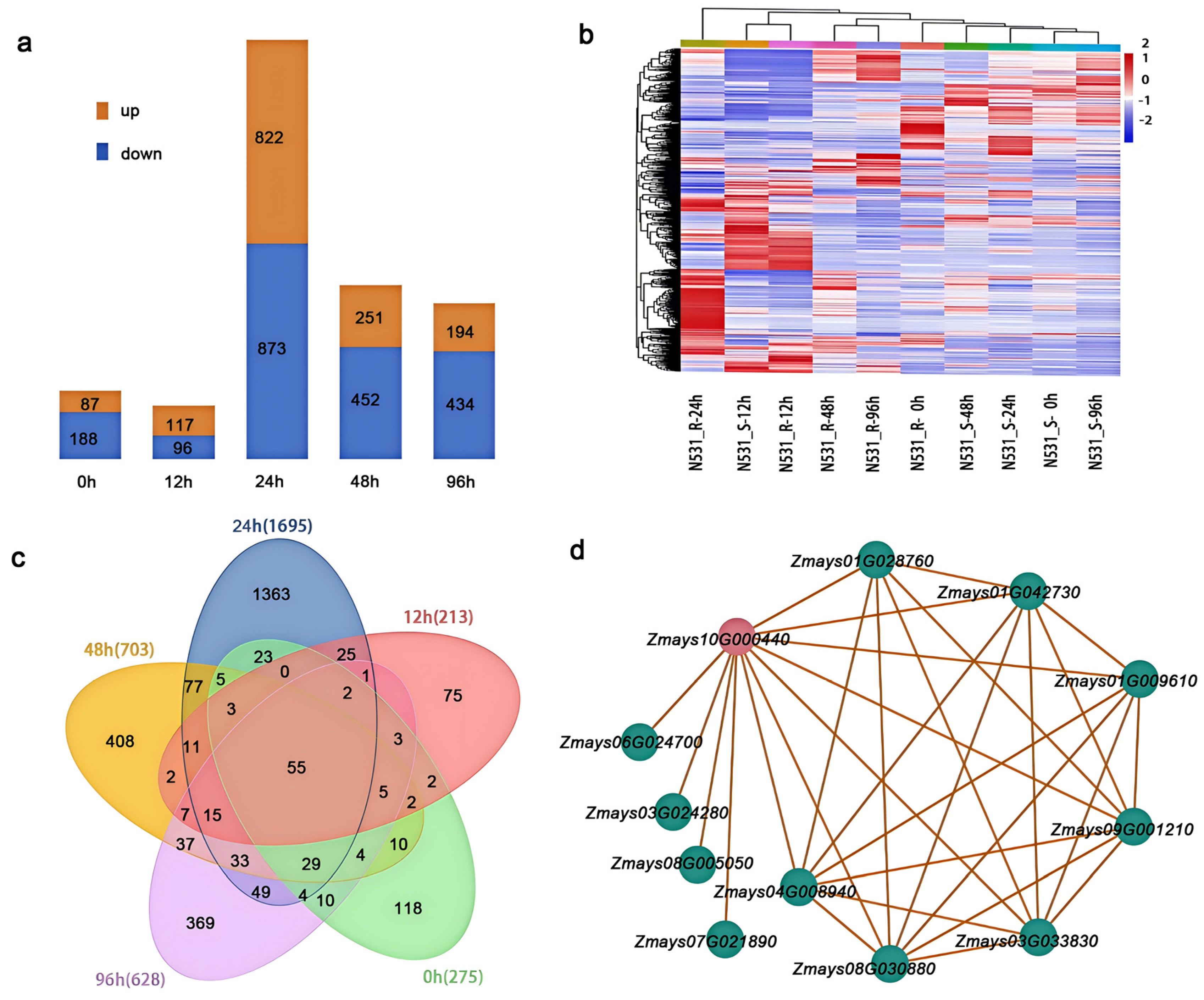
3.4. Transcriptome Expression Pattern Analysis
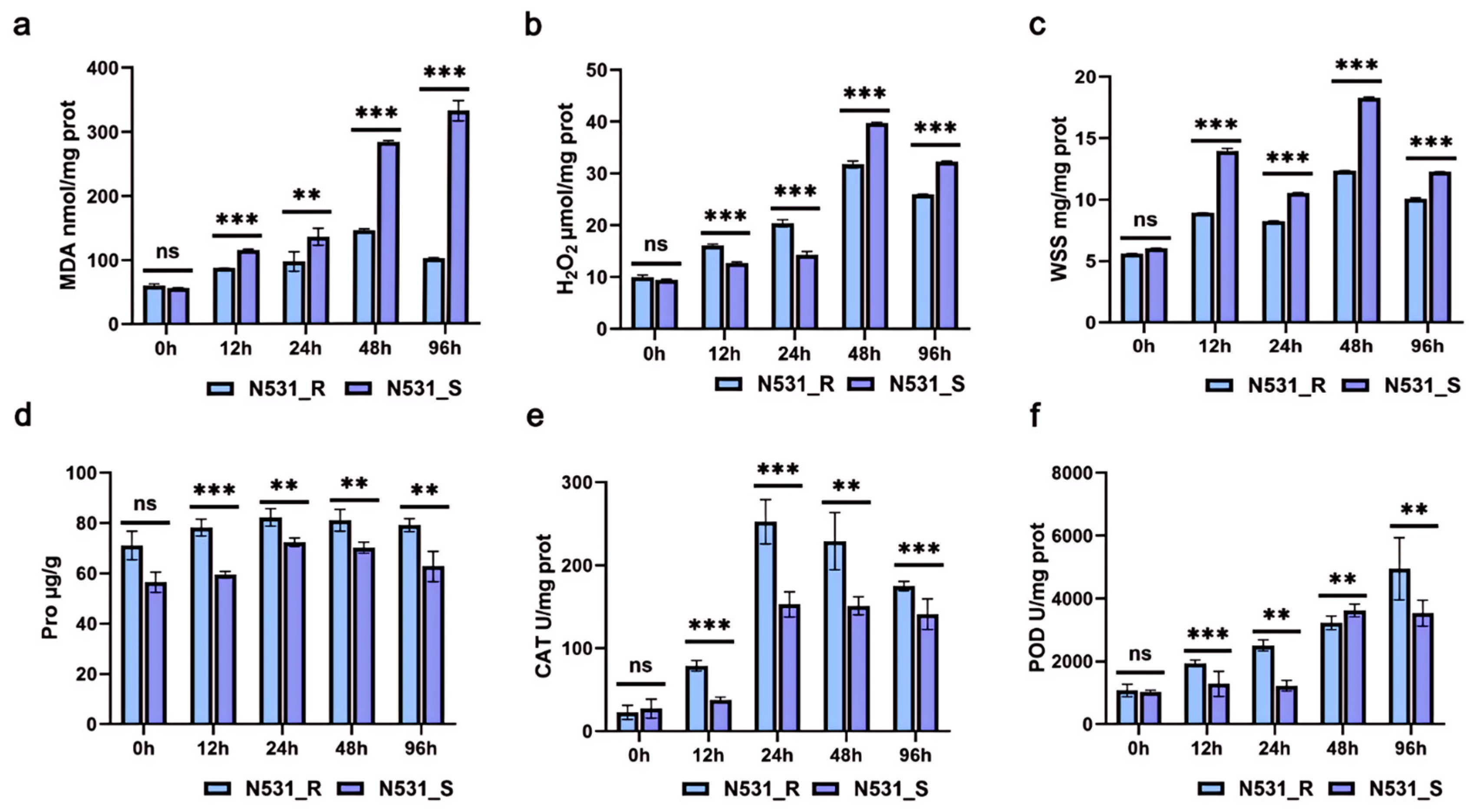
3.5. Physiological and Biochemical Indicators of Resistance and Susceptible Lines in Leaves
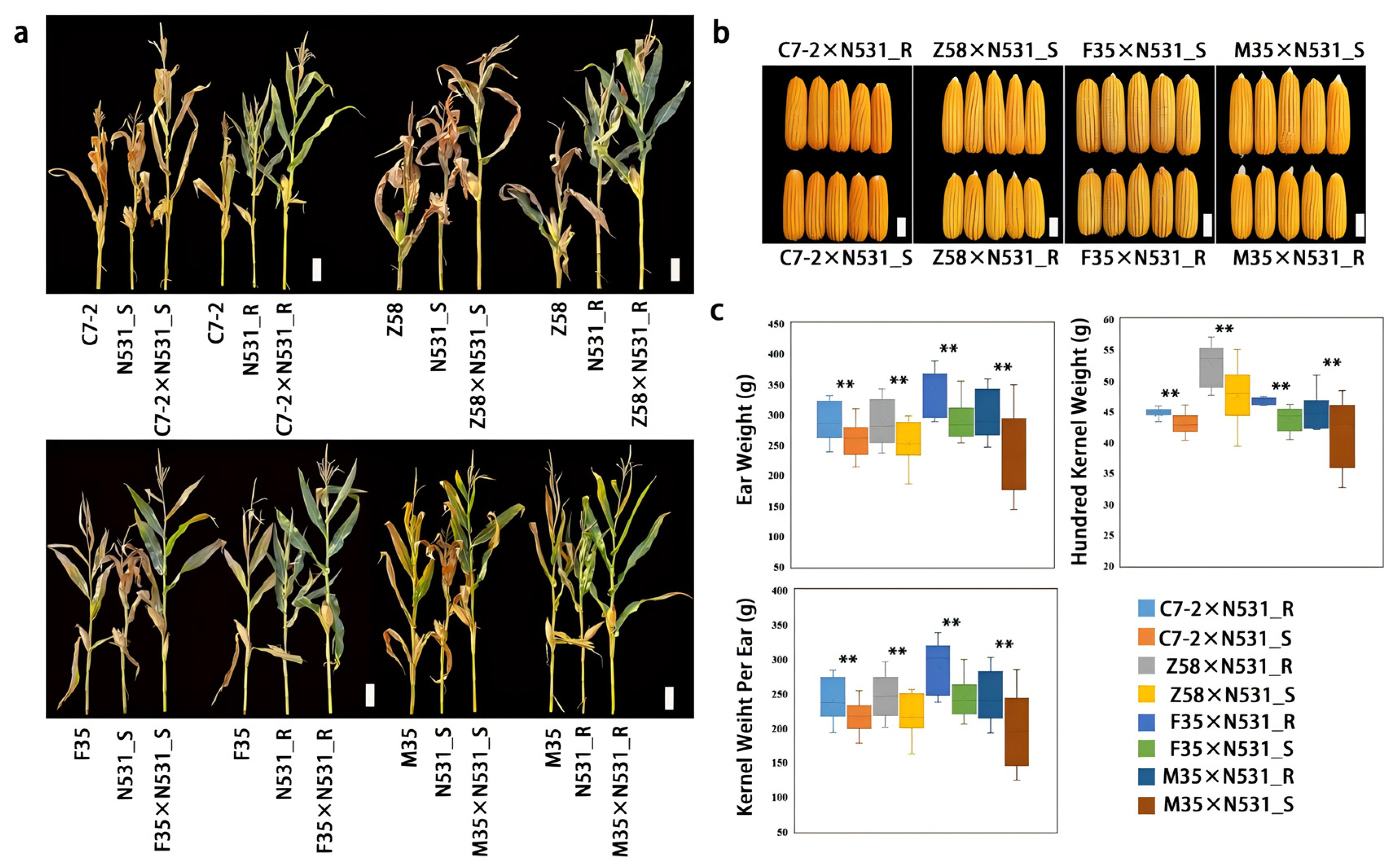
3.6. Evaluation of Breeding Potential of RppSLN
4. Discussion
4.1. Landraces Are Important Germplasm Resources for Maize Improvement
4.2. Exploring Resistant Loci That Could Achieve Durable Resistance to SCR by Aggregating Multiple Resistance Genes Is Essential
4.3. Exploring Network Regulation for Resistance to SCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, L.J.; Li, L.L.; Dong, Z.K.; Zhu, J.S.; Guo, W.X.; Song, Y.Y.; Cui, H.Y.; Lv, S.H.; Sindhu, L.; Men, X.Y. EIRP model driven by machine learning for predicting the occurrence risk of southern corn rust (Puccinia polysora Underw.) in northern China. Agric. For. Meteorol. 2024, 356, 110149. [Google Scholar] [CrossRef]
- Li, J.L.; Cheng, D.H.; Guo, S.W.; Chen, C.; Wang, Y.W.; Zhong, Y.; Qi, X.L.; Liu, Z.K.; Wang, D.; Wang, Y.D.; et al. Genome-wide association and genomic prediction for resistance to southern corn rust in DH and testcross populations. Front. Plant Sci. 2023, 14, 1109116. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Deng, C.; Li, X.Y.; Zhao, X.D.; Li, H.M.; Li, Z.M.; Tian, Z.Q.; Leonard, A.; Jaqueth, J.; Li, B.L.; et al. Identification and fine-mapping of RppCML496, a major QTL for resistance to Puccinia polysora in maize. Plant Genome 2021, 14, e20062. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.Y.; Li, L.F.; Guo, F.F.; Zhang, K.Y.; Dong, J.Y.; Luo, Y.; Ma, Z.H. Southern corn rust caused by Underw: A review. Phytopathol. Res. 2021, 3, 25. [Google Scholar] [CrossRef]
- Shu, G.P.; Wang, A.F.; Wang, X.C.; Ding, J.Q.; Chen, R.J.; Gao, F.; Wang, A.F.; Li, T.; Wang, Y.B. Identification of southern corn rust resistance QTNs in Chinese summer maize germplasm via multi-locus GWAS and post-GWAS analysis. Front. Plant Sci. 2023, 14, 1221395. [Google Scholar] [CrossRef]
