Small Sized Yet Powerful: Nuclear Distribution C Proteins in Plants
Abstract
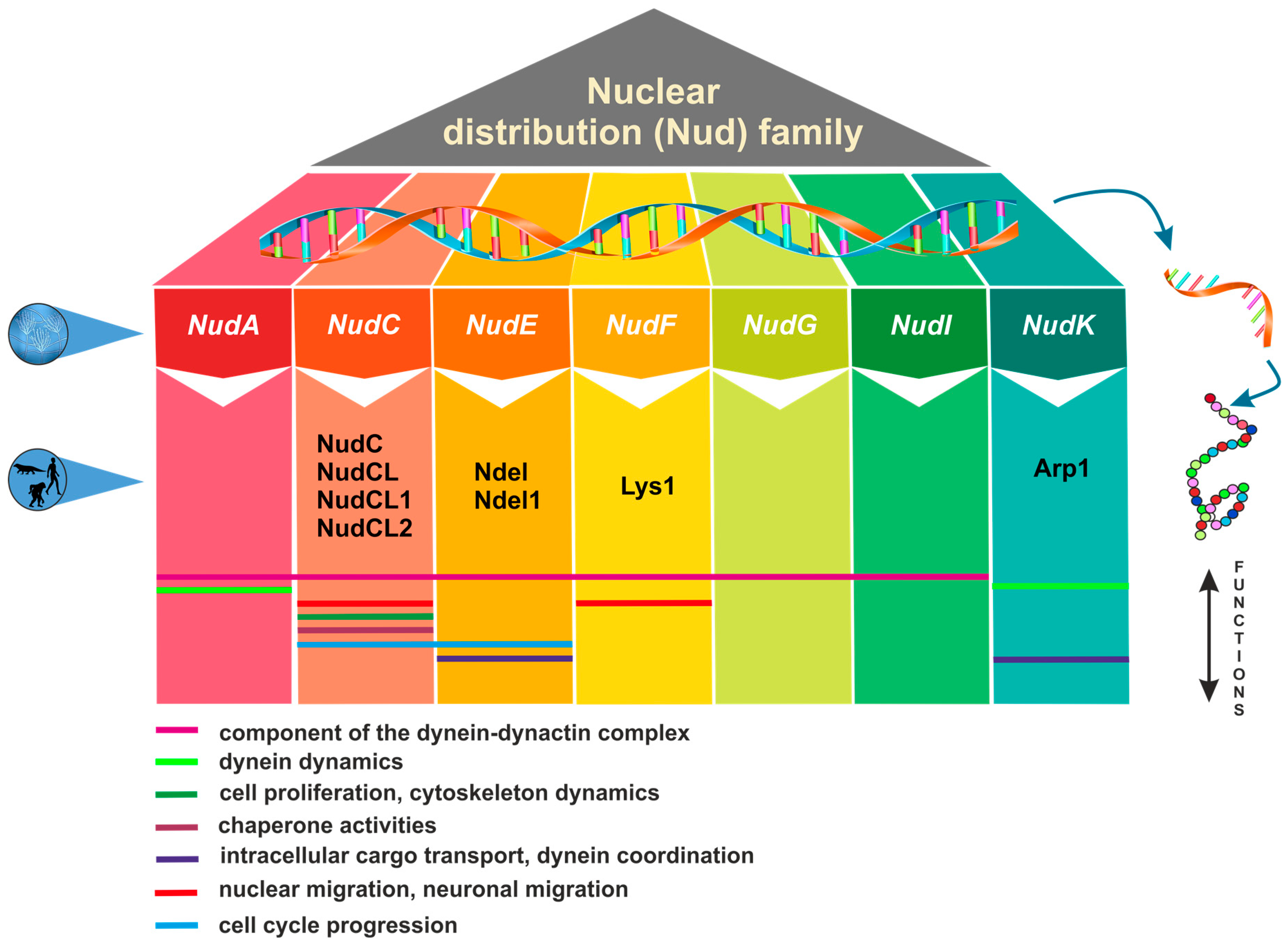
1. Nuclear Distribution Genes and Their Functional Significance across Eukaryotes
2. Structural and Functional Features of NudC Proteins
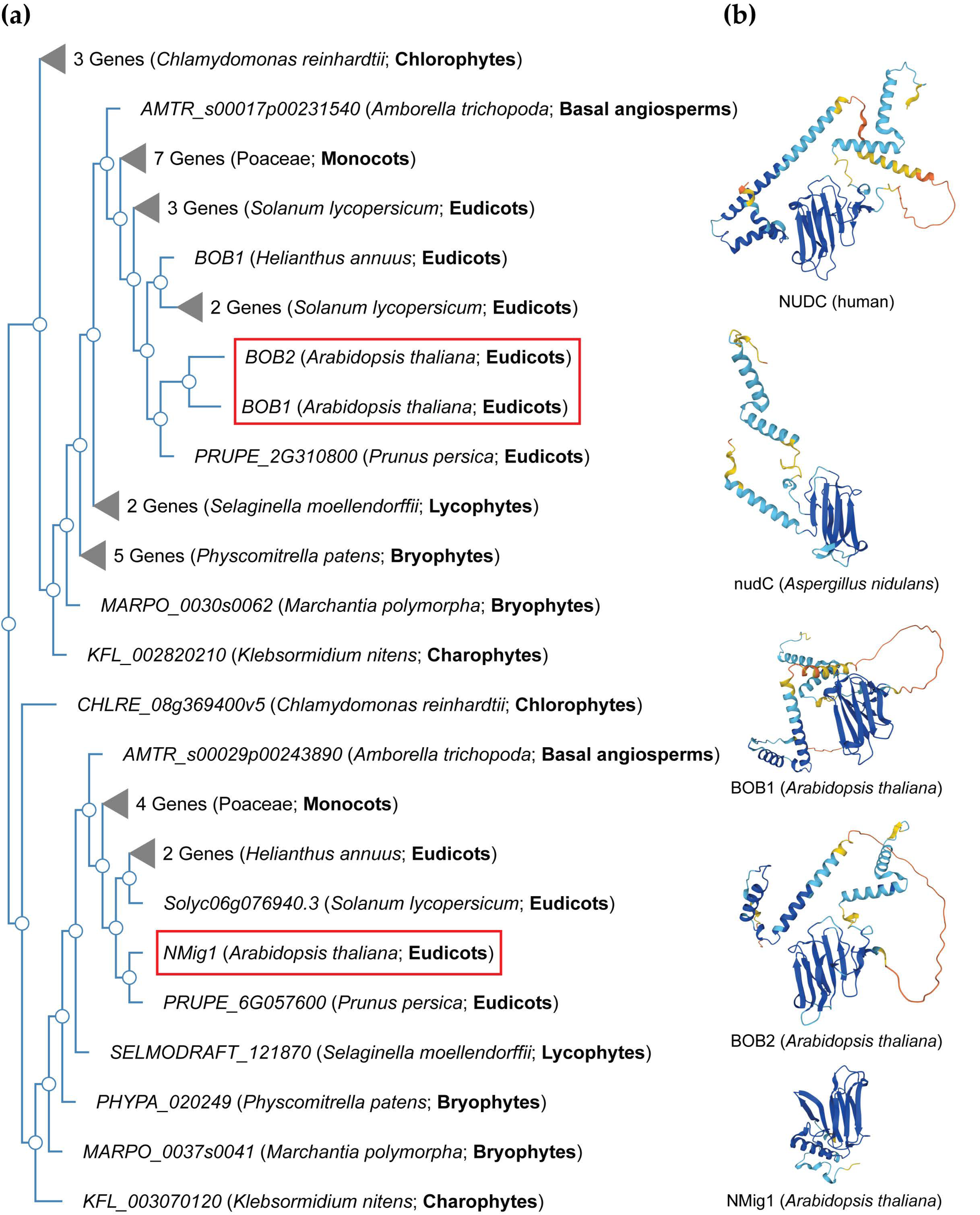
3. NudC Proteins in Plants
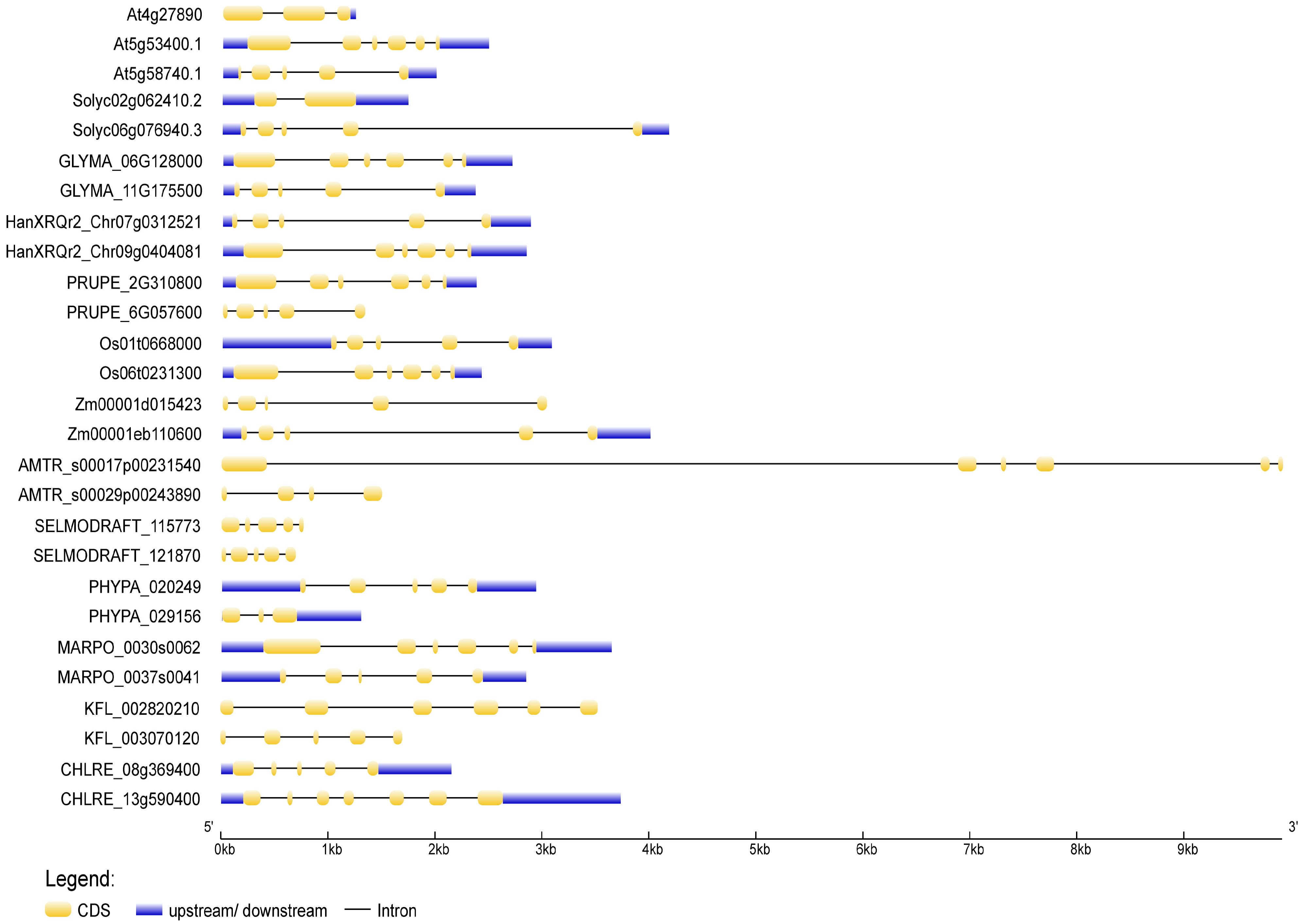
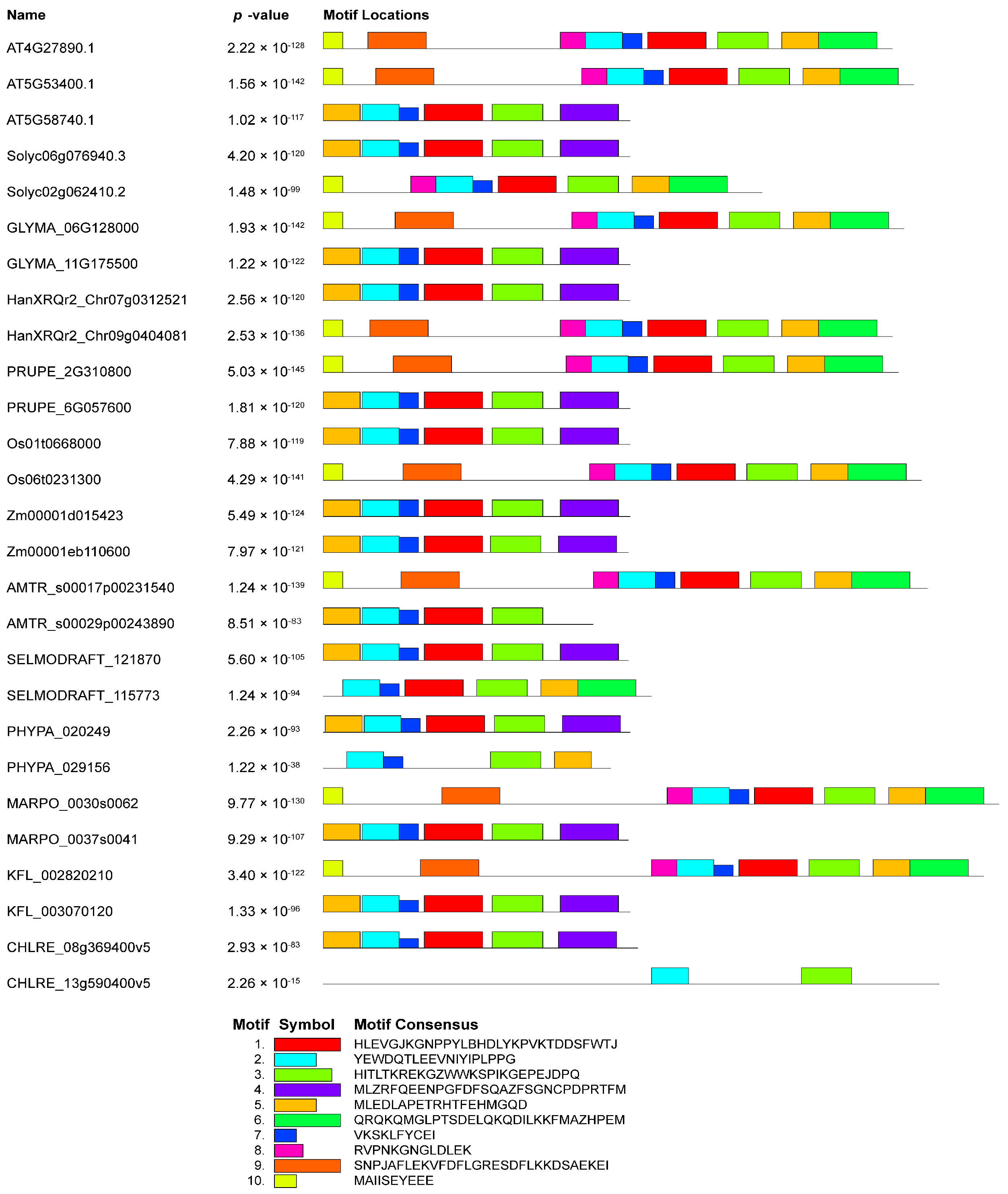
3.1. Structural Features of Plant NudC Members
3.2. Developmental Role of Plant NudC Proteins
3.3. Unveiling the Role of NudC Proteins in Plant Stress Response and Resilience
4. Future Avenues of Research on NudC Proteins in Plants
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Morris, N.R. Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 2000, 148, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Fischer, R.; Xiang, X. Accumulation of cytoplasmic dynein and dynactin at microtubule plus ends in Aspergillus nidulans is kinesin dependent. Mol. Biol. Cell 2003, 14, 1479–1488. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Beckwith, S.M.; Morris, N.R. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1994, 91, 2100–2104. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; Xiang, X.; Dawe, A.L.; Morris, N.R. Deletion of nudC, a nuclear migration gene of Aspergillus nidulans, causes morphological and cell wall abnormalities and is lethal. Mol. Biol. Cell 1997, 8, 1735–1749. [Google Scholar] [CrossRef] [PubMed]
- Xiang, X.; Roghi, C.; Morris, N.R. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1995, 92, 9890–9894. [Google Scholar] [CrossRef]
- Osmani, A.H.; Osmani, S.A.; Morris, N.R. The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J. Cell Biol. 1990, 111, 543–551. [Google Scholar] [CrossRef]
- Fath, K.R.; Trimbur, G.M.; Burgess, D.R. Molecular motors and a spectrin matrix associate with Golgi membranes in vitro. J. Cell Biol. 1997, 139, 1169–1181. [Google Scholar] [CrossRef]
- Kamal, A.; Goldstein, L.S. Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 2002, 14, 63–68. [Google Scholar] [CrossRef]
- Aniento, F.; Emans, N.; Griffiths, G.; Gruenberg, J. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes. J. Cell Biol. 1993, 123, 1373–1387. [Google Scholar] [CrossRef] [PubMed]
