An Efficient Agrobacterium-Mediated Genetic Transformation Method for Solanum betaceum Cav. Embryogenic Callus
Abstract
1. Introduction
2. Results and Discussion
2.1. Effect of Antibacterial Antibiotics on Agrobacterium
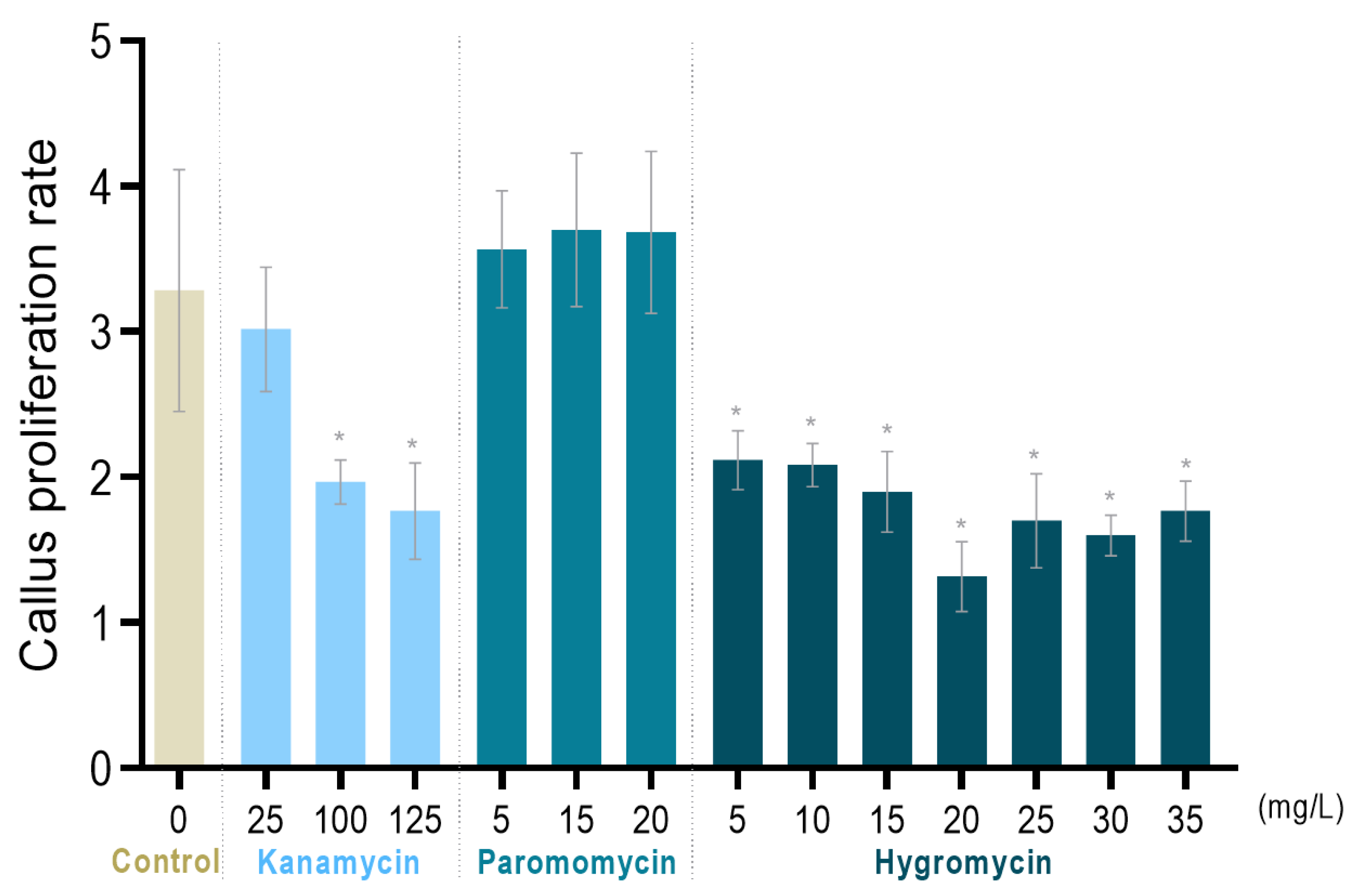
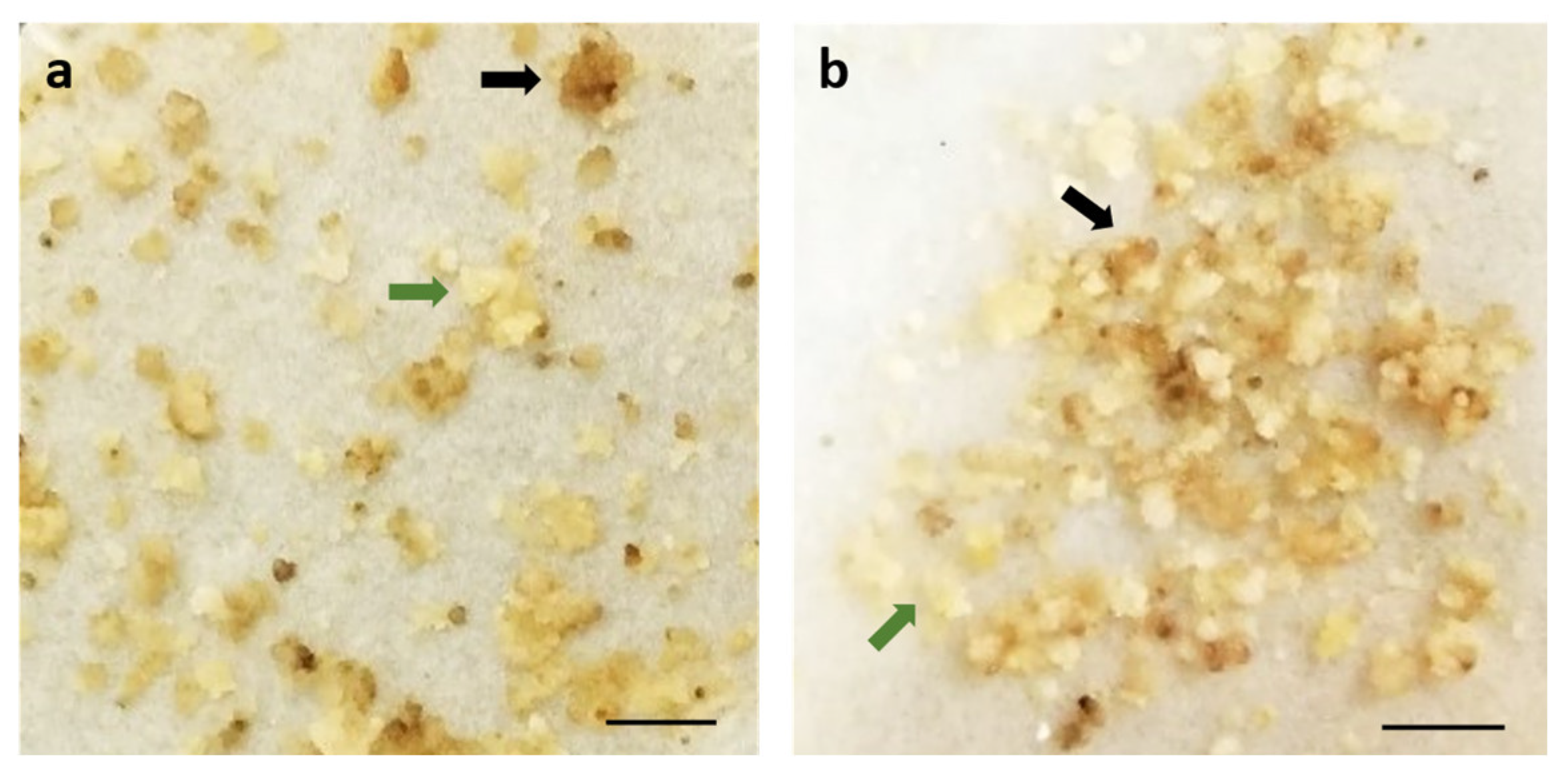
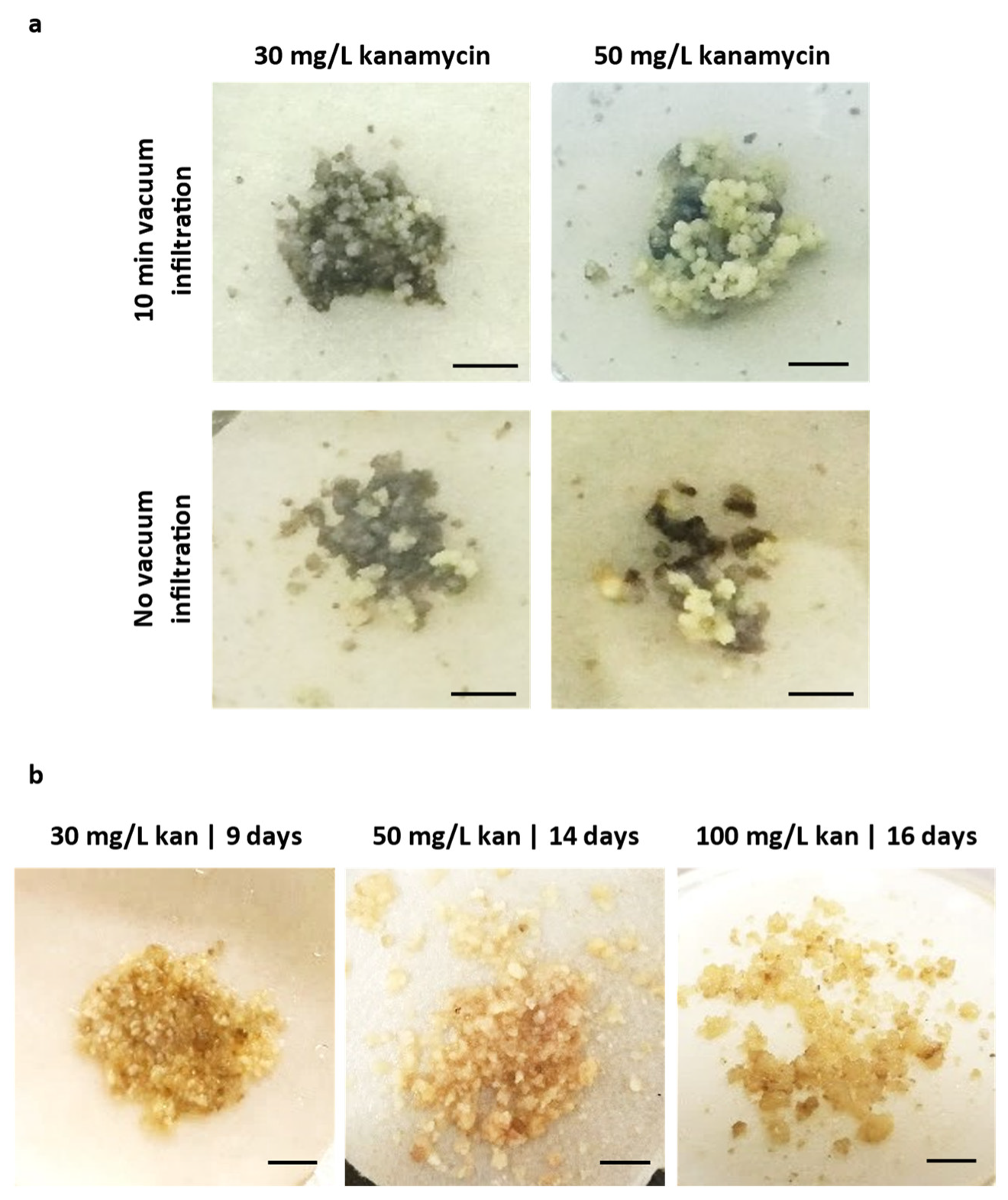
2.2. Determination of the Most Selective Antibiotic
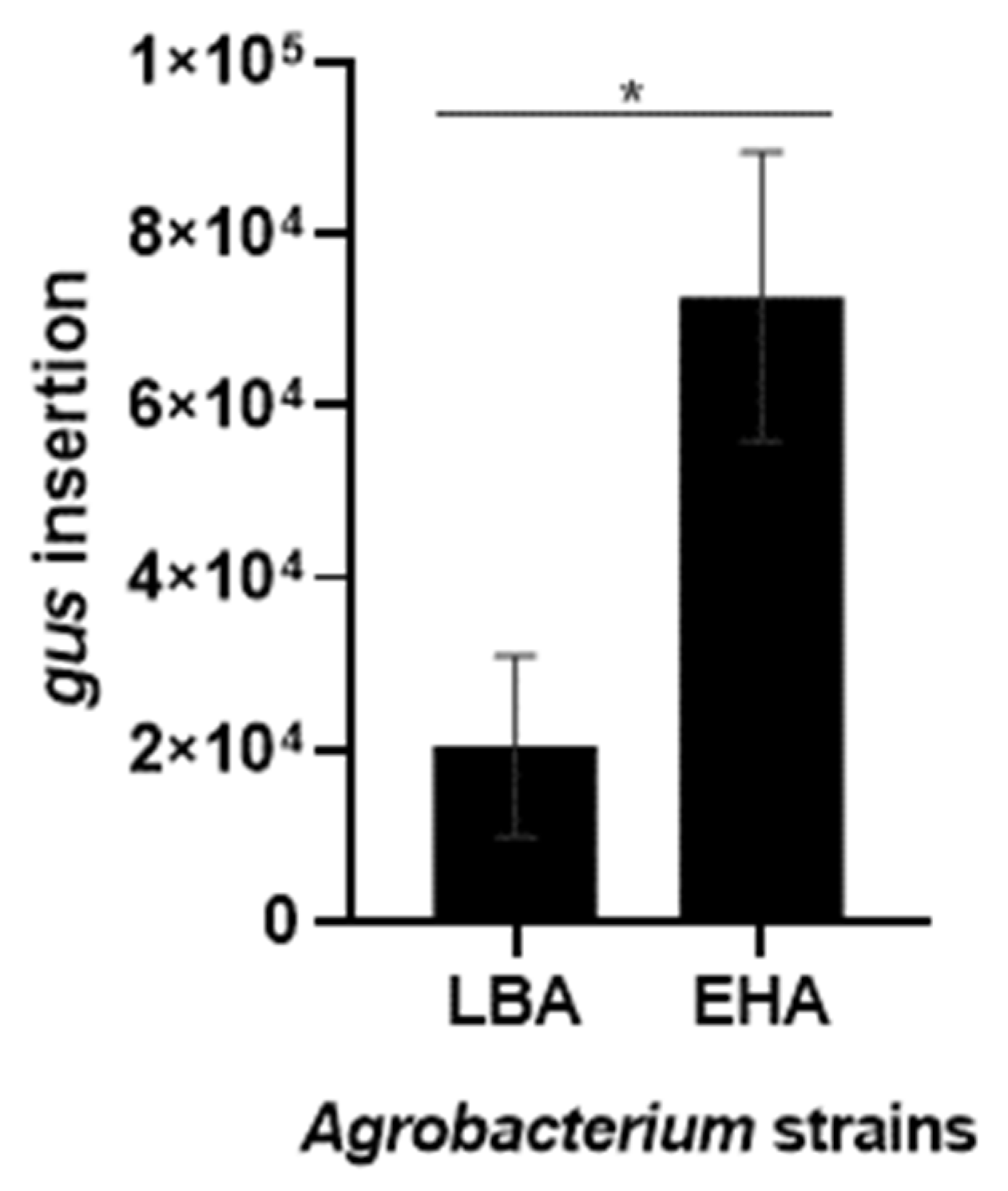
2.3. Agrobacterium-Mediated Tamarillo Callus Transformation
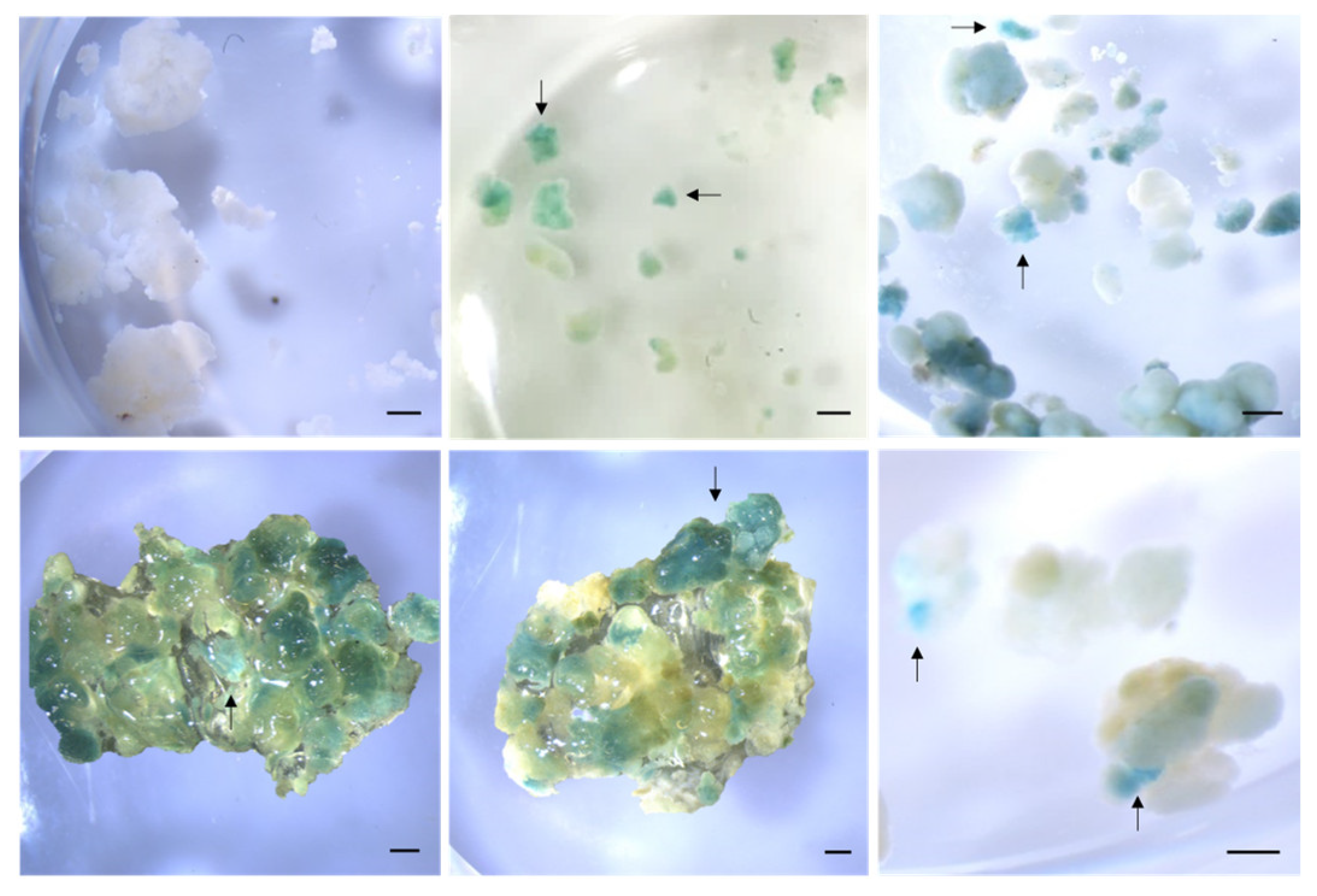
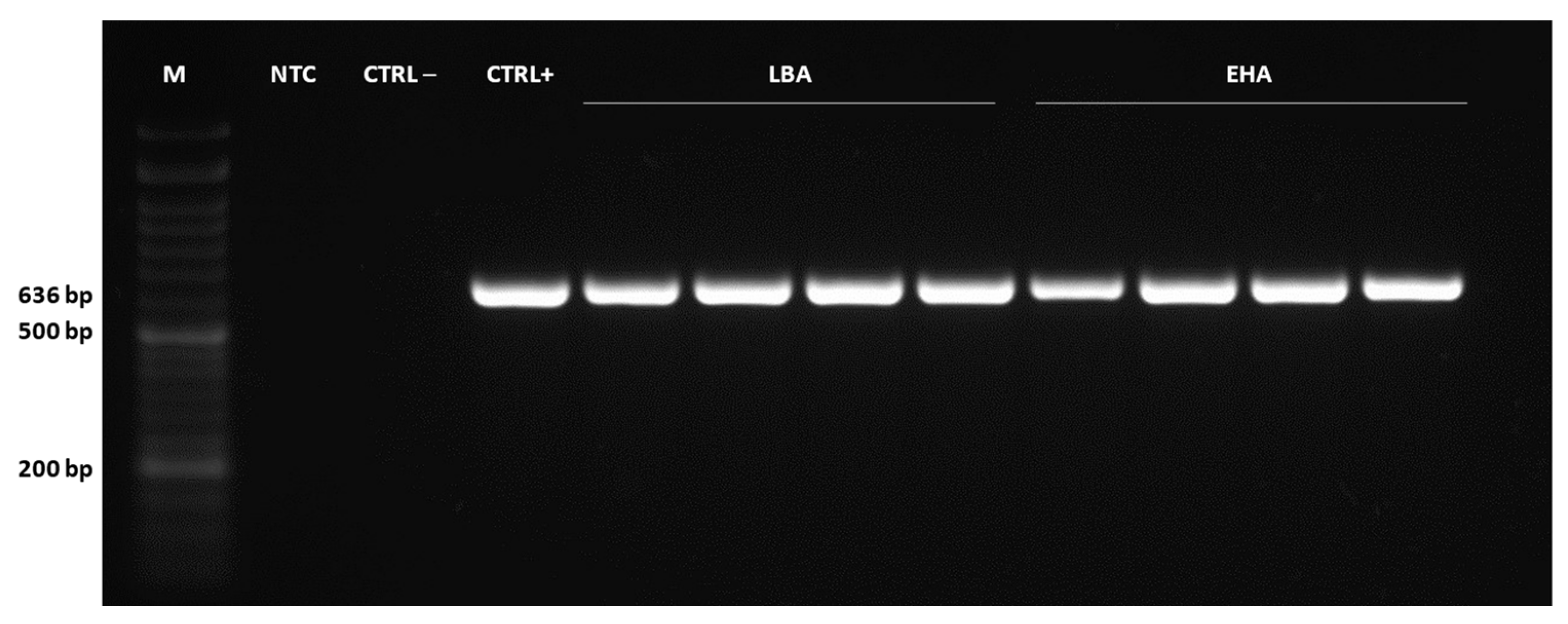
2.4. Detection of Genetically Transformed Cells
3. Materials and Methods
3.1. Induction and Proliferation of Embryogenic Callus Cultures
3.2. Agrobacterium Strains and Vector
3.3. Effect of Antibacterial Antibiotics on Agrobacterium Growth
3.4. Sensitivity Screening of Embryogenic Callus to Antibiotics
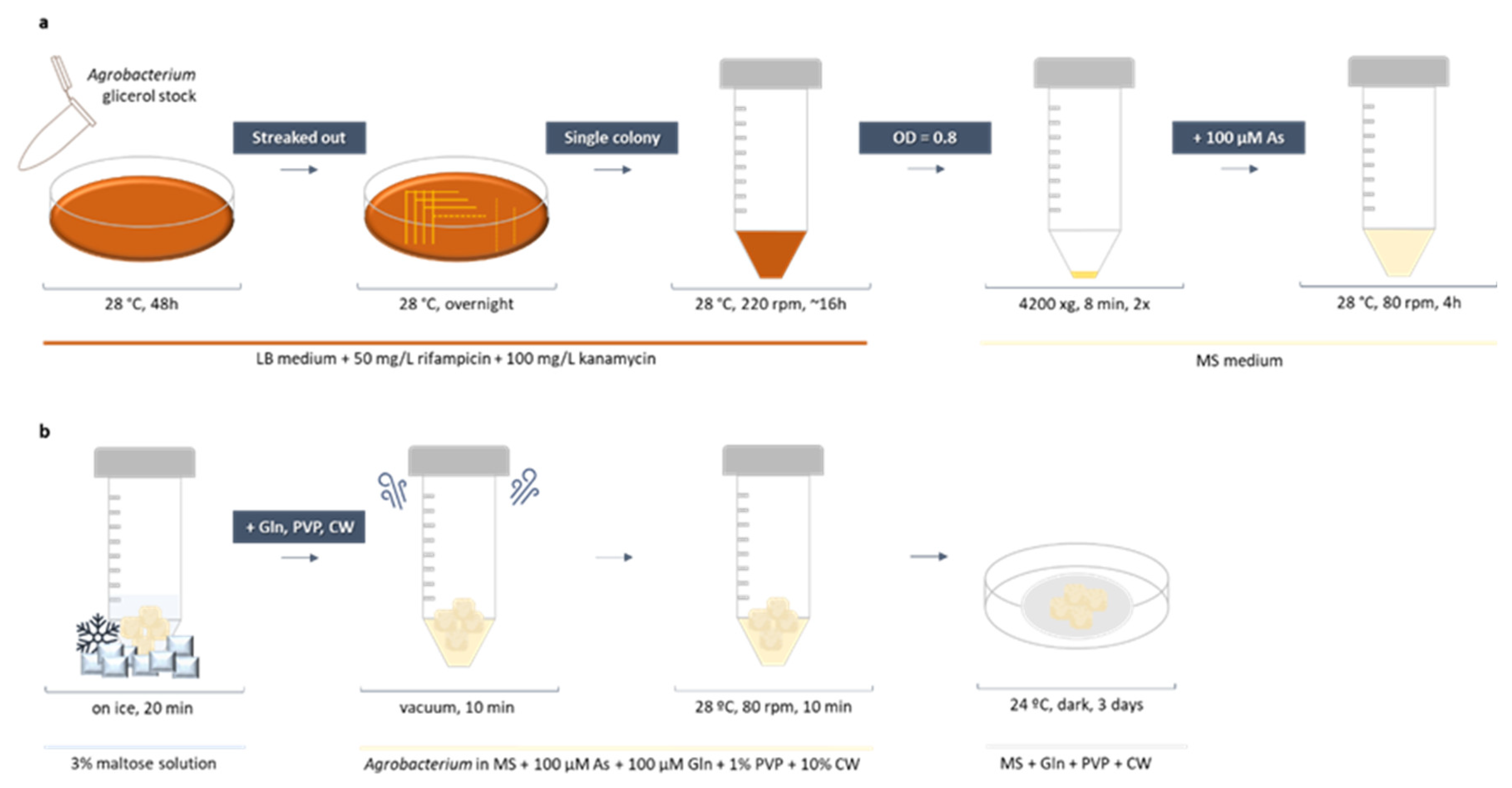
3.5. Agrobacterium Culture Preparation
3.6. Agrobacterium-Mediated Callus Transformation and Co-Cultivation
3.7. Selection of Transformed Cells
3.8. Staining for GUS Activity
3.9. PCR and qPCR Analysis of Transformants
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Song, G.Q.; Prieto, H.; Orbovic, V. Agrobacterium-mediated transformation of tree fruit crops: Methods, progress, and challenges. Front. Plant Sci. 2019, 10, 226. [Google Scholar] [CrossRef]
- Basso, M.F.; Arraes, F.B.M.; Grossi-de-Sa, M.; Moreira, V.J.V.; Alves-Ferreira, M.; Grossi-de-Sa, M.F. Insights into genetic and molecular elements for transgenic crop development. Front. Plant Sci. 2020, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P.; et al. Advancing crop transformation in the era of genome editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef]