- Li, W.R.; Liao, C.J.; Bluhm, B.H.; Mengiste, T.; Woloshuk, C.P. A maize (Zea mays L.) BIK1-like receptor-like cytoplasmic kinase contributes to disease resistance. Plant Mol. Biol. Rep. 2022, 40, 28–42. [Google Scholar] [CrossRef]
- Hao, Y.F.; Hu, Y.; Jaqueth, J.; Lin, J.G.; He, C.; Lin, G.F.; Zhao, M.X.; Ren, J.; Tamang, T.M.; Park, S.; et al. Genetic and transcriptomic dissection of host defense to Goss’s bacterial wilt and leaf blight of maize. G3-Genes Genom. Genet. 2023, 13, jkad197. [Google Scholar] [CrossRef]
- Osdaghi, E.; Robertson, A.E.; Jackson-Ziems, T.A.; Abachi, H.; Li, X.; Harveson, R.M. Clavibacter nebraskensis causing Goss’s wilt of maize: Five decades of detaining the enemy in the New World. Mol. Plant Pathol. 2023, 24, 675–692. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef]
- Corwin, J.A.; Kliebenstein, D.J. Quantitative resistance: More than just perception of a pathogen. Plant Cell. 2017, 29, 655–665. [Google Scholar] [CrossRef]
- Bokore, F.E.; Knox, R.E.; Hiebert, C.W.; Cuthbert, R.D.; Depauw, R.M.; Meyer, B.; N’diaye, A.; Pozniak, C.J.; Mccallum, B.D. A combination of leaf rust resistance genes, including Lr34 and Lr46, is the key to the durable resistance of the Canadian Wheat Cultivar, Carberry. Front. Plant Sci. 2022, 12, 775383. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pruitt, R.N.; Nürnberger, T.; Wang, Y.C. Evasion of plant immunity by microbial pathogens. Nat. Rev. Microbiol. 2022, 20, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Brewbaker, J.L.; Kim, S.K.; So, Y.S.; Logroño, M.; Moon, H.G.; Ming, R.; Lu, X.W.; Josue, A.D. General resistance in maize to southern rust (Puccinia polysora Underw). Crop Sci. 2011, 51, 1393–1409. [Google Scholar] [CrossRef]
- Li, M.S.; Zhang, S.H.; Li, X.H.; Pan, G.L.; Bai, L.; Peng, Z.B. Study on heterotic groups among maize inbred lines based on SCA. Sci. Agric. Sin. 2002, 35, 600–605. [Google Scholar]
