Comparative Transcriptome Analysis Reveals the Effect of miR156a Overexpression on Mineral Nutrient Homeostasis in Nicotiana tabacum
Abstract
1. Introduction
2. Results
2.1. Overexpression of Nta-miR156a in Tobacco Leads to Significant Changes in Biological Traits
2.2. Overexpression of Nta-miR156a Decreased Chlorophyll, Proline, Phenolic Compounds, and Flavonoid Content
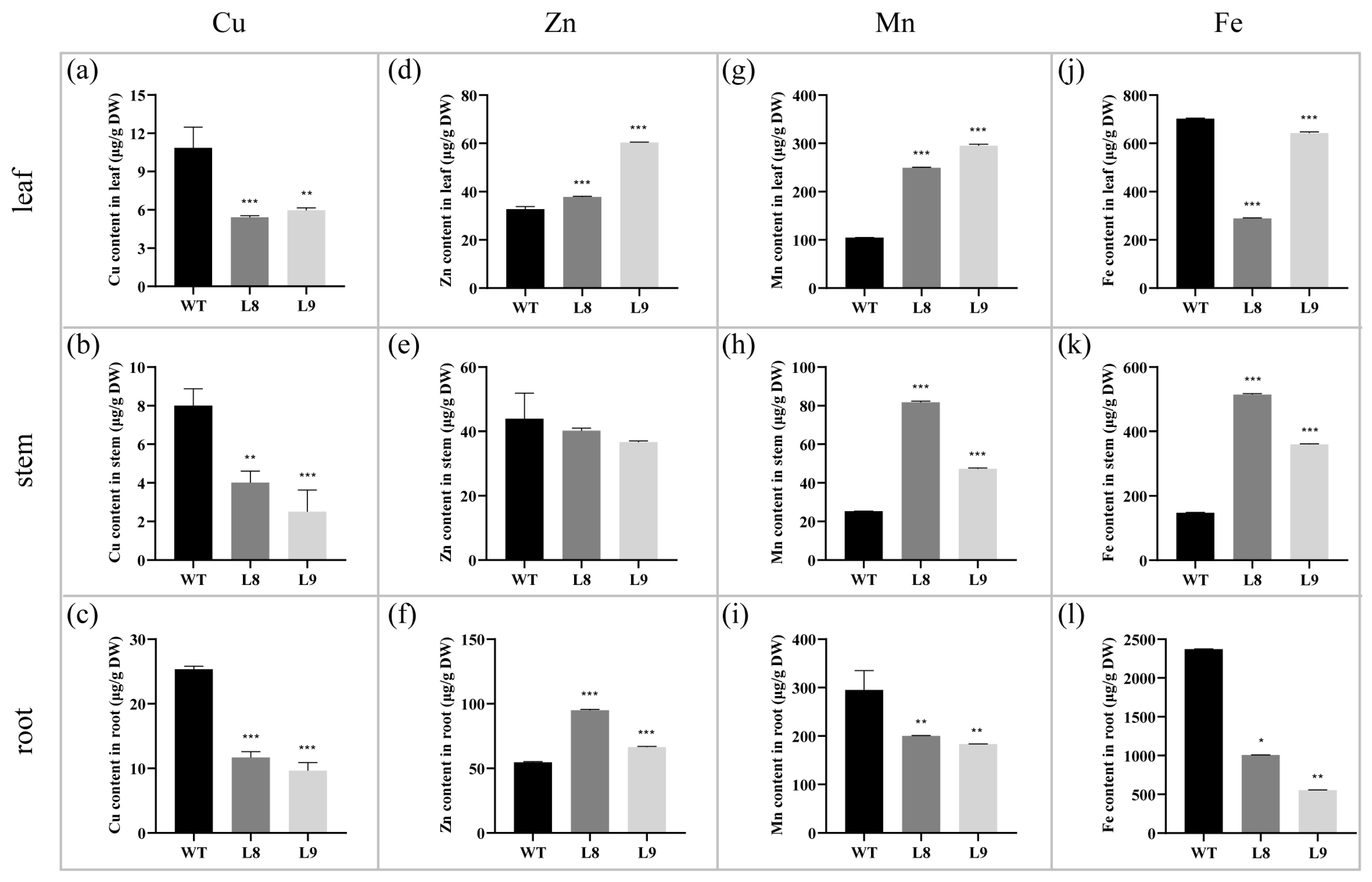
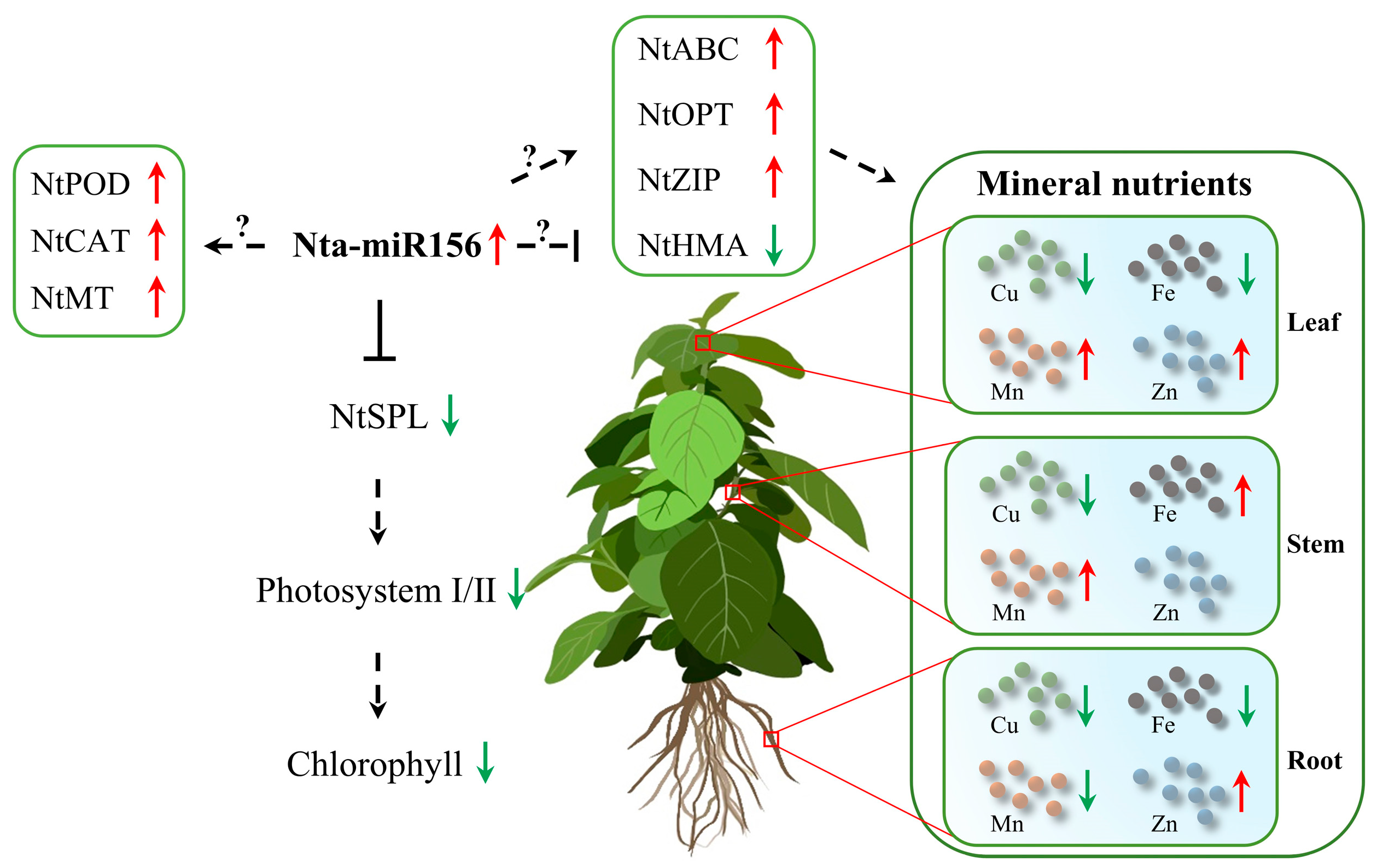
2.3. Overexpression of Nta-miR156a Disrupts Mineral Nutrient Homeostasis
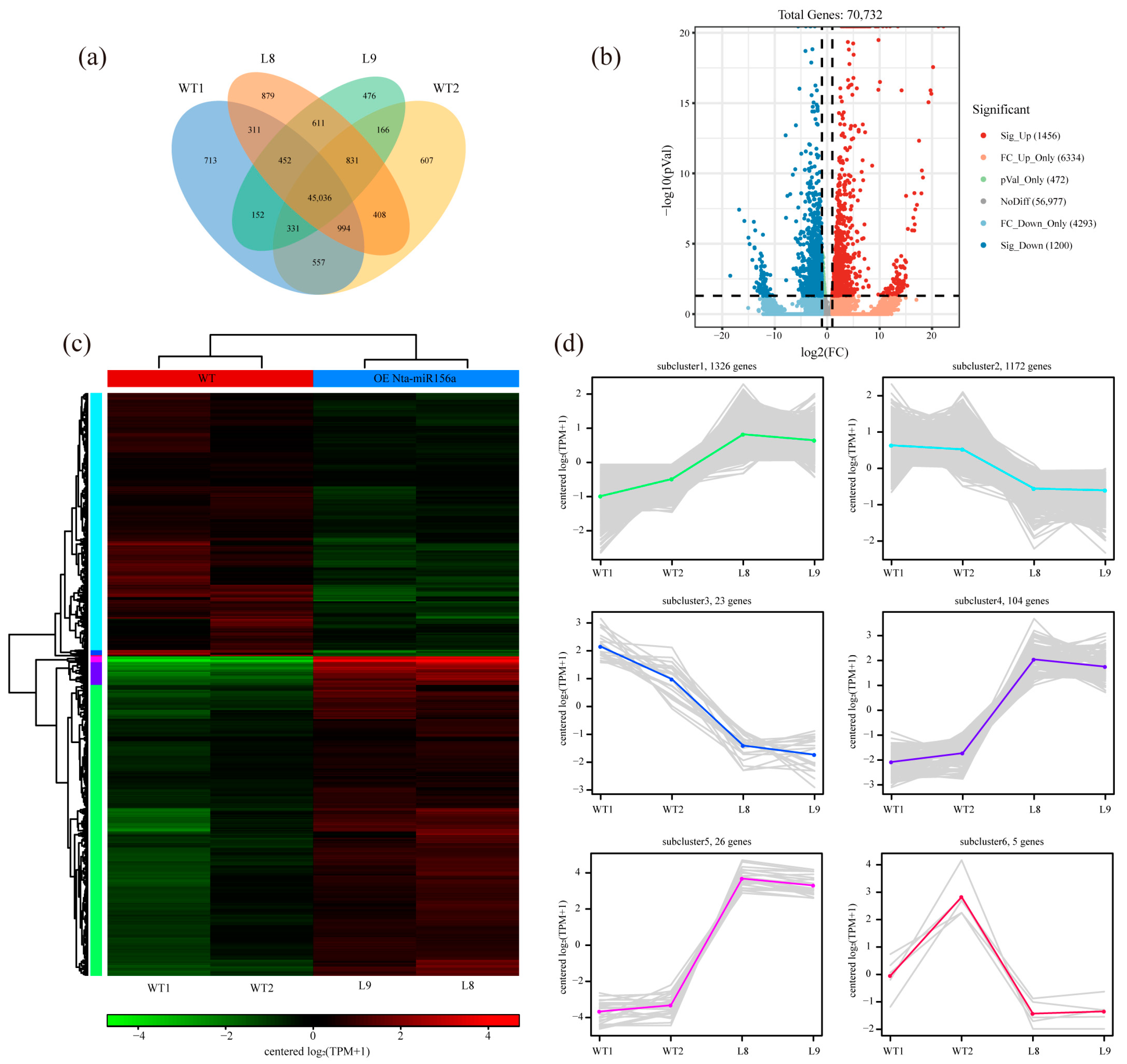
2.4. Changes in the Global Gene Expression Levels of Nta-miR156a-Overexpressing Tobacco
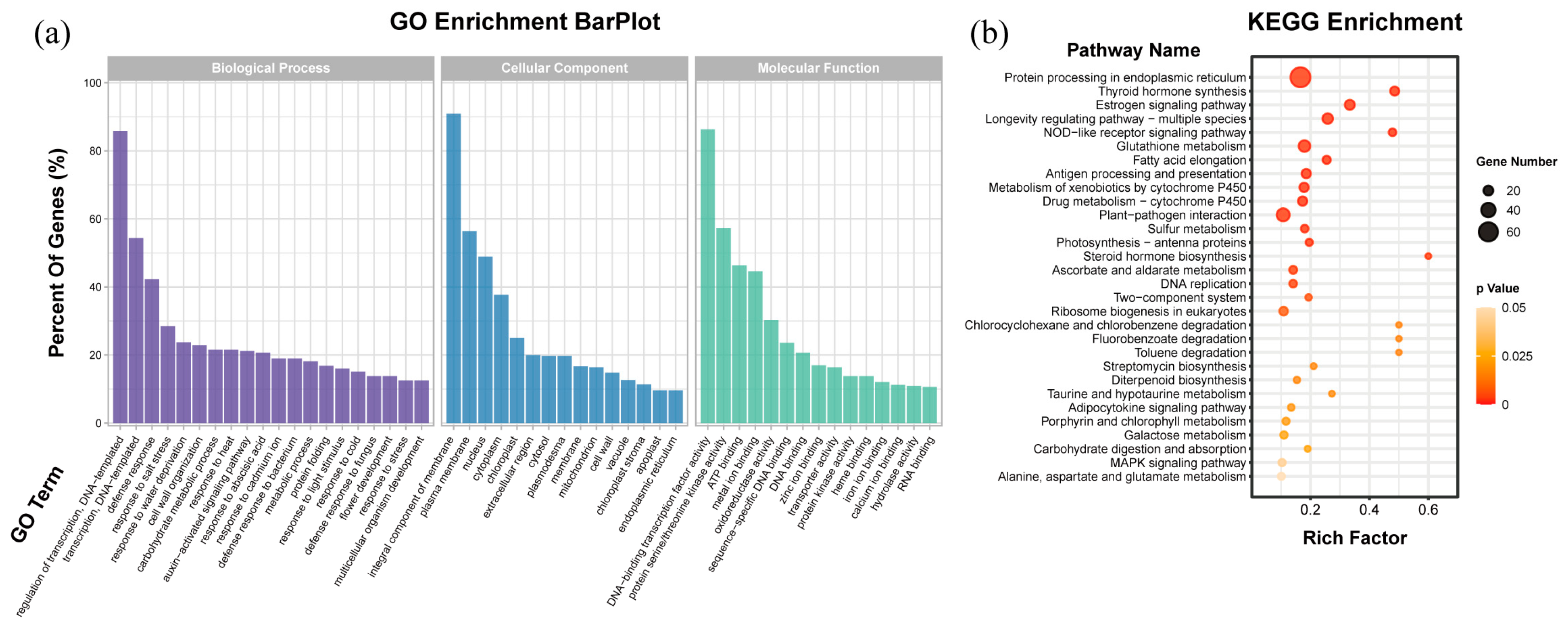
2.5. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
2.6. Comparative Analysis of Differentially Expressed Genes
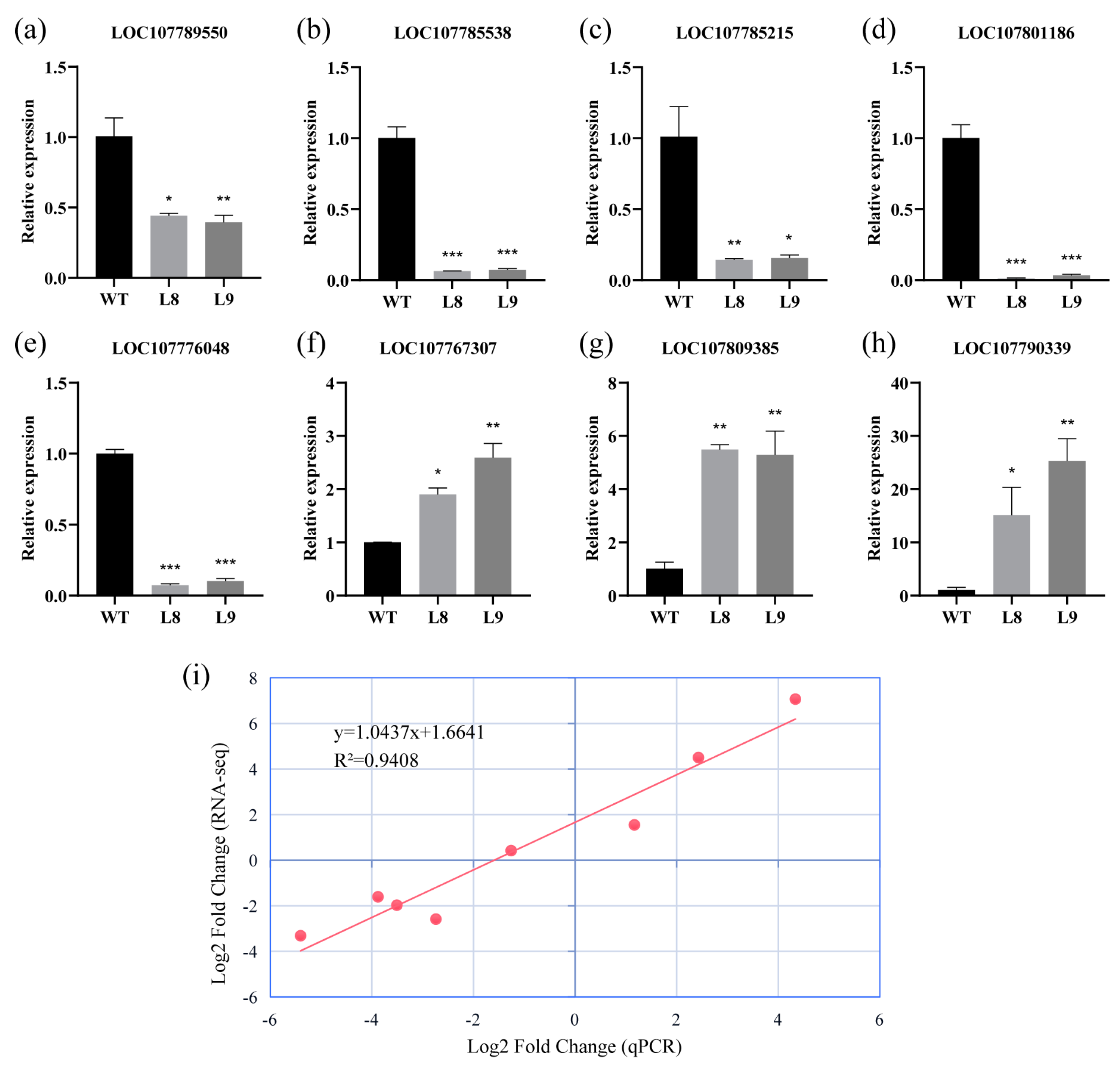
2.7. qRT-PCR Validation of RNA-seq Results
3. Discussion
4. Materials and Methods
4.1. Overexpression Vector Construction
4.2. Plant Materials and Genetic Transformation
4.3. Determination of Stress Markers
4.4. Metal Content Determination
4.5. RNA Sequencing
4.6. Sequencing Data Cleaning and Reference Genome Alignment
4.7. Expression level Analysis and Enrichment Analysis
4.8. Total RNA Isolation and qRT-PCR
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prasad, R. Micro Mineral Nutrient Deficiencies in Humans, Animals and Plants and Their Amelioration. Proc. Natl. Acad. Sci. India, Sect. B Biol. Sci. 2012, 82, 225–233. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Bindraban, P.S. Fortification of Micronutrients for Efficient Agronomic Production: A Review. Agron. Sustain. Dev. 2016, 36, 7. [Google Scholar] [CrossRef]
- Connolly, E.L.; Guerinot, M.L. Iron Stress in Plants. Genome Biol. 2002, 3, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Shenker, M.; Chen, Y. Increasing Iron Availability to Crops: Fertilizers, Organo-Fertilizers, and Biological Approaches. Soil Sci. Plant Nutr. 2005, 51, 1–17. [Google Scholar] [CrossRef]