- Mushtaq, R.; Shahzad, K.; Shah, Z.H.; Alsamadany, H.; Alzahrani, H.A.S.; Alzahrani, Y.; Mujtaba, T.; Ahmed, Z.; Mansoor, S.; Bashir, A. Isolation of biotic stress resistance genes from cotton (Gossypium arboreum) and their analysis in model plant tobacco (Nicotiana tabacum) for resistance against cotton leaf curl disease complex. J. Virol. Methods 2020, 276, 113760. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, M.; Siemiatkowska, B.; Toleco, M.R.; Jing, Y.; Strotmann, V.; Zhang, J.; Stahl, Y.; Fernie, A.R. A highly efficient Agrobacterium-mediated method for transient gene expression and functional studies in multiple plant species. Plant Commun. 2020, 1, 100028. [Google Scholar] [CrossRef] [PubMed]
- Travella, S.; Ross, S.M.; Harden, J.; Everett, C.; Snape, J.W.; Harwood, W.A. A comparison of transgenic barley lines produced by particle bombardment and Agrobacterium-mediated techniques. Plant Cell Rep. 2005, 23, 780–789. [Google Scholar] [CrossRef]
- Tzfira, T.; Hohn, B.; Gelvin, S.B. Transfer of genetic information from Agrobacterium to plants. In Reference Module in Life Sciences; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Ribeiro, T.P.; Basso, M.F.; de Carvalho, M.H.; de Macedo, L.L.P.; da Silva, D.M.L.; Lourenço-Tessutti, I.T.; de Oliveira-Neto, O.B.; de Campos-Pinto, E.R.; Lucena, W.A.; da Silva, M.C.M.; et al. Stability and tissue-specific Cry10Aa overexpression improves cotton resistance to the cotton boll weevil. Biotechnol. Res. Innov. 2019, 3, 27–41. [Google Scholar] [CrossRef]
- Wang, K. Agrobacterium Protocols; Wang, K., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1224, ISBN 978-1-4939-1657-3. [Google Scholar]