- Duan, D.; He, H. Description of a rust Puccinia polysora on corn in Hainan Island. Acta Mycol. 1984, 3, 125–126. [Google Scholar]
- Jiang, K.; Du, Q.; Qin, Z.H.; Chen, M.G.; Li, S.C.; Sun, S.L.; Wu, X.F.; Guo, Y.Y.; Shi, Y.S.; Lin, X.H.; et al. Identification of resistance to southern corn rust (Puccinia Polysora Underw) in maize germplasm. J. Plant Genet. Resour. 2013, 14, 711–714. [Google Scholar]
- Deng, C.; Leonard, A.; Cahill, J.; Lv, M.; Li, Y.R.; Thatcher, S.; Li, X.Y.; Zhao, X.D.; Du, W.J.; Li, Z.; et al. The RppC-AvrRppC NLR-effector interaction mediates the resistance to southern corn rust in maize. Mol. Plant. 2022, 15, 904–912. [Google Scholar] [CrossRef]
- An, Y.X.; Chen, L.; Li, Y.X.; Li, C.H.; Shi, Y.S.; Song, Y.C.; Zhang, D.F.; Li, Y.; Wang, T.Y. Candidate loci for the kernel row number in maize revealed by a combination of transcriptome analysis and regional association mapping. BMC Plant Biol. 2019, 19, 201. [Google Scholar] [CrossRef]
- Zhao, P.F.; Zhang, G.B.; Wu, X.J.; Li, N.; Shi, D.Y.; Zhang, D.F.; Ji, C.F.; Xu, M.L.; Wang, S.C. Fine mapping of RppP25, a southern rust resistance gene in maize. J. Integr. Plant Biol. 2013, 55, 462–472. [Google Scholar] [CrossRef]
- Deng, C.; Li, H.M.; Li, Z.M.; Tian, Z.Q.; Chen, J.F.; Chen, G.S.; Zhang, X.C.; Ding, J.Q.; Chang, Y.X. New QTL for resistance to Puccinia polysora Underw in maize. J. Appl. Genet. 2019, 60, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, J.A.; Shi, X.L.; Chen, L.; Qin, J.; Zhang, M.C.; Yang, C.Y.; Song, Q.J.; Yan, L. Development of SNP marker panels for genotyping by target sequencing (GBTS) and its application in soybean. Mol. Breed. 2023, 43, 26. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; Depristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.H.; Zhang, L.Y.; Wang, J.K. