Artificial Neural Network and Response Surface-Based Combined Approach to Optimize the Oil Content of Ocimum basilicum var. thyrsiflora (Thai Basil)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Station
2.2. Extraction of Essential Oil and Quantification
2.3. Quantitative Analysis of Soil
2.4. Data Exploration
2.5. Data Splitting
2.6. Artificial Neural Network Model Development
2.7. Model Evaluation and Selection
2.8. Variable Importance
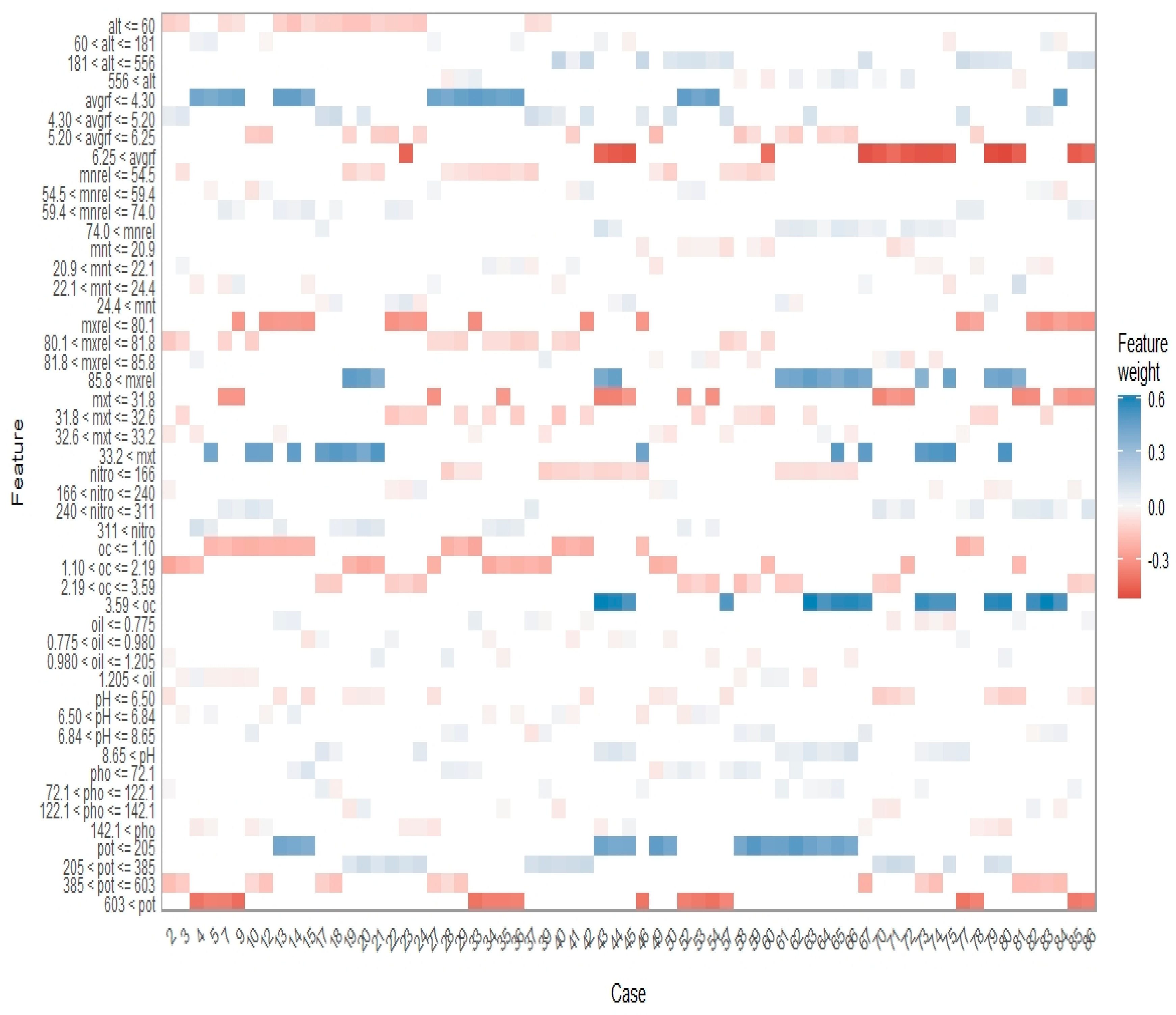
2.9. Partial Dependence Plots and Faceted Heatmap
2.10. Sensitivity Analysis
3. Results and Discussion
3.1. Model Evaluation and Selection
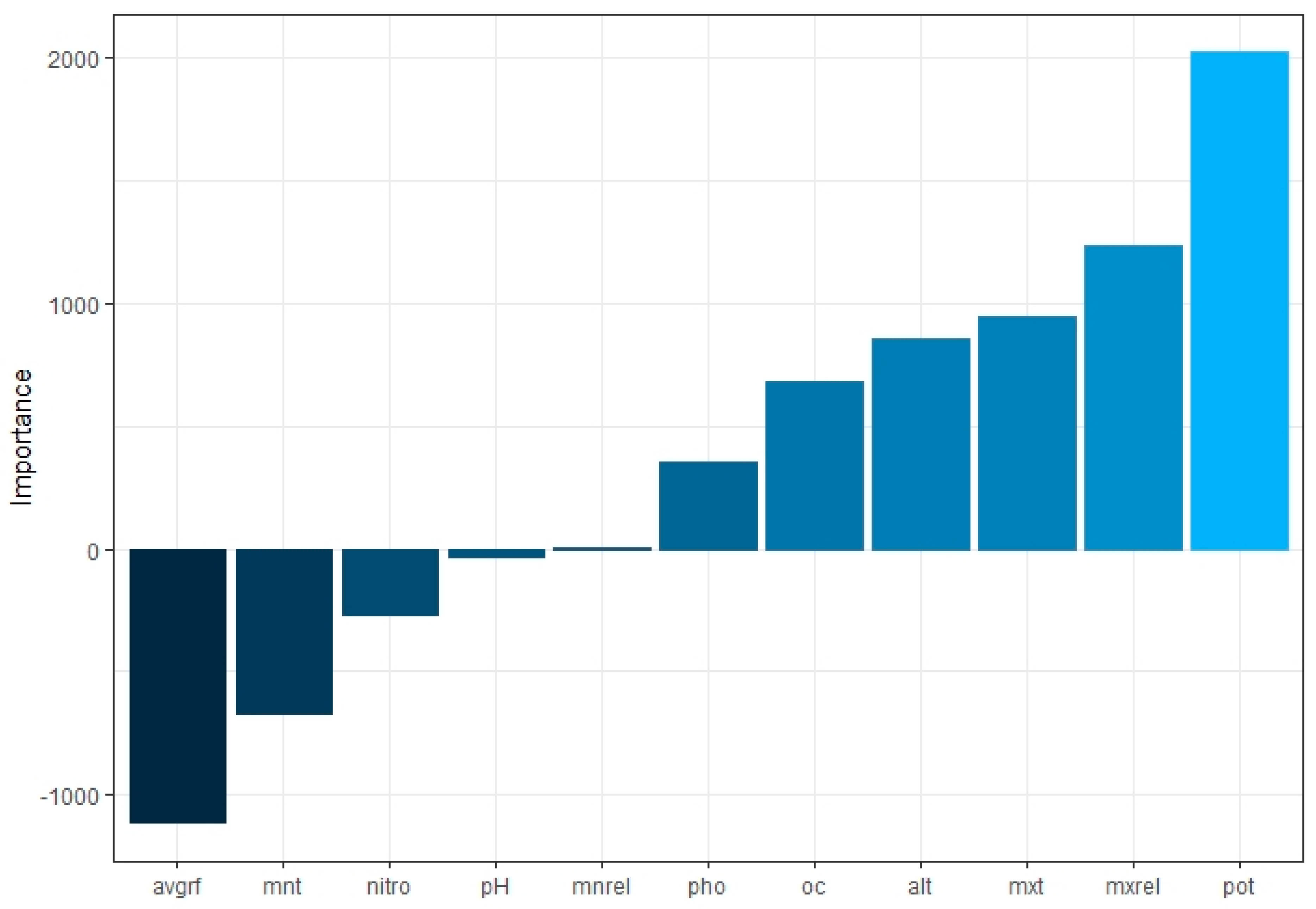
3.2. Significant Predictor Identification
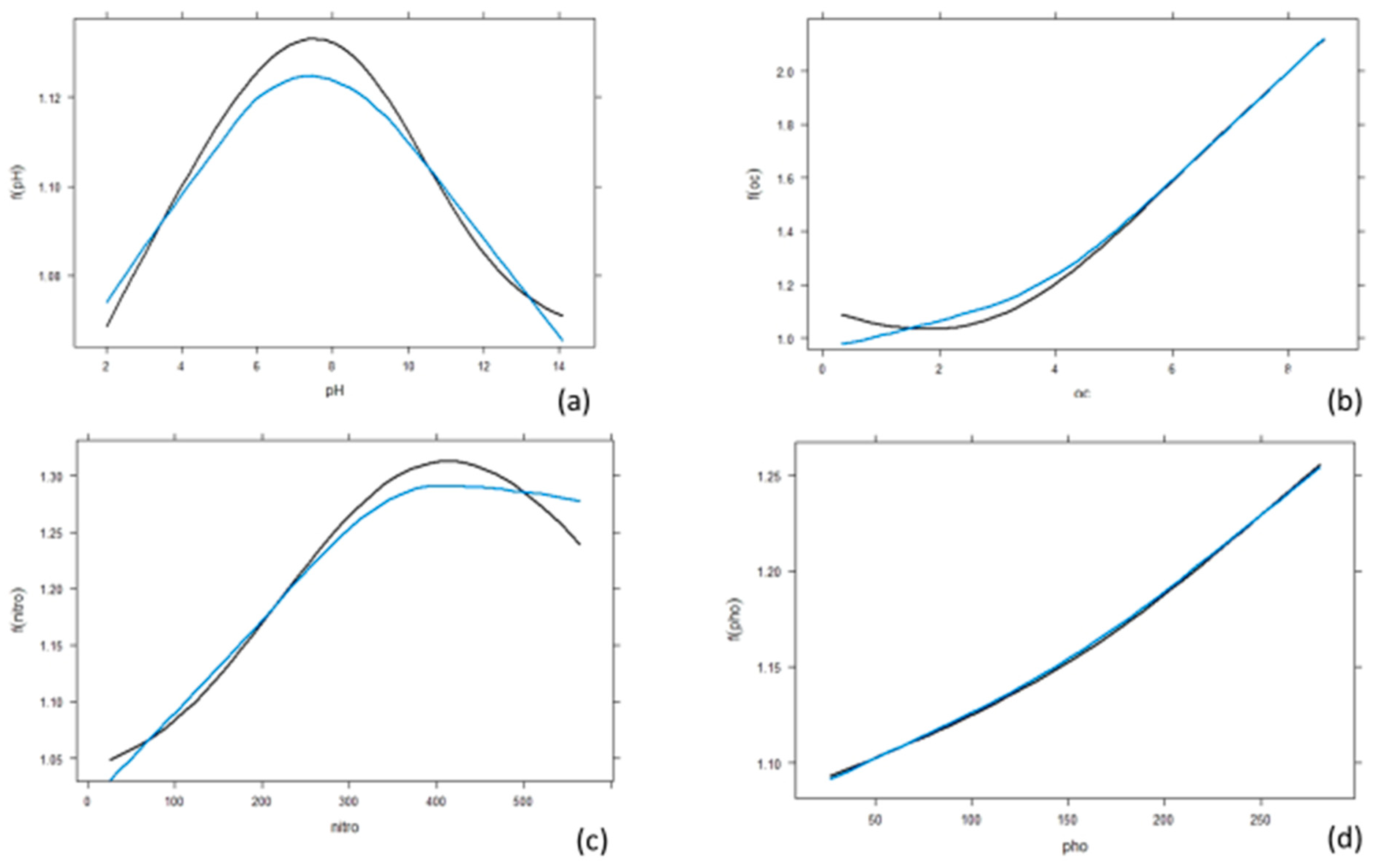
3.3. Effect of Individual Predictors on Essential Oil Content
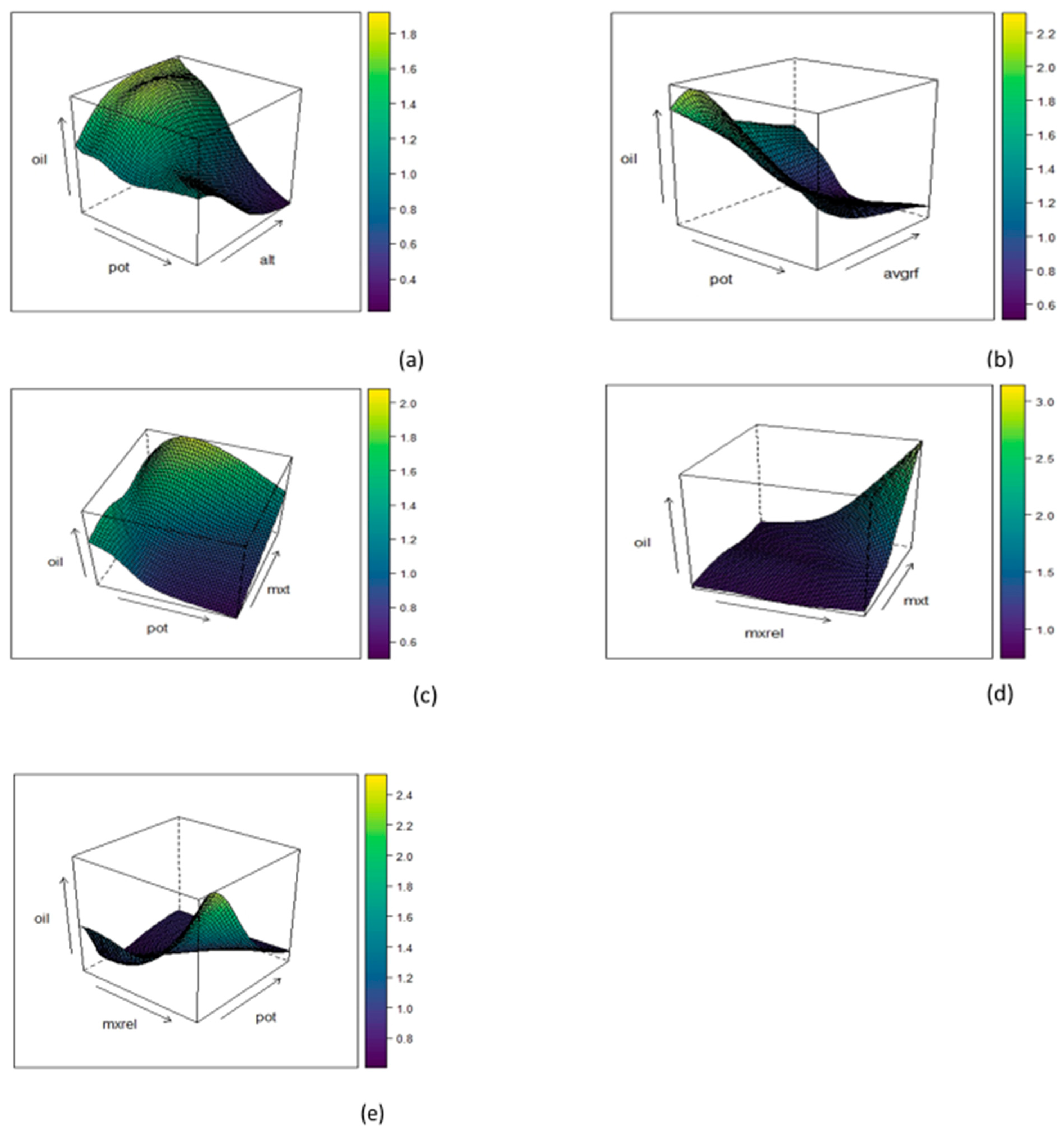
3.4. Mutual Effect of Two Predictors on Response
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Sample Code
- library(readxl)
- library(caret)
- data<-as.data.frame(read_xlsx(“tb.xlsx”))
- #calculate standard deviation
- library(mlbench)
- library(“openxlsx”)
- # calculate standard deviation for all attributes
- sd.table<-sapply(data[,1:12], sd)
- sd.table<-as.data.frame(sd.table)
- write.xlsx(sd.table,”Standarddeviation.xlsx”)
- ######Correlations analysis
- correlations <- cor(data[,1:10])
- # display the correlation matrix
- cor.table<-print(correlations)
- cor.table
- write.xlsx(cor.table,”Correlations.xlsx”)
- #creating panel plot
- library(psych)
- pairs.panels(data[c(“pH”, “oc”, “nitro”, “pho”,”pot”,”mxrel”,”mnrel”,”avgrf”,”mxt”,”mnt”,”alt”)])
- #############data splitting ###########
- set.seed(212)
- trainIndex<- createDataPartition(data$oil, p=0.8, list=FALSE)
- train <- data[ trainIndex,] ####80% of total data including validation
- test<- data[-trainIndex,]
- validIndex<- createDataPartition(data$oil, p=0.9, list=FALSE)
- train <- data[ validIndex,] ####70% of total data
- valid<- data[-validIndex,] ######10% of total data
- #######ANN model development######
- trainControl <- trainControl(method=“cv”, number=15)
- tunegrid<- expand.grid(layer1 = 16, layer2 =15, layer3=4)
- metric <- “MAE”
- set.seed(123)
- fit.ann<-train(oil~., data = train, method=“neuralnet”, metric = metric, preProcess=c(“range”), tuneGrid = tunegrid, act.fct=“logistic”,trControl = trainControl,learningrate = 0.02, linear.output=T)
- fit.ann
- ####Plotting the final model#####
- library(NeuralNetTools)
- plotnet(fit.ann$finalModel)
- ####Predicting for test set####
- pr.ann <- predict(fit.ann, newdata=test)
- pr.ann<-as.data.frame(pr.ann)
- library(“openxlsx”)
- output.table<-cbind(test$oil,pr.ann)
- output.table
- write.xlsx(output.table,’testVsPred.xlsx’)
- #####Predicting for validation set
- valid<-as.data.frame(read_xlsx(“valid.xlsx”))
- val.ann <- predict(fit.ann, newdata=valid)
- val.ann<-as.data.frame(val.ann)
- ########Variable Importance
- library(NeuralNetTools)
- olden(fit.ann)
- #####partial dependecnce plot
- #####Single variable
- library(pdp)
- library(ggplot2)
- partial(pred.var = “alt”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(alt)))
- partial(pred.var = “mnt”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(mnt)))