- Pfarr, C.M.; Coue, M.; Grissom, P.M.; Hays, T.S.; Porter, M.E.; McIntosh, J.R. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature 1990, 345, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Beckwith, S.M.; Roghi, C.H.; Liu, B.; Morris, N.R. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J. Cell Biol. 1998, 143, 1239–1247. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Huang, N.N.; Clawson, G.A.; Osmani, S.A.; Pan, W.; Xin, P.; Razzaque, M.S.; Miller, B.A. Involvement of the fungal nuclear migration gene nudC human homolog in cell proliferation and mitotic spindle formation. Exp. Cell Res. 2002, 273, 73–84. [Google Scholar] [CrossRef]
- Willins, D.A.; Xiang, X.; Morris, N.R. An alpha tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics 1995, 141, 1287–1298. [Google Scholar] [CrossRef]
- Willins, D.A.; Liu, B.; Xiang, X.; Morris, N.R. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation of Aspergillus nidulans. Mol. Gen. Genet. 1997, 255, 194–200. [Google Scholar] [CrossRef]
- Morris, N.R.; Efimov, V.P.; Xiang, X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998, 8, 467–470. [Google Scholar] [CrossRef]
- Dobyns, W.B.; Reiner, O.; Carrozzo, R.; Ledbetter, D.H. Lissencephaly. A human brain malformation associated with deletion of the LIS1 gene located at chromosome 17p13. JAMA 1993, 270, 2838–2842. [Google Scholar] [CrossRef] [PubMed]
- Hirotsune, S.; Fleck, M.W.; Gambello, M.J.; Bix, G.J.; Chen, A.; Clark, G.D.; Ledbetter, D.H.; McBain, C.J.; Wynshaw-Boris, A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 1998, 19, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Faulkner, N.E.; Dujardin, D.L.; Tai, C.Y.; Vaughan, K.T.; O’Connell, C.B.; Wang, Y.L.; Vallee, R.B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000, 2, 784–791. [Google Scholar] [CrossRef] [PubMed]
- Siller, K.H.; Serr, M.; Steward, R.; Hays, T.S.; Doe, C.Q. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell 2005, 16, 5127–5140. [Google Scholar] [CrossRef] [PubMed]
- Yingling, J.; Youn, Y.H.; Darling, D.; Toyo-Oka, K.; Pramparo, T.; Hirotsune, S.; Wynshaw-Boris, A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell 2008, 132, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.W.; Lian, W.N.; Kemal, S.; Kriegstein, A.R.; Vallee, R.B. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 2010, 13, 1463–1471. [Google Scholar] [CrossRef]
- Efimov, V.P.; Morris, N.R. The Lis1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 2000, 150, 681–688. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tsai, M.Y.; Wang, S.; Lu, B.; Chen, R.; Iii, J.R.; Zhu, X.; Zheng, Y. Requirement for Nudel and dynein for assembly of the lamin B spindle matrix. Nat. Cell Biol. 2009, 11, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Stehman, S.A.; Chen, Y.; McKenney, R.J.; Vallee, R.B. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 2007, 178, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Walsh, C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron 2004, 44, 279–293. [Google Scholar] [CrossRef]