- Tvrdý, V.; Hrubša, M.; Jirkovský, E.; Biedermann, D.; Kutý, M.; Valentová, K.; Křen, V.; Mladěnka, P. Silymarin Dehydroflavonolignans Chelate Zinc and Partially Inhibit Alcohol Dehydrogenase. Nutrients 2021, 13, 4238. [Google Scholar] [CrossRef] [PubMed]
- Bruscalupi, G.; Di Micco, P.; Failla, C.M.; Pascarella, G.; Morea, V.; Saliola, M.; De Paolis, A.; Venditti, S.; Mauro, M.L. Arabidopsis Thaliana Sirtuins Control Proliferation and Glutamate Dehydrogenase Activity. Plant Physiol. Biochem. 2023, 194, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Huo, C.; He, L.; Yu, T.; Ji, X.; Li, R.; Zhu, S.; Zhang, F.; Xie, H.; Liu, W. The Superoxide Dismutase Gene Family in Nicotiana tabacum: Genome-Wide Identification, Characterization, Expression Profiling and Functional Analysis in Response to Heavy Metal Stress. Front. Plant Sci. 2022, 13, 904105. [Google Scholar] [CrossRef]
- Hafeez, B. Role of Zinc in Plant Nutrition- A Review. AJEA 2013, 3, 374–391. [Google Scholar] [CrossRef]
- Yruela, I. Copper in Plants. Braz. J. Plant Physiol. 2005, 17, 145–156. [Google Scholar] [CrossRef]
- La Torre, A.; Iovino, V.; Caradonia, F. Copper in Plant Protection: Current Situation and Prospects. Phytopathol. Mediterr. 2018, 57, 201–236. [Google Scholar]
- Alejandro, S.; Höller, S.; Meier, B.; Peiter, E. Manganese in Plants: From Acquisition to Subcellular Allocation. Front. Plant Sci. 2020, 11, 300. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.B.; Jensen, P.E.; Husted, S. Manganese Deficiency in Plants: The Impact on Photosystem II. Trends Plant Sci. 2016, 21, 622–632. [Google Scholar] [CrossRef]
- Rubiya, S.; Dhriti, K.; Bhat, A.A. Heavy Metal Toxicity in Plants: A Review. Plant Arch. 2018, 18, 1229–1238. [Google Scholar]