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Flynn, J.M.; Hubley, R.; Goubert, C.; Rosen, J.; Clark, A.G.; Feschotte, C.; Smit, A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 2020, 117, 9451–9457. [Google Scholar] [CrossRef] [PubMed]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2009, 4, 4.10.11–14.10.14. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.W.; He, L.M.; Lai, J.S.; Dooner, H.K.; Du, C.G. HelitronScanner uncovers a large overlooked cache of transposons in many plant genomes. Proc. Natl. Acad. Sci. USA 2014, 111, 10263–10268. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.S.; Zhang, B.; Ding, J.Q.; Wang, H.Z.; Deng, C.; Wang, J.L.; Yang, Q.H.; Pi, Q.Y.; Zhang, R.Y.; Zhai, H.Y.; et al. Cloning southern corn rust resistant gene RppK and its cognate gene AvrRppK from Puccinia polysora. Nat. Commun. 2022, 13, 4392. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2013, 25, 402–408. [Google Scholar] [CrossRef]
- Kim, D.; Landmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Dai, X.K.; Zhang, M.L.; Liu, T.; Chen, X.Y.; Zhu, T.H. Brown leaf spot of Cunninghamia lanceolata caused by Colletotrichum kahawae in Sichuan Province, China. Plant Dis. 2023, 107, 2548. [Google Scholar] [CrossRef]
- Kumar, A.; Sharma, S.; Chunduri, V.; Kaur, A.; Kaur, S.; Malhotra, N.; Kumar, A.; Kapoor, P.; Kumari, A.; Kaur, J.; et al. Genome-wide identification and characterization of heat shock protein family reveals role in development and stress conditions in Triticum aestivum L. Sci. Rep. 2020, 10, 7858. [Google Scholar] [CrossRef]
- Cao, L.R.; Wang, G.R.; Fahim, A.M.; Pang, Y.Y.; Zhang, Q.J.; Zhang, X.; Wang, Z.H.; Lu, X.M. Comprehensive analysis of the DnaJ/HSP40 gene family in maize (Zea mays L.) reveals that ZmDnaJ96 enhances abiotic stress tolerance. J. Plant Growth Regul. 2024, 43, 1548–1569. [Google Scholar] [CrossRef]