- Sasaki, S.; Mori, D.; Toyo-Oka, K.; Chen, A.; Garrett-Beal, L.; Muramatsu, M.; Miyagawa, S.; Hiraiwa, N.; Yoshiki, A.; Wynshaw-Boris, A.; et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 2005, 25, 7812–7827. [Google Scholar] [CrossRef]
- Alkuraya, F.S.; Cai, X.; Emery, C.; Mochida, G.H.; Al-Dosari, M.S.; Felie, J.M.; Hill, R.S.; Barry, B.J.; Partlow, J.N.; Gascon, G.G.; et al. Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am. J. Med. Genet. 2011, 88, 536–547. [Google Scholar] [CrossRef]
- McKenney, R.J.; Vershinin, M.; Kunwar, A.; Vallee, R.B.; Gross, S.P. LIS1 and NudE induce a persistent dynein force-producing state. Cell 2010, 141, 304–314. [Google Scholar] [CrossRef]
- Feng, Y.; Olson, E.C.; Stukenberg, P.T.; Flanagan, L.A.; Kirschner, M.W.; Walsh, C.A. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron 2000, 28, 665–679. [Google Scholar] [CrossRef]
- Xiang, X.; Zuo, W.; Efimov, V.P.; Morris, N.R. Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr. Genet. 1999, 35, 626–630. [Google Scholar] [CrossRef]
- Xiang, X.; Han, G.; Winkelmann, D.A.; Zuo, W.; Morris, N.R. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein Arp1. Curr. Biol. 2000, 10, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; Morris, N.R. Extragenic suppressors of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans. Genetics 1995, 141, 453–464. [Google Scholar] [CrossRef] [PubMed]
- Chiu, Y.H.; Morris, N.R. Genetic and molecular analysis of a tRNALeu missense suppressor of nudC3, a mutation that blocks nuclear migration in Aspergillus nidulans. Genetics 1997, 145, 707–714. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wang, W.; Zhou, T.; Yang, Y. Emerging roles of NudC family: From molecular regulation to clinical implications. Sci. China Life Sci. 2016, 59, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Dawe, A.L.; Caldwell, K.A.; Harris, P.M.; Morris, N.R.; Caldwell, G.A. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev. Genes Evol. 2001, 211, 434–441. [Google Scholar] [CrossRef] [PubMed]
- Cunniff, J.; Chiu, Y.H.; Morris, N.R.; Warrior, R. Characterization of DnudC, the Drosophila homolog of an Aspergillus gene that functions in nuclear motility. Mech Dev. 1997, 66, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Moreau, N.; Aumais, J.P.; Prudhomme, C.; Morris, S.M.; Yu-Lee, L.Y. NUDC expression during amphibian development. Int. J. Dev. Biol. 2001, 45, 839–843. [Google Scholar] [PubMed]