- partial(pred.var = “mxt”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(mxt)))
- partial(pred.var = “mxrel”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(mxrelhum)))
- partial(pred.var = “avgrf”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(avgrainfall)))
- partial(pred.var = “mnrel”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(mnrel)))
- partial(pred.var = “pot”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(pot)))
- partial(pred.var = “pho”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(pho)))
- partial(pred.var = “nitro”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(nitro)))
- partial(pred.var = “oc”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(oc)))
- partial(pred.var = “pH”) %>%
- plotPartial(smooth = T, lwd = 2, ylab = expression(f(pH)))
- ####without smoothing
- fit.ann %>% # the %>% operator is read as “and then”
- partial(pred.var = “alt”) %>%
- plotPartial(smooth = F, lwd = 2, ylab = expression(f(alt)))
- ######with two predictors#######
- ######pot mxrel
- par.mxrel.pot <- partial(fit.ann, pred.var = c(“mxrel”, “pot”))
- plotPartial(par.mxrel.pot)
- # Add contour lines and use a different color palette
- rwb <- colorRampPalette(c(“red”, “white”, “blue”))
- pdp2 <- plotPartial(par.mxrel.pot, contour = TRUE, col.regions = rwb)
- pdp2
- # 3-D surface
- pdp3 <- plotPartial(par.mxrel.pot,rug=T,levelplot = FALSE,number=4, zlab = “oil”, drape = TRUE,colorkey = TRUE, screen = list(z =−40, x = −60))
- pdp3
- ###Maximum rel humidity and Max. avg. temperature
- par.mxrel.mxt <- partial(fit.ann, pred.var = c(“mxrel”, “mxt”))
- plotPartial(par.mxrel.mxt)
- # Add contour lines and use a different color palette
- rwb <- colorRampPalette(c(“red”, “white”, “blue”))
- pdp.mxrel.mxt.2d <- plotPartial(par.mxrel.mxt, contour = TRUE, col.regions = rwb)
- pdp.mxrel.mxt.2d
- # 3-D surface
- pdp.mxrel.mxt.3d <- plotPartial(par.mxrel.mxt,rug=T,levelplot = FALSE,number=4, zlab = “oil”, drape = TRUE,colorkey = TRUE, screen = list(z = −20, x = −60))
- pdp.mxrel.mxt.3d
- ###pot mxt
- par.pot.mxt <- partial(fit.ann, pred.var = c(“pot”, “mxt”))
- plotPartial(par.pot.mxt)
- # Add contour lines and use a different color palette
- rwb <- colorRampPalette(c(“red”, “white”, “blue”))
- pdp.pot.mxt.2d <- plotPartial(par.pot.mxt, contour = TRUE, col.regions = rwb)
- pdp.pot.mxt.2d
- # 3-D surface
- pdp.pot.mxt.3d <- plotPartial(par.pot.mxt,rug=T,levelplot = FALSE,number=4, zlab = “oil”, drape = TRUE,colorkey = TRUE, screen = list(z = −20, x = −40))
- pdp.pot.mxt.3d
- ######pot avgrf
- par.pot.avgrf <- partial(fit.ann, pred.var = c(“pot”, “avgrf”))
- plotPartial(par.pot.avgrf)
- # Add contour lines and use a different color palette
- rwb <- colorRampPalette(c(“red”, “white”, “blue”))
- pdp.pot.avgrf.2d <- plotPartial(par.pot.avgrf, contour = TRUE, col.regions = rwb)
- pdp.pot.avgrf.2d
- # 3-D surface
- pdp.pot.avgrf.3d <- plotPartial(par.pot.avgrf,rug=T,levelplot = FALSE,number=4, zlab = “oil”, drape = TRUE,colorkey = TRUE, screen = list(z = −40, x = −70))
- pdp.pot.avgrf.3d
- ######pot alt
- par.pot.alt <- partial(fit.ann, pred.var = c(“pot”, “alt”))
- plotPartial(par.pot.alt)