- Sasaki, A.; Yamaji, N.; Ma, J.F. Transporters Involved in Mineral Nutrient Uptake in Rice. EXBOTJ 2016, 67, 3645–3653. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Li, Y.; Liang, G. FIT and BHLH Ib Transcription Factors Modulate Iron and Copper Crosstalk in Arabidopsis. Plant Cell Environ. 2021, 44, 1679–1691. [Google Scholar] [CrossRef]
- Kehr, J. Systemic Regulation of Mineral Homeostasis by Micro RNAs. Front. Plant Sci. 2013, 4, 145. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Kawakami, Y.; Bhullar, N.K. Molecular Analysis of Iron Deficiency Response in Hexaploid Wheat. Front. Sustain. Food Syst. 2019, 3, 67. [Google Scholar] [CrossRef]
- Cai, Y.; Liang, G. CITF1 Functions Downstream of SPL7 to Specifically Regulate Cu Uptake in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 7239. [Google Scholar] [CrossRef]
- Pilon, M. The Copper MicroRNAs. New Phytol 2017, 213, 1030–1035. [Google Scholar] [CrossRef]
- Sun, K.; Wang, H.; Xia, Z. The Maize BHLH Transcription Factor BHLH105 Confers Manganese Tolerance in Transgenic Tobacco. Plant Sci. 2019, 280, 97–109. [Google Scholar] [CrossRef]
- Zlobin, I.E. Current Understanding of Plant Zinc Homeostasis Regulation Mechanisms. Plant Physiol. Biochem. 2021, 162, 327–335. [Google Scholar] [CrossRef]
- Kong, W.W.; Yang, Z.M. Identification of Iron-Deficiency Responsive MicroRNA Genes and Cis-Elements in Arabidopsis. Plant Physiol. Biochem. 2010, 48, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Islam, W.; Tauqeer, A.; Waheed, A.; Zeng, F. MicroRNA Mediated Plant Responses to Nutrient Stress. Int. J. Mol. Sci. 2022, 23, 2562. [Google Scholar] [CrossRef]
- Valdés-López, O.; Yang, S.S.; Aparicio-Fabre, R.; Graham, P.H.; Reyes, J.L.; Vance, C.P.; Hernández, G. MicroRNA Expression Profile in Common Bean (Phaseolus Vulgaris) under Nutrient Deficiency Stresses and Manganese Toxicity. New Phytol. 