- Jurkuta, R.J.; Kaplinsky, N.J.; Spindel, J.E.; Barton, M.K. Partitioning the apical domain of the Arabidopsis embryo requires the BOBBER1 NudC domain protein. Plant Cell 2009, 21, 1957–1971. [Google Scholar] [CrossRef][Green Version]
- Perez, D.E.; Hoyer, J.S.; Johnson, A.I.; Moody, Z.R.; Lopez, J.; Kaplinsky, N.J. BOBBER1 is a noncanonical Arabidopsis small heat shock protein required for both development and thermotolerance. Plant Physiol. 2009, 151, 241–252. [Google Scholar] [CrossRef]
- Velinov, V.; Vaseva, I.; Zehirov, G.; Zhiponova, M.; Georgieva, M.; Vangheluwe, N.; Beeckman, T.; Vassileva, V. Overexpression of the NMig1 gene encoding a NudC domain protein enhances root growth and abiotic stress tolerance in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 815. [Google Scholar] [CrossRef]
- Riera, J.; Lazo, P.S. The mammalian NudC-like genes: A family with functions other than regulating nuclear distribution. Cell. Mol. Life Sci. 2009, 66, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Axtell, S.M.; Truong, T.M.; O’Neal, K.D.; Yu-Lee, L.Y. Characterization of a prolactin-inducible gene, clone 15, in T cells. Mol. Endocrinol. 1995, 9, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.A.; Zhang, M.Y.; Gocke, C.D.; De Souza, C.; Osmani, A.H.; Lynch, C.; Davies, J.; Bell, L.; Osmani, S.A. A homolog of the fungal nuclear migration gene nudC is involved in normal and malignant human hematopoiesis. Exp. Hematol. 1999, 27, 742–750. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Ledbetter, D.H. Molecular cloning and characterization of the human NUDC gene. Hum Genet. 1999, 104, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Tseng, H.C.; Sapir, T.; Stern, P.; Zhou, Y.; Sanada, K.; Fischer, A.; Coquelle, F.M.; Reiner, O.; Tsai, L.H. Doublecortin-like kinase controls neurogenesis by regulating mitotic spindles and M phase progression. Neuron 2006, 49, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zimmerman, W.; Liu, X.; Erikson, R.L. A mammalian NudC-like protein essential for dynein stability and cell viability. Proc. Natl. Acad. Sci. USA 2006, 103, 9039–9044. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yan, X.; Cai, Y.; Lu, Y.; Si, J.; Zhou, T. NudC-like protein 2 regulates the LIS1/dynein pathway by stabilizing LIS1 with Hsp90. Proc. Natl. Acad. Sci. USA 2010, 107, 3499–3504. [Google Scholar] [CrossRef]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef]