- # Add contour lines and use a different color palette
- rwb <- colorRampPalette(c(“red”, “white”, “blue”))
- pdp.pot.alt.2d <- plotPartial(par.pot.alt, contour = TRUE, col.regions = rwb)
- pdp.pot.alt.2d
- # 3-D surface
- pdp.pot.alt.3d <- plotPartial(par.pot.alt,rug=T,levelplot = FALSE,number=4, zlab = “oil”, drape = TRUE,colorkey = TRUE, screen = list(z = −40, x = −60))
- pdp.pot.alt.3d
- ##### Faceted heatmap#########
- library(lime)
- lime.fit<-lime(train,fit.ann)
- exp.fit<-explain(train,lime.fit,n_features=10)
- plot_explanations(exp.fit)
- plot_features(exp.fit)
References
- Dhama, K.; Sharun, K.; Gugjoo, M.B.; Tiwari, R.; Alagawany, M.; Yatoo, M.I.; Thakur, P.; Iqbal, H.M.; Chaicumpa, W.; Michalak, I.; et al. A Comprehensive Review on Chemical Profile and Pharmacological Activities of Ocimum basilicum. Food Rev. Int. 2021, 24, 1–29. [Google Scholar] [CrossRef]
- Purushothaman, B.; Srinivasan, R.P.; Suganthi, P.; Ranganathan, B.; Gimbun, J.; Shanmugam, K. A Comprehensive Review on Ocimum basilicum. J. Nat. Remedies 2018, 18, 71–85. [Google Scholar] [CrossRef]
- Bora, K.S.; Arora, S.; Shri, R. Role of Ocimum basilicum L. in prevention of ischemia and reperfusion-induced cerebral damage, and motor dysfunctions in mice brain. J. Ethnopharmacol. 2011, 137, 1360–1365. [Google Scholar] [CrossRef] [PubMed]
- Łyczko, J.; Masztalerz, K.; Lipan, L.; Lech, K.; Carbonell-Barrachina, Á.A.; Szumny, A. Chemical determinants of dried Thai basil (O. basilicum var. thyrsiflora) aroma quality. Ind. Crops Prod. 2020, 155, 112769. [Google Scholar] [CrossRef]
- Chiang, L.-C.; Ng, L.-T.; Cheng, P.-W.; Chiang, W.; Lin, C.-C. Antiviral activities of extracts and selected pure constituents of Ocimum basilicum. Clin. Exp. Pharmacol. Physiol. 2005, 32, 811–816. [Google Scholar] [CrossRef]
- Selvakkumar, C.; Gayathri, B.; Vinaykumar, K.S.; Lakshmi, B.S.; Balakrishnan, A. Potential Anti-inflammatory Properties of Crude Alcoholic Extract of Ocimum basilicum L. in Human Peripheral Blood Mononuclear Cells. J. Health Sci. 2007, 53, 500–505. [Google Scholar] [CrossRef]
- Bravo, E.; Amrani, S.; Aziz, M.; Harnafi, H.; Napolitano, M. Ocimum basilicum ethanolic extract decreases cholesterol synthesis and lipid accumulation in human macrophages. Fitoterapia 2008, 79, 515–523. [Google Scholar] [CrossRef]
- Berić, T.; Nikolić, B.; Stanojević, J.; Vuković-Gačić, B.; Knežević-Vukčević, J. Protective effect of basil (Ocimum basilicum L.) against oxidative DNA damage and mutagenesis. Food Chem. Toxicol. 2008, 46, 724–732. [Google Scholar] [CrossRef]
- Tohti, I.; Tursun, M.; Umar, A.; Turdi, S.; Imin, H.; Moore, N. Aqueous extracts of Ocimum basilicum L. (sweet basil) decrease platelet aggregation induced by ADP and thrombin in vitro and rats arterio–venous shunt thrombosis in vivo. Thromb. Res. 2006, 118, 733–739. [Google Scholar] [CrossRef]
- Harnafi, H.; Caid, H.S.; Bouanani, N.E.H.; Aziz, M.; Amrani, S. Hypolipemic activity of polyphenol-rich extracts from Ocimum basilicum in Triton WR-1339-induced hyperlipidemic mice. Food Chem. 2008, 108, 205–212. [Google Scholar] [CrossRef]
- Available online: https://connect2india.com/global/Essential-Oil-Basil-export-from-india/1 (accessed on 25 May 2022).