2010, 187, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal Regulation of Shoot Development in Arabidopsis Thaliana by MiR156 and Its Target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, M.; Yu, J.; Guo, C. SPL9 Mediates Freezing Tolerance by Directly Regulating the Expression of CBF2 in Arabidopsis Thaliana. BMC Plant Biol. 2022, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 Improves Drought Stress Tolerance in Alfalfa (Medicago sativa) by Silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef]
- Li, X.; Hou, Y.; Xie, X.; Li, H.; Li, X.; Zhu, Y.; Zhai, L.; Zhang, C.; Bian, S. A Blueberry MIR156a–SPL12 Module Coordinates the Accumulation of Chlorophylls and Anthocyanins during Fruit Ripening. J. Exp. Bot. 2020, 71, 5976–5989. [Google Scholar] [CrossRef]
- Yin, H.; Hong, G.; Li, L.; Zhang, X.; Kong, Y.; Sun, Z.; Li, J.; Chen, J.; He, Y. MiR156/SPL9 Regulates Reactive Oxygen Species Accumulation and Immune Response in Arabidopsis Thaliana. Phytopathology® 2019, 109, 632–642. [Google Scholar] [CrossRef]
- Zhang, L.; Ding, H.; Jiang, H.; Wang, H.; Chen, K.; Duan, J.; Feng, S.; Wu, G. Regulation of Cadmium Tolerance and Accumulation by MiR156 in Arabidopsis. Chemosphere 2020, 242, 125168. [Google Scholar] [CrossRef]
- He, L.; Peng, X.; Cao, H.; Yang, K.; Xiang, L.; Li, R.; Zhang, F.; Liu, W. The NtSPL Gene Family in Nicotiana Tabacum: Genome-Wide Investigation and Expression Analysis in Response to Cadmium Stress. Genes 2023, 14, 183. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Mangrauthia, S.K.; Sarla, N. Expression Profiling of Iron Deficiency Responsive MicroRNAs and Gene Targets in Rice Seedlings of Madhukar x Swarna Recombinant Inbred Lines with Contrasting Levels of Iron in Seeds. Plant Soil. 2015, 396, 137–150. [Google Scholar] [CrossRef]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of Copper Homeostasis by Micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef] [PubMed]