- Garcia-Ranea, J.A.; Mirey, G.; Camonis, J.; Valencia, A. p23 and HSP20/alpha-crystallin proteins define a conserved sequence domain present in other eukaryotic protein families. FEBS Lett. 2002, 529, 162–167. [Google Scholar] [CrossRef]
- Zheng, M.; Cierpicki, T.; Burdette, A.J.; Utepbergenov, D.; Janczyk, P.; Derewenda, U.; Stukenberg, P.T.; Caldwell, K.A.; Derewenda, Z.S. Structural features and chaperone activity of the NudC protein family. J. Mol. Biol. 2011, 409, 722–741. [Google Scholar] [CrossRef]
- Gocke, C.D.; Osmani, S.A.; Miller, B.A. The human homologue of the Aspergillus nuclear migration gene nudC is preferentially expressed in dividing cells and ciliated epithelia. Histochem. Cell Biol. 2000, 114, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Aumais, J.P.; Liu, X.; Yu-Lee, L.Y.; Erikson, R.L. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev. Cell 2003, 5, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Aumais, J.P.; Williams, S.N.; Luo, W.; Nishino, M.; Caldwell, K.A.; Caldwell, G.A.; Lin, S.H.; Yu-Lee, L.Y. Role for NudC, a dynein-associated nuclear movement protein, in mitosis and cytokinesis. J. Cell Sci. 2003, 116, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Chuang, C.; Pan, J.; Hawke, D.H.; Lin, S.-H.; Yu-Lee, L. NudC deacetylation regulates mitotic progression. PLoS ONE 2013, 8, e73841. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, W.; Lu, Y.; Yan, X.; Yan, X.; Zhu, X.; Liu, W.; Yang, Y.; Zhou, T. NudC regulates actin dynamics and ciliogenesis by stabilizing cofilin 1. Cell Res. 2016, 26, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.M.; Albrecht, U.; Reiner, O.; Eichele, G.; Yu-Lee, L.Y. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr. Biol. 1998, 8, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Jacob, J.; Michowski, W.; Nowotny, M.; Kuznicki, J.; Chazin, W.J. Human Sgt1 binds HSP90 through the CHORD-Sgt1 domain and not the tetratricopeptide repeat domain. J. Biol. Chem. 2004, 279, 16511–16517. [Google Scholar] [CrossRef]
- Botër, M.; Amigues, B.; Peart, J.; Breuer, C.; Kadota, Y.; Casais, C.; Moore, G.; Kleanthous, C.; Ochsenbein, F.; Shirasu, K.; et al. Structural and functional analysis of SGT1 reveals that its interaction with HSP90 is required for the accumulation of Rx, an R protein involved in plant immunity. Plant Cell 2007, 19, 3791–3804. [Google Scholar] [CrossRef]
- Echtenkamp, F.J.; Zelin, E.; Oxelmark, E.; Woo, J.I.; Andrews, B.J.; Garabedian, M.; Freeman, B.C. Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol. Cell 2011, 43, 229–241. [Google Scholar] [CrossRef]