- Rawat, R.; Tiwari, V.; Singh, R.; Bisht, I. Assessment of the Essential Oil Composition in Ocimum species of Uttarakhand. J. Essent. Oil Bear. Plants 2017, 20, 1331–1341. [Google Scholar] [CrossRef]
- Radusiene, J.; Stanius, Z.; Cirak, C.; Odabas, M. Quantitative effects of temperature and light intensity on the accumulation of bioactive compounds in St. John’s worth. In XXVIII International Horticultural Congress on Science and Horticulture for People (IHC2010): A New Look at Medicinal and Aromatic Plants Seminar; International Society for Horticultural Science: Angers, France, 2010; pp. 135–140. [Google Scholar]
- Saffariha, M.; Jahani, A.; Jahani, R. A comparison of artificial intelligence techniques for predicting hyperforin content in Hypericum perforatum L. in different ecological habitats. Plant Direct 2021, 5, e363. [Google Scholar] [CrossRef] [PubMed]
- Gopal, P.M.; Bhargavi, R. A novel approach for efficient crop yield prediction. Comput. Electron. Agric. 2019, 165, 104968. [Google Scholar] [CrossRef]
- Torkashvand Moradabadi, M. Sensitivity Analysis and Reexamination of the Techniques for Evaluating Adult Death Registration. Ph.D. Thesis, Tehran University, Tehran, Iran, 2017. [Google Scholar]
- Besalatpour, A.; Ayoubi, S.; Hajabbasi, M.; Mosaddeghi, M.; Schulin, R. Estimating wet soil aggregate stability from easily available properties in a highly mountainous watershed. Catena 2013, 111, 72–79. [Google Scholar] [CrossRef]
- Alam, M.A.; Naik, P.K. Impact of Soil Nutrients and Environmental Factors on Podophyllotoxin Content among 28 Podophyllum Hexandrum Populations of Northwestern Himalayan Region Using Linear and Nonlinear Approaches. Commun. Soil Sci. Plant Anal. 2009, 40, 2485–2504. [Google Scholar] [CrossRef]
- Padhiari, B.M.; Ray, A.; Champati, B.B.; Jena, S.; Sahoo, A.; Kuanar, A.; Halder, T.; Ghosh, B.; Naik, P.K.; Patnaik, J.; et al. Artificial neural network (ANN) model for prediction and optimization of bacoside A content in Bacopa monnieri: A statistical approach and experimental validation. Plant Biosyst.—Int. J. Deal. All Asp. Plant Biol. 2022, 156, 1346–1357. [Google Scholar] [CrossRef]
- Olsen, S.; Sommers, L. Phosphorus. In Methods of Soil Analysis, 2nd ed.; Page, L.A., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy: Madison, WI, USA, 1982; pp. 403–430. [Google Scholar]
- Jackson, M.L. Soil Chemical Analysis; Prentice-Hall of India Pvt. Ltd.: New Delhi, India, 1973; pp. 38–204. [Google Scholar] [CrossRef]
- Subbaiah, B. A rapid procedure for estimation of available nitrogen in soil. Curr. Sci. 1956, 25, 259–260. [Google Scholar]
- Friedrich, L. mlbench: Machine Learning Benchmark Problems. R package version 2.1-3. Available online: https://rdrr.io/cran/mlbench/ (accessed on 1 January 2023).
- Revelle, W. psych: Procedures for Personality and Psychological Research; Northwestern University: Evanston, IL, USA, 2021. [Google Scholar]
- Kuhn, M. Caret: Classification and Regression Training. R Package Version 6.0-88. 2021. Available online: https://CRAN.R-project.org/package=caret (accessed on 1 January 2022).
- Beck, M.W. NeuralNetTools: Visualization and Analysis Tools for Neural Networks. J. Stat. Softw. 2018, 85, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Olden, J.D.; Joy, M.K.; Death, R.G. An accurate comparison of methods for quantifying variable importance in artificial neural networks using simulated data. Ecol. Model. 2004, 178, 389–397. [Google Scholar] [CrossRef]
- Brandon, M.G. pdp: An R Package for Constructing Partial Dependence Plots. R J. 2017, 9, 421–436. [Google Scholar] [CrossRef]
- Muller, H.-G. Weighted Local Regression and Kernel Methods for Nonparametric Curve Fitting. J. Am. Stat. Assoc. 1987, 82, 231. [Google Scholar] [CrossRef]
- Cleveland, W.S.; Devlin, S.J. Locally Weighted Regression: An Approach to Regression Analysis by Local Fitting. J. Am. Stat. Assoc. 1988, 83, 596–610. [Google Scholar] [CrossRef]
- Lin, P.T.; Michaël, B. lime: Local Interpretable Model-Agnostic Explanations. R Package Version 0.5.2. 2021. Available online: https://CRAN.R-project.org/package=lime (accessed on 1 January 2022).
- Schober, P.; Boer, C.; Schwarte, L.A. Correlation Coefficients: Appropriate Use and Interpretation. Anesth. Analg. 2018, 126, 1763–1768. [Google Scholar] [CrossRef] [PubMed]
- Nicodemus, K.K.; Malley, J.D. Predictor correlation impacts machine learning algorithms: Implications for genomic studies. Bioinformatics 2009, 25, 1884–1890. [Google Scholar] [CrossRef] [PubMed]
- De Veaux, R.D.; Ungar, L.H. Multicollinearity: A tale of two nonparametric regressions. In Selecting Models from Data: Artificial Intelligence and Statistics IV; Springer: New York, NY, USA, 1994; pp. 393–402. [Google Scholar] [CrossRef]
- Obite, C.P.; Olewuezi, N.P.; Ugwuanyim, G.U.; Bartholomew, D.C. Multicollinearity Effect in Regression Analysis: A Feed Forward Artificial Neural Network Approach. Asian J. Probab. Stat. 2020, 6, 22–33. [Google Scholar] [CrossRef]
- Khajeh, M.; Moghaddam, M.G.; Shakeri, M. Application of artificial neural network in predicting the extraction yield of essential oils of Diplotaenia cachrydifolia by supercritical fluid extraction. J. Supercrit. Fluids 2012, 69, 91–96. [Google Scholar] [CrossRef]
- Ghoreishi, S.; Heidari, E. Extraction of Epigallocatechin-3-gallate from green tea via supercritical fluid technology: Neural network modeling and response surface optimization. J. Supercrit. Fluids 2013, 74, 128–136. [Google Scholar] [CrossRef]
- Lashkarbolooki, M.; Shafipour, Z.S.; Hezave, A.Z. Trainable cascade-forward back-propagation network modeling of spearmint oil extraction in a packed bed using SC-CO2. J. Supercrit. Fluids 2013, 73, 108–115. [Google Scholar] [CrossRef]
- Pilkington, J.L.; Preston, C.; Gomes, R.L. Comparison of response surface methodology (RSM) and artificial neural networks (ANN) towards efficient extraction of artemisinin from Artemisia annua. Ind. Crops Prod. 2014, 58, 15–24. [Google Scholar] [CrossRef]