- Perea-García, A.; Andrés-Bordería, A.; Huijser, P.; Peñarrubia, L. The Copper-MicroRNA Pathway Is Integrated with Developmental and Environmental Stress Responses in Arabidopsis Thaliana. Int. J. Mol. Sci. 2021, 22, 9547. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for MiR156-SPL Module in Arabidopsis Thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Busch, M.A.; Bomblies, K.; Weigel, D. Activation of a Floral Homeotic Gene in Arabidopsis. Science 1999, 285, 585–587. [Google Scholar] [CrossRef]
- Zhu, Q.-H.; Helliwell, C.A. Regulation of Flowering Time and Floral Patterning by MiR172. J. Exp. Bot. 2011, 62, 487–495. [Google Scholar] [CrossRef]
- Gou, J.; Tang, C.; Chen, N.; Wang, H.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Allen, R.D.; et al. SPL7 and SPL8 Represent a Novel Flowering Regulation Mechanism in Switchgrass. New Phytol. 2019, 222, 1610–1623. [Google Scholar] [CrossRef]
- Wang, L.; Ming, L.; Liao, K.; Xia, C.; Sun, S.; Chang, Y.; Wang, H.; Fu, D.; Xu, C.; Wang, Z.; et al. Bract Suppression Regulated by the MiR156/529-SPLs-NL1-PLA1 Module Is Required for the Transition from Vegetative to Reproductive Branching in Rice. Mol. Plant 2021, 14, 1168–1184. [Google Scholar] [CrossRef]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.; Matts, J.; Wolf, J.; Mann, D.G.J.; Stewart, C.N.; Tang, Y.; et al. Overexpression of MiR156 in Switchgrass (Panicum Virgatum L.) Results in Various Morphological Alterations and Leads to Improved Biomass Production. Plant Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. MicroRNA156 as a Promising Tool for Alfalfa Improvement. Plant Biotechnol. J. 2015, 13, 779–790. [Google Scholar] [CrossRef]
- Hu, T.; Manuela, D.; Xu, M. SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 and 13 Repress BLADE-ON-PETIOLE 1 and 2 Directly to Promote Adult Leaf Morphology in Arabidopsis. J. Exp. Bot. 2023. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Rojas, C.H.; Rocha, G.H.B.; Polverari, L.; Pinheiro Brito, D.A.; Batista, D.S.; Notini, M.M.; da Cruz, A.C.F.; Morea, E.G.O.; Sabatini, S.; Otoni, W.C.; et al. MiR156-Targeted SPL10 Controls Arabidopsis Root Meristem Activity and Root-Derived de Novo Shoot Regeneration via Cytokinin Responses. J. Exp. Bot. 2020, 71, 934–950. [Google Scholar] [CrossRef] [PubMed]