- Prohens, J.; Nuez, F. The tamarillo (Cyphomandra betacea). Small Fruits Rev. 2001, 1, 43–68. [Google Scholar] [CrossRef]
- Acosta-Quezada, P.G.; Raigón, M.D.; Riofrío-Cuenca, T.; García-Martínez, M.D.; Plazas, M.; Burneo, J.I.; Figueroa, J.G.; Vilanova, S.; Prohens, J. Diversity for chemical composition in a collection of different varietal types of tree tomato (Solanum betaceum Cav.), an andean exotic fruit. Food Chem. 2015, 169, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, F. Tamarillo (Solanum betaceum): Chemical composition, biological properties, and product innovation. Trends Food Sci. Technol. 2020, 95, 45–58. [Google Scholar] [CrossRef]
- Correia, S.I.; Canhoto, J.M. Biotechnology of tamarillo (Cyphomandra betacea): From in vitro cloning to genetic transformation. Sci. Hortic. 2012, 148, 161–168. [Google Scholar] [CrossRef]
- Correia, S.; Lopes, M.L.; Canhoto, J.M. Somatic embryogenesis induction system for cloning an adult Cyphomandra betacea (Cav.) Sendt. (tamarillo). Trees-Struct. Funct. 2011, 25, 1009–1020. [Google Scholar] [CrossRef]
- Correia, S.I.; Lopes, M.L.; Canhoto, J.M. Somatic embryogenesis in tamarillo (Cyphomandra betacea): Recent advances. Acta Hortic. 2009, 839, 157–164. [Google Scholar] [CrossRef]
- Correia, S.; Cunha, A.E.; Salgueiro, L.; Canhoto, J.M. Somatic embryogenesis in tamarillo (Cyphomandra betacea): Approaches to increase efficiency of embryo formation and plant development. Plant Cell Tissue Organ. Cult. 2012, 109, 143–152. [Google Scholar] [CrossRef]
- Cordeiro, D.; Rito, M.; Borges, F.; Canhoto, J.; Correia, S. Selection and validation of reference genes for qPCR analysis of miRNAs and their targets during somatic embryogenesis in tamarillo (Solanum betaceum Cav.). Plant Cell Tissue Organ. Cult. 2020, 143, 109–120. [Google Scholar] [CrossRef]