- Zheng, P.P.; Cao, L.; Zhang, C.; Pan, W.C.; Wang, W.; Yu, X.; Li, Y.P.; Fan, T.T.; Miao, M.; Tang, X.F.; et al. MYB43 as a novel substrate for CRL4PRL1 E3 ligases negatively regulates cadmium tolerance through transcriptional inhibition of HMAs in Arabidopsis. New Phytol. 2022, 234, 884–901. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Luo, J.; Rouse, M.N.; Hua, L.; Li, H.N.; Li, B.S.; Li, T.Y.; Zhang, W.J.; Gao, C.X.; Wang, Y.P.; Dubcovsky, J.; et al. Identification and characterization of Sr22b, a new allele of the wheat stem rust resistance gene Sr22 effective against the Ug99 race group. Plant Biotechnol. J. 2022, 20, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.C.; Cheng, Y.K.; Hao, W.H.; Bai, B.; Fu, L.P.; Ren, Y.; Hao, Y.F.; Wang, F.J.; Lin, R.M.; Si, H.Q.; et al. Identification of stripe rust resistance gene YrBDT in Chinese landrace wheat Baidatou using BSE-seq and BSR-seq. Theor. Appl. Genet. 2024, 137, 199. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef]
- Mondragon-Palomino, M.; Gaut, B.S. Gene conversion and the evolution of three leucine-rich repeat gene families in Arabidopsis thaliana. Mol. Biol. Evol. 2005, 22, 2444–2456. [Google Scholar] [CrossRef]
- Sinapidou, E.; Williams, K.; Nott, L.; Bahkt, S.; Tör, M.; Crute, I.; Bittner-Eddy, P.; Beynon, J. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004, 38, 898–909. [Google Scholar] [CrossRef]
- Ashikawa, I.; Hayashi, N.; Yamane, H.; Kanamori, H.; Wu, J.Z.; Matsumoto, T.; Ono, K.; Yano, M. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Geneti. 2008, 180, 2267–2276. [Google Scholar] [CrossRef]
- Narusaka, M.; Shirasu, K.; Noutoshi, Y.; Kubo, Y.; Shiraishi, T.; Iwabuchi, M.; Narusaka, Y. RRS1 and RRS4 provide a dual Resistence-gene system against fungal and bacterial pathogens. Plant J. 2009, 60, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Meunier, E.; Broz, P. Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. 2017, 38, 744–757. [Google Scholar] [CrossRef]
- Zhang, T.; Li, C.; Zhu, J.; Li, Y.; Wang, Z.; Tong, C.Y.; Xi, Y.; Han, Y.; Koiwa, H.; Peng, X.; et al. Structured 3′ UTRs destabilize mRNAs in plants. Genome Biol. 2024, 25, 54. [Google Scholar] [CrossRef]