- Cui, N.; Sun, X.; Sun, M.; Jia, B.; Duanmu, H.; Lv, D.; Duan, X.; Zhu, Y. Overexpression of OsmiR156k Leads to Reduced Tolerance to Cold Stress in Rice (Oryza Sativa). Mol. Breed. 2015, 35, 1–11. [Google Scholar] [CrossRef]
- Feyissa, B.A.; Arshad, M.; Gruber, M.Y.; Kohalmi, S.E.; Hannoufa, A. The Interplay between MiR156/SPL13 and DFR/WD40–1 Regulate Drought Tolerance in Alfalfa. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef]
- Girgžde, E.; Samsone, I.; Gailis, A. Peroxidase, Polyphenol Oxidase Activity and Total Phenolic Concentration in Birch (Betula Pendula) in Vitro Shoots during Rejuvenation. Environ. Exp. Biol. 2019, 17, 15–19. [Google Scholar]
- Wu, Q.; Yang, J.; Cheng, N.; Hirschi, K.D.; White, F.F.; Park, S. Glutaredoxins in Plant Development, Abiotic Stress Response, and Iron Homeostasis: From Model Organisms to Crops. Environ. Exp. Bot. 2017, 139, 91–98. [Google Scholar] [CrossRef]
- GUJJAR, R.S.; PATHAK, A.D.; KARKUTE, S.G.; SUPAIBULWATANA, K. Multifunctional Proline Rich Proteins and Their Role in Regulating Cellular Proline Content in Plants under Stress. Biol. Plant. 2019, 63, 448–454. [Google Scholar] [CrossRef]
- Beyene, G.; Foyer, C.H.; Kunert, K.J. Two New Cysteine Proteinases with Specific Expression Patterns in Mature and Senescent Tobacco (Nicotiana tabacum L.) Leaves*. J. Exp. Bot. 2006, 57, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Ducos, E.; Fraysse, A.S.; Boutry, M. NtPDR3, an Iron-Deficiency Inducible ABC Transporter in Nicotiana tabacum. FEBS Lett. 2005, 579, 6791–6795. [Google Scholar] [CrossRef]
- Palusińska, M.; Barabasz, A.; Kozak, K.; Papierniak, A.; Maślińska, K.; Antosiewicz, D.M. Zn/Cd Status-Dependent Accumulation of Zn and Cd in Root Parts in Tobacco Is Accompanied by Specific Expression of ZIP Genes. BMC Plant Biol. 2020, 20, 1–19. [Google Scholar] [CrossRef]