- Casimiro, B.; Mota, I.; Veríssimo, P.; Canhoto, J.; Correia, S. Enhancing the production of hydrolytic enzymes in elicited tamarillo (Solanum betaceum Cav.) cell suspension cultures. Plants 2023, 12, 190. [Google Scholar] [CrossRef]
- Correia, S.; Canhoto, J.M. Somatic embryogenesis of tamarillo (Solanum betaceum Cav.). In Step Wise Protocols for Somatic Embryogenesis of Important Woody Plants; Jain, S.M., Gupta, P.K., Eds.; Springer: Cham, Switzerland, 2018; pp. 171–179. [Google Scholar]
- Dhekney, S.A.; Sessions, S.K.; Brungart-Rosenberg, M.; Claflin, C.; Li, Z.T.; Gray, D.J. Genetic modification of grapevine embryogenic cultures. Methods Mol. Biol. 2019, 1864, 191–201. [Google Scholar] [CrossRef]
- Ganapathi, T.R.; Higgs, N.S.; Balint-Kurti, P.J.; Arntzen, C.J.; May, G.; van Eck, J.M. Agrobacterium-mediated transformation of embryogenic cell suspensions of the banana cultivar Rasthali (AAB). Plant Cell Rep. 2001, 20, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Leelavathi, S.; Sunnichan, V.G.; Kumria, R.; Vijaykanth, G.P.; Bhatnagar, R.K.; Reddy, V.S. A simple and rapid Agrobacterium-mediated transformation protocol for cotton (Gossypium hirsutum L.): Embryogenic calli as a source to generate large numbers of transgenic plants. Plant Cell Rep. 2004, 22, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Ratjens, S.; Mortensen, S.; Kumpf, A.; Bartsch, M.; Winkelmann, T. Embryogenic callus as target for efficient transformation of Cyclamen persicum enabling gene function studies. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rajewski, A.C.; Elkins, K.B.; Henry, A.; van Eck, J.; Litt, A. In vitro plant regeneration and Agrobacterium tumefaciens–mediated transformation of Datura stramonium (solanaceae). Appl. Plant Sci. 2019, 7, e01220. [Google Scholar] [CrossRef] [PubMed]