- Zhang, D.L.; Gao, Z.Y.; Zhang, H.; Yang, Y.Z.; Yang, X.X.; Zhao, X.F.; Guo, H.L.; Nagalakshmi, U.; Li, D.W.; Dinesh-Kumar, S.P.; et al. The MAPK-Alfin-like 7 module negatively regulates ROS scavenging genes to promote NLR-mediated immunity. Proc. Natl. Acad. Sci. USA 2023, 120, e2214750120. [Google Scholar] [CrossRef]
- Milne, G.L.; Yin, H.Y.; Hardy, K.D.; Davies, S.S.; Roberts, L.J. Isoprostane generation and function. Chem. Rev. 2011, 111, 5973–5996. [Google Scholar] [CrossRef] [PubMed]

| F2 Population | Total | Resistant | Susceptible | Segregation Ratio | χ2 | p-Value |
|---|---|---|---|---|---|---|
| N531_R×F35 | 183 | 128 | 55 | 2.7 | 0.89 | 0.35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Ma, S.; Zhang, D.; Li, C.; Chen, L.; Tang, B.; An, Y.; Liu, X.; He, G.; Shi, Y.; et al. Identification of RppSLN from an Elite Landrace: A Major Locus Conferring Resistance to Southern Corn Rust in Maize (Zea mays L.). Plants 2024, 13, 3227. https://doi.org/10.3390/plants13223227
Wang Y, Ma S, Zhang D, Li C, Chen L, Tang B, An Y, Liu X, He G, Shi Y, et al. Identification of RppSLN from an Elite Landrace: A Major Locus Conferring Resistance to Southern Corn Rust in Maize (Zea mays L.). Plants. 2024; 13(22):3227. https://doi.org/10.3390/plants13223227
Chicago/Turabian StyleWang, Yufei, Shuai Ma, Dengfeng Zhang, Chunhui Li, Lin Chen, Bin Tang, Yixin An, Xuyang Liu, Guanhua He, Yunsu Shi, and et al. 2024. "Identification of RppSLN from an Elite Landrace: A Major Locus Conferring Resistance to Southern Corn Rust in Maize (Zea mays L.)" Plants 13, no. 22: 3227. https://doi.org/10.3390/plants13223227
APA StyleWang, Y., Ma, S., Zhang, D., Li, C., Chen, L., Tang, B., An, Y., Liu, X., He, G., Shi, Y., Li, Y., Wang, T., Yang, D., & Li, Y. (2024). Identification of RppSLN from an Elite Landrace: A Major Locus Conferring Resistance to Southern Corn Rust in Maize (Zea mays L.). Plants, 13(22), 3227. https://doi.org/10.3390/plants13223227