- Ma, Y.; Wei, N.; Wang, Q.; Liu, Z.; Liu, W. Genome-Wide Identification and Characterization of the Heavy Metal ATPase (HMA) Gene Family in Medicago Truncatula under Copper Stress. Int. J. Biol. Macromol. 2021, 193, 893–902. [Google Scholar] [CrossRef]
- Takahashi, R.; Bashir, K.; Ishimaru, Y.; Nishizawa, N.K.; Nakanishi, H. The Role of Heavy-Metal ATPases, HMAs, in Zinc and Cadmium Transport in Rice. Plant Signal. Behav. 2012, 7, 1605–1607. [Google Scholar] [CrossRef] [PubMed]
- Hermand, V.; Julio, E.; Dorlhac de Borne, F.; Punshon, T.; Ricachenevsky, F.K.; Bellec, A.; Gosti, F.; Berthomieu, P. Inactivation of Two Newly Identified Tobacco Heavy Metal ATPases Leads to Reduced Zn and Cd Accumulation in Shoots and Reduced Pollen Germination. Metallomics 2014, 6, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; He, L.; Huo, C.; Jiang, X.; Chen, H.; Wang, R.; Tang, M.; Dong, L.; Chen, J.; Li, Y.; et al. Genome-Wide Identification and Expression Analysis of Heavy Metal Stress–Responsive Metallothionein Family Genes in Nicotiana Tabacum. Plant Mol. Biol. Rep. 2021, 39, 443–454. [Google Scholar] [CrossRef]
- Li, R.; Yang, Y.; Cao, H.; Peng, X.; Yu, Q.; He, L.; Chen, J.; Xiang, L.; Liu, W. Heterologous Expression of the Tobacco Metallothionein Gene NtMT2F Confers Enhanced Tolerance to Cd Stress in Escherichia Coli and Arabidopsis Thaliana. Plant Physiol. Biochem. 2023, 195, 247–255. [Google Scholar] [CrossRef]
- Guo, Z.; Kuang, Z.; Zhao, Y.; Deng, Y.; He, H.; Wan, M.; Tao, Y.; Wang, D.; Wei, J.; Li, L.; et al. PmiREN2.0: From Data Annotation to Functional Exploration of Plant MicroRNAs. Nucleic Acids Res. 2022, 50, D1475–D1482. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Campos, A.L.; Gutiérrez-Ortega, A. Agrobacterium-Mediated Transformation of Nicotiana tabacum Cv. Xanthi Leaf Explants. Bio-Protocol 2019, e3150. [Google Scholar] [CrossRef]