- Matveeva, T.V.; Lutova, L.A. Horizontal gene transfer from Agrobacterium to plants. Front. Plant Sci. 2014, 5, 326. [Google Scholar] [CrossRef] [PubMed]
- Niedbała, G.; Niazian, M.; Sabbatini, P. Modeling Agrobacterium-mediated gene transformation of tobacco (Nicotiana tabacum)-a model plant for gene transformation studies. Front. Plant Sci. 2021, 12, 1454. [Google Scholar] [CrossRef]
- Swartwood, K.; van Eck, J. Development of plant regeneration and Agrobacterium tumefaciens-mediated transformation Methodology for Physalis Pruinosa. Plant Cell Tissue Organ. Cult. 2019, 137, 465–472. [Google Scholar] [CrossRef]
- Nava, E.; Dávila, Y.; Arellano, J.; Ortiz, A.; Alvárez, L.; Marquina, S.; Villarreal, M.L. Genetic transformation of Solanum chrysotrichum by Agrobacterium tumefaciens and the production of antifungal saponins. Vitr. Cell. Dev. Biol.-Plant 2011, 47, 650–657. [Google Scholar] [CrossRef]
- Kumar, A.; Miller, M.; Whitty, P.; Lyon, J.; Davie, P. Agrobacterium-mediated transformation of five wild Solanum species using in vitro microtubers. Plant Cell Rep. 1995, 14, 324–328. [Google Scholar] [CrossRef]
- Curtis, I.S.; Power, J.B.; Hedden, P.; Phillips, A.; Lowe, K.C.; Ward, D.A.; Davey, M.R. Transformation and characterization of transgenic plants of Solanum dulcamara L.-incidence of transgene silencing. Ann. Bot. 2000, 86, 63–71. [Google Scholar] [CrossRef]
- Van Eck, J.; Keen, P.; Tjahjadi, M. Agrobacterium tumefaciens-mediated transformation of tomato. Methods Mol. Biol. 2019, 1864, 225–234. [Google Scholar] [CrossRef]