- Alvarez, R. Predicting average regional yield and production of wheat in the Argentine Pampas by an artificial neural network approach. Eur. J. Agron. 2009, 30, 70–77. [Google Scholar] [CrossRef]
- Sodeifian, G.; Sajadian, S.A.; Ardestani, N.S. Optimization of essential oil extraction from Launaea acanthodes Boiss: Utilization of supercritical carbon dioxide and cosolvent. J. Supercrit. Fluids 2016, 116, 46–56. [Google Scholar] [CrossRef]
- Akbar, A.; Kuanar, A.; Joshi, R.K.; Sandeep, I.S.; Mohanty, S.; Naik, P.K.; Mishra, A.; Nayak, S. Development of Prediction Model and Experimental Validation in Predicting the Curcumin Content of Turmeric (Curcuma longa L.). Front. Plant Sci. 2016, 7, 1507. [Google Scholar] [CrossRef] [PubMed]
- Akbar, A.; Kuanar, A.; Patnaik, J.; Mishra, A.; Nayak, S. Application of Artificial Neural Network modeling for optimization and prediction of essential oil yield in turmeric (Curcuma longa L.). Comput. Electron. Agric. 2018, 148, 160–178. [Google Scholar] [CrossRef]
- Garson, G.D. Interpreting neural network connection weights. Artif. Intell. Expert 1991, 6, 46–51. [Google Scholar] [CrossRef]
- Goh, A. Back-propagation neural networks for modeling complex systems. Artif. Intell. Eng. 1995, 9, 143–151. [Google Scholar] [CrossRef]
- Zhang, Z.; Beck, M.W.; Winkler, D.A.; Huang, B.; Sibanda, W.; Goyal, H.; on behalf of AME Big-Data Clinical Trial Collaborative Group. Opening the black box of neural networks: Methods for interpreting neural network models in clinical applications. Ann. Transl. Med. 2018, 6, 216. [Google Scholar] [CrossRef]






| SL. No. | Agroclimatic Zones | Districts | Accession No. | Latitude | Longitude | Altitude |
|---|---|---|---|---|---|---|
| 1. | East and South East Coastal Plain | Jagatsingpur | T1 | 20.2549° N | 86.1706° E | 46 |
| T2 | 20.2553° N | 86.1735° E | 43 | |||
| T3 | 20.2555° N | 86.1740° E | 41 | |||
| Khurda | T4 | 20.1869° N | 86.1737° E | 75 | ||
| T5 | 20.1863° N | 85.6223° E | 181 | |||
| T6 | 20.1850° N | 85.6215° E | 195 | |||
| Puri | T7 | 20.1868° N | 85.6234° E | 0.1 | ||
| T8 | 19.8135° N | 85.8312° E | 74 | |||
| T9 | 19.8120° N | 85.8340° E | 85 | |||
| Nayagarh | T10 | 19.8142° N | 85.8318° E | 178 | ||
| T11 | 20.1231° N | 85.1038° E | 14 | |||
| T12 | 20.1251° N | 85.1045° E | 55 | |||
| 2. | North Eastern Coastal Plain | Bhadrak | T13 | 21.0574° N | 86.4963° E | 23 |
| T14 | 21.0570° N | 86.4968° E | 23.6 | |||
| T15 | 21.0566° N | 86.4970° E | 24 | |||
| Balasore | T16 | 21.4934° N | 86.9135° E | 16 | ||
| T17 | 21.4940° N | 86.9140° E | 16.3 | |||
| T18 | 21.4944° N | 86.9145° E | 17 | |||
| Jajpur | T19 | 20.8341° N | 86.3326° E | 8 | ||
| T20 | 20.8348° N | 86.3330° E | 9 | |||
| T21 | 20.8352° N | 86.3338° E | 10 | |||
| 3. | North Eastern Ghat | Ganjam | T22 | 19.3874° N | 85.0515° E | 3 |
| T23 | 19.3870° N | 85.0520° E | 568 | |||
| T24 | 19.3866° N | 85.0525° E | 570 | |||
| Gajapati | T25 | 19.1912° N | 84.1857° E | 180.5 | ||
| T26 | 19.1918° N | 84.1860° E | 180.7 | |||
| T27 | 19.1924° N | 84.1863° E | 181 | |||
| Kandhamal | T28 | 20.1342° N | 84.0167° E | 700 | ||
| T29 | 20.1348° N | 84.0170° E | 591 | |||
| T30 | 20.1354° N | 84.0173° E | 550 | |||
| 4. | Mid Central Table Land | Angul | T31 | 20.8444° N | 85.1511° E | 876 |
| T32 | 20.8450° N | 85.1520° E | 218.3 | |||
| T33 | 20.8456° N | 85.1526° E | 224 | |||
| Dhenkanal | T34 | 20.6505° N | 85.5981° E | 80 | ||
| T35 | 20.6510° N | 85.5988° E | 79.6 | |||
| T36 | 20.6515° N | 85.5995° E | 79 | |||
| Cuttack | T37 | 20.4625° N | 85.8830° E | 36 | ||
| T38 | 20.4630° N | 85.8840° E | 36 | |||
| T39 | 20.4635° N | 85.8850° E | 37 | |||
| 5. | Western Central Table Land | Boudh | T40 | 20.8418° N | 84.3200° E | 218 |
| T41 | 20.8420° N | 84.3202° E | 221 | |||
| T42 | 20.8422° N | 84.3204° E | 226 | |||
| Bargarh | T43 | 21.3470° N | 83.6320° E | 171 | ||
| T44 | 21.3472° N | 83.6322° E | 170 | |||
| T45 | 21.3474° N | 83.6324° E | 169 | |||
| Jharsuguda | T46 | 21.8554° N | 84.0062° E | 218 | ||
| T47 | 21.8562° N | 84.0065° E | 216 | |||
| T48 | 21.8560° N | 84.0065° E | 214 | |||
| 6. | Eastern Ghat High Land | Nawarangpur | T49 | 19.2281° N | 82.5470° E | 557 |
| T50 | 19.2288° N | 82.5478° E | 553 | |||
| T51 | 19.2295° N | 82.5484° E | 548 | |||
| Rayagada | T52 | 19.1712° N | 83.4163° E | 207 | ||
| T53 | 19.1718° N | 83.4169° E | 217 | |||
| T54 | 19.1724° N | 83.4174° E | 227 | |||
| Koraput (East) | T55 | 18.8561° N | 82.7347° E | 218 | ||
| T56 | 18.8570° N | 82.7355° E | 218 | |||
| T57 | 18.8579° N | 82.7362° E | 219 | |||
| 7. | North Central Plateau | Mayurbhanj (South) | T58 | 22.0087° N | 86.4187° E | 559 |
| T59 | 22.0090° N | 86.4193° E | 564 | |||
| T60 | 22.0093° N | 86.4197° E | 568 | |||
| Keonjhar (North) | T61 | 21.6289° N | 85.5817° E | 596 | ||
| T62 | 21.6287° N | 85.5815° E | 593 | |||
| T63 | 21.6285° N | 85.5813° E | 590 | |||
| Mayurbhanj (North) | T64 | 22.0087° N | 86.4187° E | 570 | ||
| T65 | 22.0091° N | 86.4196° E | 596 | |||
| T66 | 22.0095° N | 86.4205° E | 610 | |||
| 8. | South Eastern Ghat | Keonjhar (South) | T67 | 21.6289° N | 85.5817° E | 193 |
| T68 | 21.6285° N | 85.5813° E | 193 | |||
| T69 | 21.6281° N | 85.5810° E | 193 | |||
| Koraput (South-East) | T70 | 18.8561° N | 82.7347° E | 870 | ||
| T71 | 18.8566° N | 82.7354° E | 356 | |||
| T72 | 18.8572° N | 82.7359° E | 110 | |||
| Malkangiri | T73 | 18.3436° N | 81.8825° E | 178 | ||
| T74 | 18.3441° N | 81.8821° E | 170 | |||
| T75 | 18.349° N | 81.8817° E | 162 | |||
| 9. | North Western Plateau | Sundargarh | T76 | 22.1240° N | 84.0432° E | 233 |
| T77 | 22.1248° N | 84.0437° E | 231 | |||
| T78 | 22.1256° N | 84.0442° E | 229 | |||
| Deogarh | T79 | 21.5383° N | 84.7289° E | 254 | ||
| T80 | 21.5388° N | 84.7293° E | 253 | |||
| T81 | 21.5392° N | 84.7297° E | 252 | |||