- Lopez-Hidalgo, C.; Meijon, M.; Lamelas, L.; Valledor, L. The Rainbow Protocol: A Sequential Method for Quantifying Pigments, Sugars, Free Amino Acids, Phenolics, Flavonoids and MDA from a Small Amount of Sample. Plant Cell Environ. 2021, 44, 1977–1986. [Google Scholar] [CrossRef]
- Tüzen, M. Determination of Heavy Metals in Soil, Mushroom and Plant Samples by Atomic Absorption Spectrometry. Microchem. J. 2003, 74, 289–297. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]



| Sample | WT1 | WT2 | L8 | L9 |
|---|---|---|---|---|
| Raw reads | 51,615,914 | 50,783,586 | 58,896,570 | 40,957,114 |
| Clean reads | 48,983,642 | 48,352,528 | 56,810,132 | 38,606,522 |
| Raw bases | 7,742,387,100 | 7,617,537,900 | 8,834,485,500 | 6,143,567,100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Ji, X.; Cao, H.; Huo, C.; He, L.; Peng, X.; Yang, Y.; Yang, F.; Xiong, S. Comparative Transcriptome Analysis Reveals the Effect of miR156a Overexpression on Mineral Nutrient Homeostasis in Nicotiana tabacum. Plants 2023, 12, 1739. https://doi.org/10.3390/plants12091739
Liu W, Ji X, Cao H, Huo C, He L, Peng X, Yang Y, Yang F, Xiong S. Comparative Transcriptome Analysis Reveals the Effect of miR156a Overexpression on Mineral Nutrient Homeostasis in Nicotiana tabacum. Plants. 2023; 12(9):1739. https://doi.org/10.3390/plants12091739
Chicago/Turabian StyleLiu, Wanhong, Xue Ji, Hanping Cao, Chunsong Huo, Linshen He, Xiang Peng, Ya Yang, Fang Yang, and Shu Xiong. 2023. "Comparative Transcriptome Analysis Reveals the Effect of miR156a Overexpression on Mineral Nutrient Homeostasis in Nicotiana tabacum" Plants 12, no. 9: 1739. https://doi.org/10.3390/plants12091739
APA StyleLiu, W., Ji, X., Cao, H., Huo, C., He, L., Peng, X., Yang, Y., Yang, F., & Xiong, S. (2023). Comparative Transcriptome Analysis Reveals the Effect of miR156a Overexpression on Mineral Nutrient Homeostasis in Nicotiana tabacum. Plants, 12(9), 1739. https://doi.org/10.3390/plants12091739