- Khatun, M.; Borphukan, B.; Alam, I.; Keya, C.A.; Khan, H.; Reddy, M.K.; Salimullah, M. An improved Agrobacterium mediated transformation and regeneration protocol for successful genetic engineering and genome editing in eggplant. Sci. Hortic. 2022, 293, 110716. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Gardner, R.C. Agrobacterium-mediated transformation of pepino and regeneration of transgenic plants. Plant Cell Rep. 1991, 10, 208–212. [Google Scholar] [CrossRef]
- Peng, W.; Wang, X.; Wei, H.; Zhang, Z.; Teng, C.; Li, Q.; Lyu, K.; Lyu, S.; Fan, Y. Development of Agrobacterium tumefaciens- mediated transformation of Solanum nigrum and expression of AcMYB110 in S. Nigrum. Research Square 2022. preprint. [Google Scholar] [CrossRef]
- Ducreux, L.J.M.; Morris, W.L.; Taylor, M.A.; Millam, S. Agrobacterium-mediated transformation of Solanum phureja. Plant Cell Rep. 2005, 24, 10–14. [Google Scholar] [CrossRef]
- Ugandhar, T.; Odelu, G.; Parvathi, D.; Anitha, U.D.; Venkateshwarlu, M. Agrobacterium-mediated genetic transformation and regeneration from leaf explants of Solanum thorvum (Swartz) a medicinally important plant. Int. J. Adv. Res. 2017, 5, 896–904. [Google Scholar] [CrossRef]
- Shilpha, J.; Jayashre, M.; Joe Virgin Largia, M.; Ramesh, M. Direct shoot organogenesis and Agrobacterium tumefaciens mediated transformation of Solanum trilobatum L. Turk. J. Biol. 2016, 40, 866–877. [Google Scholar] [CrossRef]
- Bruce, M.A.; Shoup Rupp, J.L. Agrobacterium-mediated transformation of Solanum tuberosum L., potato. Methods Mol. Biol. 2019, 1864, 203–223. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Gardner, R.C. Regeneration of transgenic tamarillo plants. Plant Cell Rep. 1993, 12, 347–351. [Google Scholar] [CrossRef]