| Sambalpur | T82 | 21.4669° N | 83.9812° E | 135 | ||
| T83 | 21.4673° N | 83.9818° E | 252 | |||
| T84 | 21.4677° N | 83.9822° E | 312 | |||
| 10. | Western Undulating Zone | Kalahandi | T85 | 19.9137° N | 83.1649° E | 355 |
| T86 | 19.9141° N | 83.1653° E | 352 | |||
| T87 | 19.9146° N | 83.1657° E | 349 | |||
| Bolangir | T88 | 20.7011° N | 83.4846° E | 383 | ||
| T89 | 20.7017° N | 83.4848° E | 556 | |||
| T90 | 20.7023° N | 83.4850° E | 615 | |||
| Nuapada | T91 | 20.8060° N | 82.5361° E | 1200 | ||
| T92 | 20.8068° N | 82.5368° E | 1202 | |||
| T93 | 20.8076° N | 82.5375° E | 1204 |
| SL. No. | Agroclimatic Zones | Districts | Accession No. | pH | Max. Rel. Humidity | Min. Rel. Humidity | Avg. Rainfall | Max. Avg. Temp. | Min. Avg. Temp. | Altitude |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. | East and South East Coastal Plain | Jagatsingpur | T1 | 6.5 | 81.7 | 54.3 | 4.8 | 32.9 | 21.9 | 46 |
| T2 | 6.4 | 81.2 | 55.1 | 4.6 | 32.4 | 21.5 | 43 | |||
| T3 | 6.3 | 81 | 55 | 4.5 | 32.2 | 21.2 | 40 | |||
| Khurda | T4 | 6.74 | 82.1 | 56.8 | 3.2 | 32.8 | 22.8 | 75 | ||
| T5 | 6.9 | 81 | 59.7 | 5.4 | 33.9 | 21.6 | 181 | |||
| T6 | 6.99 | 80.98 | 59.9 | 5.9 | 4.1 | 21.1 | 190 | |||
| Puri | T7 | 6.84 | 80.2 | 62.9 | 3.4 | 30.6 | 23.4 | 0.1 | ||
| T8 | 8.4 | 75.3 | 71.6 | 6.4 | 34 | 24.1 | 74 | |||
| T9 | 8.9 | 72.2 | 73.4 | 7.2 | 35.6 | 25.4 | 86 | |||
| Nayagarh | T10 | 7 | 81.3 | 59.4 | 5.6 | 33.5 | 21.7 | 178 | ||
| T11 | 5.1 | 100.3 | 25.1 | 4.9 | 39.2 | 12.1 | 14 | |||
| T12 | 5 | 102.3 | 20.8 | 4.3 | 41.2 | 10.5 | 55 | |||
| 2. | North Eastern Coastal Plain | Bhadrak | T13 | 6.5 | 78.2 | 62.3 | 3.9 | 33.1 | 22.4 | 23 |
| T14 | 6.9 | 78.6 | 62.5 | 3.5 | 33.2 | 22.8 | 23.6 | |||
| T15 | 7.1 | 79.2 | 62.9 | 3.1 | 33.4 | 23.2 | 24 | |||
| Balasore | T16 | 13.7 | 83 | 74 | 4.8 | 34.2 | 32.4 | 16 | ||
| T17 | 13.2 | 82.8 | 74.9 | 4.6 | 34.6 | 32.8 | 16.3 | |||
| T18 | 13.1 | 82.5 | 75 | 4.4 | 34.9 | 33.2 | 16.9 | |||
| Jajpur | T19 | 4.3 | 98.9 | 23.9 | 5.4 | 38.7 | 11.4 | 8 | ||
| T20 | 4.7 | 98.2 | 23.1 | 5.9 | 38.1 | 11.7 | 9 | |||
| T21 | 8 | 97.9 | 22.8 | 6.4 | 37.7 | 12.1 | 10 | |||
| 3. | North Eastern Ghat | Ganjam | T22 | 8.6 | 74.6 | 71.3 | 6.1 | 32 | 27 | 3 |
| T23 | 8.7 | 87.5 | 92.8 | 6.1 | 33.3 | 27.4 | 568 | |||
| T24 | 8.8 | 90.2 | 98.9 | 6.2 | 33.7 | 28.1 | 569 | |||
| Gajapati | T25 | 6.08 | 80.3 | 60.1 | 3.4 | 30.2 | 23.8 | 180.5 | ||
| T26 | 6.21 | 80.1 | 59.8 | 3.5 | 30.4 | 23.9 | 180.7 | |||
| T27 | 6.42 | 79.8 | 59.4 | 3.6 | 30.6 | 24 | 181 | |||
| Kandhamal | T28 | 7.3 | 80.5 | 53.8 | 4.3 | 32.8 | 22.1 | 700 | ||
| T29 | 9.2 | 97.4 | 84.3 | 5.2 | 32.3 | 24.4 | 591 | |||
| T30 | 10.1 | 106.2 | 97.8 | 6.1 | 31.7 | 26.2 | 341 | |||
| 4. | Mid Central Table Land | Angul | T31 | 7.61 | 76.2 | 50.9 | 3.82 | 31.2 | 18.1 | 876 |
| T32 | 6.79 | 79.7 | 55.7 | 5.3 | 34.2 | 19.3 | 218.3 | |||
| T33 | 6.12 | 81.2 | 60.1 | 7.5 | 37.6 | 20.2 | 216.7 | |||
| Dhenkanal | T34 | 6.75 | 81.4 | 51.8 | 4.08 | 31.8 | 21.4 | 80 | ||
| T35 | 6.79 | 81.9 | 51.4 | 4.12 | 31.6 | 21.2 | 79.6 | |||
| T36 | 7.12 | 82.3 | 50.9 | 4.16 | 31.4 | 21 | 79.1 | |||
| Cuttack | T37 | 6.82 | 81.8 | 54.3 | 4.9 | 33.2 | 21.9 | 36 | ||
| T38 | 6.86 | 81.8 | 54.3 | 4.9 | 33.2 | 21.9 | 36 | |||
| T39 | 6.91 | 82 | 54.6 | 5.1 | 32.6 | 22.2 | 36.4 | |||
| 5. | Western Central Table Land | Boudh | T40 | 6.6 | 80.7 | 58.4 | 5.1 | 32.5 | 22.3 | 218 |
| T41 | 6.45 | 80.9 | 58.7 | 4.9 | 32.1 | 22.6 | 221 | |||
| T42 | 6.23 | 81.1 | 59.2 | 4.5 | 30.9 | 22.9 | 225 | |||
| Bargarh | T43 | 10.8 | 86.7 | 82.2 | 7.7 | 29.3 | 25.4 | 171 | ||
| T44 | 11 | 87.1 | 82 | 7.5 | 29.4 | 25.6 | 170 | |||
| T45 | 11.3 | 87.8 | 81.9 | 7.3 | 29.5 | 25.9 | 169 | |||
| Jharsuguda | T46 | 6.8 | 80.1 | 55.3 | 5.1 | 34.6 | 19.5 | 218 | ||
| T47 | 6.45 | 81 | 53 | 4.5 | 33 | 19 | 216 | |||
| T48 | 6.24 | 81.8 | 51.5 | 3.9 | 32.6 | 18.5 | 214 | |||
| 6. | Eastern Ghat High Land | Nawarangpur | T49 | 6.2 | 82.4 | 54.1 | 5.4 | 32.7 | 21.9 | 557 |
| T50 | 6.3 | 83 | 54.3 | 5.5 | 32.5 | 21.6 | 553 | |||
| T51 | 6.4 | 83.4 | 54.7 | 5.9 | 32.2 | 21.3 | 550 | |||
| Rayagada | T52 | 6.83 | 82.5 | 54.9 | 3 | 31.7 | 18.9 | 207 | ||
| T53 | 6.65 | 81.5 | 53.6 | 4.6 | 32.8 | 19.1 | 217 | |||
| T54 | 6.45 | 80.5 | 52.4 | 5 | 33.3 | 19.6 | 227 | |||
| Koraput (East) | T55 | 6.51 | 81.3 | 53.1 | 4.7 | 33.1 | 19.2 | 218 | ||
| T56 | 6.54 | 81.3 | 53.1 | 4.7 | 33.1 | 19.2 | 218 | |||
| T57 | 6.98 | 81.6 | 53.8 | 5.2 | 33.8 | 19.6 | 219 | |||
| 7. | North Central Plateau | Mayurbhanj (South) | T58 | 7.1 | 81.7 | 54.3 | 6.2 | 32.1 | 18.7 | 559 |
| T59 | 7.3 | 81.4 | 54.8 | 6.5 | 32.9 | 18.2 | 564 | |||
| T60 | 7.5 | 81.1 | 55.2 | 6.7 | 333.3 | 17.8 | 569 | |||
| Keonjhar (North) | T61 | 9.2 | 98 | 85 | 5.5 | 32.8 | 24.8 | 596 | ||
| T62 | 9.1 | 97.6 | 84.6 | 5.4 | 32.5 | 24.6 | 593 | |||
| T63 | 9 | 97.1 | 84.1 | 5.3 | 32.1 | 24.4 | 590 | |||
| Mayurbhanj (North) | T64 | 8.9 | 87.8 | 93 | 5.9 | 33.1 | 27.6 | 570 | ||
| T65 | 9.3 | 98 | 85 | 5.5 | 32.8 | 24.8 | 596 | |||
| T66 | 9.6 | 98.3 | 80 | 5.1 | 32.2 | 21.2 | 605 | |||
| 8. | South Eastern Ghat | Keonjhar (South) | T67 | 7.6 | 89 | 93 | 9.4 | 38.2 | 24.2 | 193 |
| T68 | 7.3 | 89 | 93 | 9.4 | 38.2 | 24.2 | 193 | |||
| T69 | 7.9 | 90 | 94 | 9.6 | 38.4 | 24.4 | 194 | |||
| Koraput (South-East) | T70 | 5.8 | 85 | 77 | 8.7 | 29.6 | 20.6 | 870 | ||
| T71 | 4.5 | 78.4 | 64.3 | 8.59 | 25.8 | 23.7 | 356 | |||
| T72 | 4 | 72.3 | 57.7 | 8.34 | 21.4 | 26.7 | 350 | |||
| Malkangiri | T73 | 12.6 | 86 | 83 | 11.9 | 36 | 22 | 178 | ||
| T74 | 10.7 | 86.2 | 82.4 | 7.6 | 29.5 | 25.3 | 170 | |||
| T75 | 8.6 | 86.4 | 81.5 | 6.8 | 24.8 | 27.6 | 162 | |||
| 9. | North Western Plateau | Sundargarh | T76 | 11.3 | 66 | 64 | 5.4 | 31.5 | 20.5 | 233 |
| T77 | 10.9 | 65.8 | 64.3 | 5.2 | 31.2 | 20.3 | 231 | |||
| T78 | 10.5 | 65.2 | 64.8 | 5 | 30.9 | 20.1 | 229 | |||
| Deogarh | T79 | 3.1 | 97.9 | 56.1 | 11 | 32 | 25 | 254 | ||