- Taipale, M.; Tucker, G.; Peng, J.; Krykbaeva, I.; Lin, Z.Y.; Larsen, B.; Choi, H.; Berger, B.; Gingras, A.C.; Lindquist, S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 2014, 158, 434–448. [Google Scholar] [CrossRef]
- Faircloth, L.M.; Churchill, P.F.; Caldwell, G.A.; Caldwell, K.A. The microtubule-associated protein, NUD-1, exhibits chaperone activity in vitro. Cell Stress Chaperones 2009, 14, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.J.; Liu, X.; Jin, Q.; Cai, Y.; Yang, Y.; Zhou, T. The L279P mutation of nuclear distribution gene C (NudC) influences its chaperone activity and lissencephaly protein 1 (LIS1) stability. J. Biol. Chem. 2010, 285, 29903–29910. [Google Scholar] [CrossRef] [PubMed]
- Biebl, M.M.; Delhommel, F.; Faust, O.; Zak, K.M.; Agam, G.; Guo, X.; Mühlhofer, M.; Dahiya, V.; Hillebrand, D.; Popowicz, G.M.; et al. NudC guides client transfer between the Hsp40/70 and Hsp90 chaperone systems. Mol. Cell 2022, 82, 555–569. [Google Scholar] [CrossRef]
- Biebl, M.M.; Lopez, A.; Rehn, A.; Freiburger, L.; Lawatscheck, J.; Blank, B.; Sattler, M.; Buchner, J. Structural elements in the flexible tail of the co-chaperone p23 coordinate client binding and progression of the Hsp90 chaperone cycle. Nat. Commun. 2021, 12, 828. [Google Scholar] [CrossRef] [PubMed]
- López, A.; Elimelech, A.R.; Klimm, K.; Sattler, M. The charged linker modulates the conformations and molecular interactions of Hsp90. ChemBioChem 2021, 22, 1084–1092. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, N.; Kanamaru, K.; Ueno, Y.; Kojima, S.; Kobayashi, T.; Machida, C.; Machida, Y. ASYMMETRIC-LEAVES2 and an ortholog of eukaryotic NudC domain proteins repress expression of AUXIN RESPONSE-FACTOR and class 1 KNOX homeobox genes for development of flat symmetric leaves in Arabidopsis. Biol. Open 2012, 1, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Silverblatt-Buser, E.W.; Frick, M.A.; Rabeler, C.; Kaplinsky, N.J. Genetic interactions between BOB1 and multiple 26S proteasome subunits suggest a role for proteostasis in regulating Arabidopsis development. G3-Genes Genom. Genet. 2018, 8, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, J.; Jiang, S.; Wang, H.; Gao, Y.; Zhang, H.; Li, D.; Song, F. Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2019, 20, 815–830. [Google Scholar] [CrossRef]