- Cohen, D.; van den Brink, R.C.; MacDiarmid, R.M.; Beck, D.L.; Forster, R.L.S. Resistance to tamarillo mosaic virus in transgenic tamarillos and expression of the transgenes in F1 progeny. Acta Hortic. 2000, 521, 43–49. [Google Scholar] [CrossRef]
- Correia, S.; Alhinho, A.T.; Casimiro, B.; Miguel, C.M.; Oliveira, M.; Veríssimo, P.; Canhoto, J. NEP-TC a rRNA methyltransferase involved on somatic embryogenesis of tamarillo (Solanum betaceum Cav.). Front. Plant Sci. 2019, 10, 438. [Google Scholar] [CrossRef] [PubMed]
- Aida, R.; Hirose, Y.; Kishimoto, S.; Shibata, M. Agrobacterium tumefaciens–mediated transformation of Cyclamen persicum Mill. Plant Sci. 1999, 148, 1–7. [Google Scholar] [CrossRef]
- Hood, E.E.; Gelvin, S.B.; Melchers, L.S.; Hoekema, A. New Agrobacterium helper plasmids for gene transfer to plants. Transgenic. Res. 1993, 2, 208–218. [Google Scholar] [CrossRef]
- Hoekema, A.; Hirsch, P.R.; Hooykaas, P.J.J.; Schilperoort, R.A. A binary plant vector strategy based on separation of Vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 1983, 303, 179–180. [Google Scholar] [CrossRef]
- Vancanneyt, G.; Schmidt, R.; O’Connor-Sanchez, A.; Willmitzer, L.; Rocha-Sosa, M. Construction of an intron-containing marker gene: Splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Molec. Gen. Genet. 1990, 220, 245–250. [Google Scholar] [CrossRef]
- Kole, C.; Hall, T.C. Transgenic Temperate Fruits and Nuts; Wiley-Blackwell: Chichester, UK, 2008; Volume 4. [Google Scholar]
- López, E.; Proaño, K.; Jadán, M.; Mihai, R. Callus tissue induction and analysis of GUS reporter gene expression in tomato (Solanum lycopersicum L.) Transformed with Agrobacterium tumefaciens. Rom. Biotechnol. Lett. 2015, 20, 10205–10211. [Google Scholar]