| T80 | 3.2 | 98.1 | 55.9 | 10.9 | 31.7 | 24.7 | 253 | |||
| T81 | 3.3 | 98.6 | 55.2 | 10.3 | 31.2 | 24.2 | 252 | |||
| Sambalpur | T82 | 7.9 | 79.4 | 59.1 | 4.5 | 31.3 | 20.9 | 135 | ||
| T83 | 3.4 | 98.1 | 55.9 | 10.9 | 31.8 | 24.7 | 252 | |||
| T84 | 2.9 | 99.3 | 51.2 | 16.2 | 32.5 | 27.3 | 255 | |||
| 10. | Western Undulating Zone | Kalahandi | T85 | 4.4 | 78 | 64 | 8.76 | 26 | 24 | 355 |
| T86 | 4.7 | 77.8 | 63.8 | 8.7 | 25.4 | 23.6 | 352 | |||
| T87 | 5.1 | 77.1 | 63.2 | 8.6 | 24.8 | 23.2 | 350 | |||
| Bolangir | T88 | 6.2 | 76 | 74 | 8.01 | 32.8 | 30.8 | 383 | ||
| T89 | 6.3 | 82.3 | 53.8 | 5.3 | 32.5 | 21.7 | 556 | |||
| T90 | 6.5 | 88.4 | 51.2 | 5.1 | 31.8 | 21.1 | 565 | |||
| Nuapada | T91 | 2.1 | 92 | 63 | 9.69 | 30 | 23 | 1200 | ||
| T92 | 2.4 | 91.7 | 62.8 | 9.71 | 29.8 | 21 | 1202 | |||
| T93 | 2.7 | 91.4 | 62.2 | 9.75 | 29.1 | 20 | 1201 |
| SL. No. | Agroclimatic Zones | Districts | Accession No. | Organic Carbon (%) | Nitrogen | Phosphorous | Potassium |
|---|---|---|---|---|---|---|---|
| 1. | East and South East Coastal Plain | Jagatsingpur | T1 | 1.59 | 236.2 | 79.4 | 542.2 |
| T2 | 1.61 | 235.9 | 81.4 | 538.7 | |||
| T3 | 1.63 | 235.5 | 81.8 | 532.5 | |||
| Khurda | T4 | 1.11 | 503.7 | 161.7 | 918.4 | ||
| T5 | 1.41 | 166.5 | 217.2 | 493.2 | |||
| T6 | 1.62 | 160.7 | 220.8 | 490.5 | |||
| Puri | T7 | 0.99 | 305.31 | 265.1 | 796.94 | ||
| T8 | 1.7 | 270.4 | 63.4 | 408.3 | |||
| T9 | 2 | 255.3 | 62.5 | 400.9 | |||
| Nayagarh | T10 | 0.76 | 250 | 169.05 | 491.9 | ||
| T11 | 0.83 | 250.4 | 169.02 | 491.2 | |||
| T12 | 0.91 | 250.8 | 169 | 491 | |||
| 2. | North Eastern Coastal Plain | Bhadrak | T13 | 0.87 | 352.5 | 64.7 | 201.6 |
| T14 | 0.82 | 352.1 | 64.2 | 201.2 | |||
| T15 | 0.78 | 351.8 | 63.8 | 200.8 | |||
| Balasore | T16 | 3.2 | 340.2 | 83.2 | 602.3 | ||
| T17 | 3.4 | 179.5 | 281.9 | 209.4 | |||
| T18 | 3.6 | 178.9 | 280.8 | 208.7 | |||
| Jajpur | T19 | 1.8 | 375.4 | 132.2 | 302.5 | ||
| T20 | 2 | 381.5 | 130.3 | 294.6 | |||
| T21 | 2.2 | 385.7 | 128.8 | 290.2 | |||
| 3. | North Eastern Ghat | Ganjam | T22 | 3.1 | 183.7 | 280.3 | 208.3 |
| T23 | 3.4 | 340.6 | 83.8 | 602.4 | |||
| T24 | 3.6 | 341.3 | 82.6 | 601.1 | |||
| Gajapati | T25 | 1.36 | 166.2 | 217.5 | 493.9 | ||
| T26 | 1.27 | 176 | 26.58 | 78.1 | |||
| T27 | 1.11 | 186.2 | 25.6 | 76.8 | |||
| Kandhamal | T28 | 0.52 | 141.2 | 37.2 | 519 | ||
| T29 | 0.58 | 141.3 | 37.5 | 519.7 | |||
| T30 | 1.02 | 141,5 | 37.9 | 520.2 | |||
| 4. | Mid Central Table Land | Angul | T31 | 0.94 | 162.3 | 33.2 | 771.6 |
| T32 | 0.91 | 162.7 | 33.7 | 771.8 | |||
| T33 | 0.89 | 163.2 | 34.1 | 772.1 | |||
| Dhenkanal | T34 | 1.79 | 562.5 | 132.4 | 921.6 | ||
| T35 | 1.81 | 562.1 | 132.1 | 921.3 | |||
| T36 | 1.86 | 558.5 | 131.8 | 920.8 | |||
| Cuttack | T37 | 1.5 | 251.2 | 96.3 | 306 | ||
| T38 | 3.61 | 152.4 | 72.6 | 89.1 | |||
| T39 | 3.89 | 150.8 | 70.9 | 88.1 | |||
| 5. | Western Central Table Land | Boudh | T40 | 0.32 | 125 | 127.91 | 309.12 |
| T41 | 0.35 | 123 | 128.1 | 310.1 | |||
| T42 | 0.37 | 121 | 128.7 | 310.8 | |||
| Bargarh | T43 | 5.2 | 140.3 | 142.3 | 30 | ||
| T44 | 5.4 | 140.7 | 141.9 | 32 | |||
| T45 | 5.6 | 141.1 | 141.4 | 33 | |||
| Jharsuguda | T46 | 0.94 | 112.5 | 75.5 | 603.46 | ||
| T47 | 0.91 | 112.4 | 74.9 | 602.9 | |||
| T48 | 0.88 | 112.3 | 74.4 | 602.23 | |||
| 6. | Eastern Ghat High Land | Nawarangpur | T49 | 1.14 | 175 | 26.48 | 77.95 |
| T50 | 1.01 | 113 | 75.3 | 603.8 | |||
| T51 | 1 | 110 | 97.8 | 602.4 | |||
| Rayagada | T52 | 3.27 | 316.2 | 29.3 | 924.8 | ||
| T53 | 3.98 | 164.5 | 82.4 | 93.4 | |||
| T54 | 4.02 | 163.8 | 84.7 | 94.6 | |||
| Koraput (East) | T55 | 4.61 | 381.2 | 32.4 | 961.3 | ||
| T56 | 4.74 | 380.6 | 32.1 | 960.8 | |||
| T57 | 4.82 | 380.3 | 31.8 | 960.4 | |||
| 7. | North Central Plateau | Mayurbhanj (South) | T58 | 2.23 | 175 | 50.72 | 73.92 |
| T59 | 4.01 | 164.3 | 82.6 | 93.1 | |||
| T60 | 6.32 | 160.8 | 88.9 | 96.8 | |||
| Keonjhar (North) | T61 | 3.57 | 152.7 | 72.8 | 89.3 | ||
| T62 | 3.65 | 152.5 | 72.4 | 89.1 | |||
| T63 | 3.99 | 152.3 | 72 | 88.8 | |||
| Mayurbhanj (North) | T64 | 3.98 | 164.5 | 82.4 | 93.4 | ||
| T65 | 4.72 | 383.2 | 32.7 | 959.6 | |||
| T66 | 5.88 | 386.8 | 31.6 | 960.6 | |||
| 8. | South Eastern Ghat | Keonjhar (South) | T67 | 8.4 | 261 | 145.2 | 391.2 |
| T68 | 8 | 231 | 152 | 308.7 | |||
| T69 | 7.6 | 201 | 165.9 | 300.7 | |||
| Koraput (South-East) | T70 | 2.3 | 242.3 | 132.7 | 296.3 | ||
| T71 | 3.4 | 340.4 | 83.5 | 602.4 | |||
| T72 | 4.6 | 444.4 | 82.9 | 603.9 | |||
| Malkangiri | T73 | 6.36 | 216.4 | 121.2 | 386.4 | ||
| T74 | 5.1 | 140.1 | 142.5 | 29.8 | |||
| T75 | 5 | 132.4 | 162.6 | 30 | |||
| 9. | North Western Plateau | Sundargarh | T76 | 1.06 | 285.3 | 161.2 | 603.4 |
| T77 | 1.1 | 285.1 | 161.4 | 603.1 | |||
| T78 | 1.4 | 284.8 | 161.6 | 603 | |||
| Deogarh | T79 | 8.1 | 230.4 | 152.3 | 308.5 | ||
| T80 | 8.3 | 230.5 | 152.7 | 308.8 | |||
| T81 | 8.5 | 230.6 | 153.1 | 309.1 | |||
| Sambalpur | T82 | 8.6 | 262.5 | 139.75 | 594.8 | ||
| T83 | 8.9 | 263.7 | 139.65 | 592.6 | |||
| T84 | 9.2 | 264.1 | 139.55 | 590.4 | |||
| 10. | Western Undulating Zone | Kalahandi | T85 | 2.4 | 240.3 | 76.3 | 813.7 |
| T86 | 2.5 | 240.4 | 76.2 | 813.6 | |||
| T87 | 2.6 | 240.5 | 76.1 | 813.5 | |||
| Bolangir | T88 | 8.9 | 290.4 | 89.1 | 503.1 | ||
| T89 | 8 | 230.2 | 152.7 | 308.3 | |||
| T90 | 7.2 | 229.8 | 153.8 | 307.6 | |||
| Nuapada | T91 | 1.6 | 270.3 | 63.2 | 408.1 | ||
| T92 | 1.18 | 175.4 | 26.51 | 77.81 | |||
| T93 | 1.02 | 174.3 | 25.65 | 75.75 |
| SL. No. | Agroclimatic Zones | Districts | Accession no. | Experimental Thai Basil Oil Yield (X1) | Predicted Thai Basil Oil Yield (X2) | Absolute = |X1 − X2| |
|---|---|---|---|---|---|---|
| 1. | East and South East Coastal Plain | Jagatsingpur | T1 | 1.3 | 1.29 | 0.01 |
| Khurda | T4 | 1.42 | 1.42 | 0 | ||
| T5 | 1.21 | 1.21 | 0 | |||
| T6 | 1.2 | 1.19 | 0.01 | |||
| Puri | T7 | 1.1 | 1.11 | 0.01 | ||
| T8 | 1.65 | 1.67 | 0.02 | |||