- Velinov, V.; Georgieva, M.; Zehirov, G.; Vassileva, V. NudC-like genes contribute to root growth and branching in Arabidopsis thaliana. Comptes Rendus Acad. Bulg. Sci. 2021, 74, 1767–1773. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Kaplinsky, N.J. Temperature compensation of auxin dependent developmental patterning. Plant Signal. Behav. 2009, 4, 1157–1158. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iwakawa, H.; Ueno, Y.; Semiarti, E.; Onouchi, H.; Kojima, S.; Tsukaya, H.; Hasebe, M.; Soma, T.; Ikezaki, M.; Machida, C.; et al. 2002. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002, 43, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, M.; Takahashi, H.; Iwakawa, H.; Nakagawa, A.; Ishikawa, T.; Tanaka, H.; Matsumura, Y.; Pekker, I.; Eshed, Y.; Vial-Pradel, S.; et al. Dual regulation of ETTIN (ARF3) gene expression by AS1-AS2, which maintains the DNA methylation level, is involved in stabilization of leaf adaxial-abaxial partitioning in Arabidopsis. Development 2013, 140, 1958–1969. [Google Scholar] [CrossRef] [PubMed]
- Machida, C.; Nakagawa, A.; Kojima, S.; Takahashi, H.; Machida, Y. The complex of ASYMMETRIC LEAVES (AS) proteins plays a central role in antagonistic interactions of genes for leaf polarity specification in Arabidopsis. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 655–671. [Google Scholar] [CrossRef] [PubMed]
- Studer, S.; Narberhaus, F. Chaperone activity and homo-and hetero-oligomer formation of bacterial small heat shock proteins. J. Biol. Chem. 2000, 275, 37212–37218. [Google Scholar] [CrossRef] [PubMed]
- Eisenhardt, B.D. Small heat shock proteins: Recent developments. BioMol. Concepts 2013, 4, 583–595. [Google Scholar] [CrossRef]
- Boelens, W.C.; Croes, Y.; de Ruwe, M.; de Reu, L.; de Jong, W.W. Negative charges in the C-terminal domain stabilize the αB-crystallin complex. J. Biol. Chem. 1998, 273, 28085–28090. [Google Scholar] [CrossRef]
- Haslbeck, M.; Franzmann, T.; Weinfurtner, D.; Buchner, J. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005, 12, 842–846. [Google Scholar] [CrossRef]
- Bondino, G.H.; Valle, M.E.; Ten Have, A. Evolution and functional diversification of the small heat shock protein/α-crystallin family in higher plants. Planta 2012, 235, 1299–1313. [Google Scholar] [CrossRef] [PubMed]
- Paul, A.; Rao, S.; Mathur, S. The α-crystallin domain containing genes: Identification, phylogeny and expression profiling in abiotic stress, phytohormone response and development in tomato (Solanum lycopersicum). Front. Plant Sci. 2016, 7, 426. [Google Scholar] [CrossRef] [PubMed]
- Wallace, E.W.J.; Kear-Scott, J.L.; Pilipenko, E.V.; Schwartz, M.H.; Laskowski, P.R.; Rojek, A.E.; Katanski, C.D.; Riback, J.A.; Dion, M.F.; Franks, A.M.; et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015, 162, 1286–1298. [Google Scholar] [CrossRef]
- Kirschner, M.; Winkelhaus, S.; Thierfelder, J.M.; Nover, L. Transient expression and heat-stress-induced co-aggregation of endogenous and heterologous small heat-stress proteins in tobacco protoplasts. Plant J. 2000, 24, 397–412. [Google Scholar] [CrossRef] [PubMed]
- Miroshnichenko, S.; Tripp, J.; Nieden, U.Z.; Neumann, D.; Conrad, U.; Manteuffel, R. Immunomodulation of function of small heat shock proteins prevents their assembly into heat stress granules and results in cell death at sublethal temperatures. Plant J. 2005, 41, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Heider, H.; Hohfeld, I.; Lyck, R.; Schmidt, E.; Nover, L. The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell Biol. 1998, 18, 2240–2251. [Google Scholar] [CrossRef] [PubMed]
- Siddique, M.; Gernhard, S.; von Koskull-Doring, P.; Vierling, E.; Scharf, K.D. The plant sHSP superfamily: Five new members in Arabidopsis thaliana with unexpected properties. Cell Stress Chaperones 2008, 13, 183–197. [Google Scholar] [CrossRef]
- Lawrence, R.; Kaplinsky, N. Arabidopsis Heat Shock Granules exhibit dynamic cellular behavior and can form in response to protein misfolding in the absence of elevated temperatures. MicroPubl Biol. 2020, 2020. [Google Scholar] [CrossRef]
- Shang, L.; Zhou, Y.; Wen, S.; Wang, K.; Li, Y.; Zhang, M.; Jian, H.; Lyu, D. Construction of heat stress regulation networks based on Illumina and SMRT sequencing data in potato. Front. Plant Sci. 2023, 14, 1271084. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vassileva, V.; Georgieva, M.; Todorov, D.; Mishev, K. Small Sized Yet Powerful: Nuclear Distribution C Proteins in Plants. Plants 2024, 13, 119. https://doi.org/10.3390/plants13010119
Vassileva V, Georgieva M, Todorov D, Mishev K. Small Sized Yet Powerful: Nuclear Distribution C Proteins in Plants. Plants. 2024; 13(1):119. https://doi.org/10.3390/plants13010119
Chicago/Turabian StyleVassileva, Valya, Mariyana Georgieva, Dimitar Todorov, and Kiril Mishev. 2024. "Small Sized Yet Powerful: Nuclear Distribution C Proteins in Plants" Plants 13, no. 1: 119. https://doi.org/10.3390/plants13010119
APA StyleVassileva, V., Georgieva, M., Todorov, D., & Mishev, K. (2024). Small Sized Yet Powerful: Nuclear Distribution C Proteins in Plants. Plants, 13(1), 119. https://doi.org/10.3390/plants13010119