| 2. | North Eastern Coastal Plain | Bhadrak | T13 | 0.76 | 0.76 | 0 |
| T14 | 1.45 | 1.43 | 0.02 | |||
| T15 | 0.81 | 0.79 | 0.02 | |||
| Jajpur | T19 | 0.4 | 0.40 | 0 | ||
| T20 | 0.9 | 0.88 | 0.02 | |||
| T21 | 0.98 | 1.01 | 0.03 | |||
| 3. | North Eastern Ghat | Ganjam | T22 | 0.97 | 0.98 | 0.01 |
| Gajapati | T25 | 0.81 | 0.77 | 0.04 | ||
| T26 | 0.94 | 1 | 0.06 | |||
| T27 | 1.68 | 1.68 | 0 | |||
| Kandhamal | T28 | 1.2 | 1.20 | 0 | ||
| T29 | 1.07 | 1.06 | 0.01 | |||
| T30 | 1.1 | 1.08 | 0.02 | |||
| 4. | Mid Central Table Land | Angul | T31 | 0.78 | 0.79 | 0.01 |
| T32 | 1.18 | 1.18 | 0 | |||
| Dhenkanal | T34 | 0.98 | 0.96 | 0.02 | ||
| Cuttack | T37 | 0.67 | 0.67 | 0 | ||
| T38 | 1 | 1 | 0 | |||
| T39 | 0.65 | 0.65 | 0 | |||
| 5. | Western Central Table Land | Boudh | T40 | 1.01 | 0.96 | 0.05 |
| T41 | 0.78 | 0.80 | 0.02 | |||
| Bargarh | T43 | 0.89 | 0.90 | 0.01 | ||
| T44 | 0.98 | 0.98 | 0 | |||
| Jharsuguda | T46 | 0.92 | 0.92 | 0 | ||
| T47 | 0.76 | 0.75 | 0.01 | |||
| 6. | Eastern Ghat High Land | Nawarangpur | T49 | 0.73 | 0.73 | 0 |
| T50 | 0.76 | 0.78 | 0.02 | |||
| T51 | 1.27 | 1.24 | 0.03 | |||
| Rayagada | T52 | 0.9 | 0.90 | 0 | ||
| T53 | 0.95 | 1 | 0.05 | |||
| T54 | 0.97 | 0.98 | 0.01 | |||
| Koraput (East) | T55 | 0.91 | 0.91 | 0 | ||
| T56 | 0.93 | 0.95 | 0.02 | |||
| T57 | 0.69 | 0.67 | 0.02 | |||
| 7. | North Central Plateau | Mayurbhanj (South) | T58 | 1.4 | 1.38 | 0.02 |
| T59 | 1.2 | 1.25 | 0.05 | |||
| T60 | 1.3 | 1.32 | 0.02 | |||
| Keonjhar (North) | T61 | 3.62 | 3.57 | 0.13 | ||
| T62 | 3.5 | 3.52 | 0.02 | |||
| Mayurbhanj (North) | T64 | 0.93 | 0.98 | 0.05 | ||
| T65 | 0.91 | 0.96 | 0.05 | |||
| T66 | 1.1 | 1.07 | 0.03 | |||
| 8. | South Eastern Ghat | Keonjhar (South) | T67 | 3.94 | 3.94 | 0 |
| T68 | 3.7 | 3.73 | 0.03 | |||
| Koraput (South-East) | T70 | 0.72 | 0.75 | 0.03 | ||
| T71 | 1.02 | 0.97 | 0.05 | |||
| T72 | 1.3 | 1.34 | 0.04 | |||
| Malkangiri | T73 | 0.64 | 0.61 | 0.03 | ||
| T74 | 0.76 | 0.77 | 0.01 | |||
| 9. | North Western Plateau | Sundargarh | T76 | 0.68 | 0.69 | 0.01 |
| T77 | 0.79 | 0.78 | 0.01 | |||
| Sambalpur | T82 | 1.1 | 1.12 | 0.02 | ||
| T83 | 1.25 | 1.27 | 0.02 | |||
| T84 | 1.2 | 1.20 | 0 | |||
| 10. | Western Undulating Zone | Kalahandi | T85 | 0.36 | 0.36 | 0 |
| T86 | 0.12 | 0.13 | 0.01 | |||
| Bolangir | T88 | 1.3 | 1.30 | 0 | ||
| T89 | 1.5 | 1.45 | 0.05 | |||
| T90 | 1.1 | 1.09 | 0.01 | |||
| Nuapada | T91 | 0.7 | 0.69 | 0.01 | ||
| T92 | 0.9 | 0.89 | 0.01 |
| SL. No. | Agroclimatic Zones | Districts | Accession no. | Experimental Thai Basil Oil Yield (X1) | Predicted Thai Basil Oil Yield (X2) | Absolute = |X1 − X2| |
|---|---|---|---|---|---|---|
| 1. | East and South East Coastal Plain | Jagatsingpur | T2 | 1.2 | 1.19 | 0.01 |
| T3 | 1.2 | 1.27 | 0.07 | |||
| Nayagarh | T10 | 1.3 | 1.27 | 0.030 | ||
| T11 | 0.65 | 0.77 | 0.12 | |||
| T12 | 1.39 | 1.41 | 0.02 | |||
| 2. | North Eastern Coastal Plain | Balasore | T16 | 0.81 | 0.84 | 0.03 |
| T17 | 1 | 1.29 | 0.29 | |||
| T18 | 1 | 1.28 | 0.28 | |||
| 3. | North Eastern Ghat | Ganjam | T23 | 0.94 | 0.94 | 0 |
| 4. | Mid Central Table Land | Dhenkanal | T34 | 0.75 | 0.78 | 0.03 |
| T35 | 0.69 | 0.73 | 0.04 | |||
| 5. | Western Central TableLand | Boudh | T42 | 0.86 | 0.90 | 0.04 |
| 6. | North Central Plateau | Keonjhar (North) | T61 | 3.72 | 3.16 | 0.56 |
| 7. | South Eastern Ghat | Keonjhar (South) | T67 | 4.1 | 4.46 | 0.36 |
| Malkangiri | T75 | 0.71 | 0.66 | 0.05 | ||
| 8. | North Western Plateau | Sundargarh | T78 | 0.95 | 0.96 | 0.01 |
| Deogarh | T79 | 1 | 1.28 | 0.28 | ||
| T80 | 0.75 | 0.78 | 0.03 | |||
| T81 | 0.38 | 0.34 | 0.04 | |||
| 9. | Western Undulating Zone | Kalahandi | T87 | 0.78 | 0.53 | 0.25 |
| SL. No. | Agroclimatic Zones | Districts | Accession No. | Experimental Thai Basil Oil Yield (X1) | Predicted Thai Basil Oil Yield (X2) | Absolute = |X1 − X2| |
|---|---|---|---|---|---|---|
| 1. | East and South East Coastal Plain | Puri | T9 | 0.13 | 0.14 | 0.01 |
| 2. | North Eastern Ghat | Ganjam | T24 | 0.94 | 0.97 | 0.03 |
| 3. | Mid Central Table Land | Angul | T33 | 0.78 | 0.60 | 0.18 |
| 4. | Western Central Table Land | Bargarh | T45 | 0.8 | 0.69 | 0.11 |
| Jharsuguda | T48 | 0.77 | 0.55 | 0.22 | ||
| 5. | Western Undulating Zone | Nuapada | T93 | 0.13 | 0.13 | 0 |
| Predictors | Sensitivity Value |
|---|---|
| pH | 2.32 |
| Organic Carbon | 2.31 |
| Nitrogen | 109.99 |
| Phosphorous | 65.47 |
| Potassium | 284.49 |
| Maximum relative humidity | 6.62 |
| Minimum relative humidity | 14.42 |
| Average rainfall | 2.21 |
| Maximum average temperature | 2.46 |
| Minimum average temperature | 3.66 |
| Altitude | 274.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahu, A.; Nayak, G.; Bhuyan, S.K.; Akbar, A.; Bhuyan, R.; Kar, D.; Kuanar, A. Artificial Neural Network and Response Surface-Based Combined Approach to Optimize the Oil Content of Ocimum basilicum var. thyrsiflora (Thai Basil). Plants 2023, 12, 1776. https://doi.org/10.3390/plants12091776
Sahu A, Nayak G, Bhuyan SK, Akbar A, Bhuyan R, Kar D, Kuanar A. Artificial Neural Network and Response Surface-Based Combined Approach to Optimize the Oil Content of Ocimum basilicum var. thyrsiflora (Thai Basil). Plants. 2023; 12(9):1776. https://doi.org/10.3390/plants12091776
Chicago/Turabian StyleSahu, Akankshya, Gayatree Nayak, Sanat Kumar Bhuyan, Abdul Akbar, Ruchi Bhuyan, Dattatreya Kar, and Ananya Kuanar. 2023. "Artificial Neural Network and Response Surface-Based Combined Approach to Optimize the Oil Content of Ocimum basilicum var. thyrsiflora (Thai Basil)" Plants 12, no. 9: 1776. https://doi.org/10.3390/plants12091776
APA StyleSahu, A., Nayak, G., Bhuyan, S. K., Akbar, A., Bhuyan, R., Kar, D., & Kuanar, A. (2023). Artificial Neural Network and Response Surface-Based Combined Approach to Optimize the Oil Content of Ocimum basilicum var. thyrsiflora (Thai Basil). Plants, 12(9), 1776. https://doi.org/10.3390/plants12091776