- Kumar, N.; Gulati, A.; Bhattacharya, A. L-glutamine and L-glutamic acid facilitate successful Agrobacterium infection of recalcitrant tea cultivars. Appl. Biochem. Biotechnol. 2013, 170, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Priya, A.M.; Pandian, S.K.; Manikandan, R. The effect of different antibiotics on the elimination of Agrobacterium and high frequency Agrobacterium-mediated transformation of indica rice (Oryza sativa L.). Czech J. Genet. Plant Breed. 2012, 48, 120–130. [Google Scholar] [CrossRef]
- Chu, M.; Quiñonero, C.; Akdemir, H.; Alburquerque, N.; Pedreño, M.Á.; Burgos, L. Agrobacterium-mediated transformation of Vitis Cv. Monastrell suspension-cultured cells: Determination of critical parameters. Biotechnol. Prog. 2016, 32, 725–734. [Google Scholar] [CrossRef]
- Sun, Z.L.; Li, X.; Zhou, W.; Yan, J.D.; Gao, Y.R.; Li, X.W.; Sun, J.C.; Fang, K.F.; Zhang, Q.; Xing, Y.; et al. Agrobacterium-mediated genetic transformation of chinese chestnut (Castanea mollissima Blume). Plant Cell Tissue Organ. Cult. 2020, 140, 95–103. [Google Scholar] [CrossRef]
- Ma, R.; Yu, Z.; Cai, Q.; Li, H.; Dong, Y.; Oksman-Caldentey, K.M.; Rischer, H. Agrobacterium-mediated genetic transformation of the medicinal plant Veratrum dahuricum. Plants 2020, 9, 191. [Google Scholar] [CrossRef]
- Zhang, W.J.; Dewey, R.E.; Boss, W.; Phillippy, B.Q.; Qu, R. Enhanced Agrobacterium-mediated transformation efficiencies in monocot cells is associated with attenuated defense responses. Plant Mol. Biol. 2013, 81, 273–286. [Google Scholar] [CrossRef]
- Carvalho, C.H.; Zehr, U.B.; Gunaratna, N.; Anderson, J.; Kononowicz, H.H.; Hodges, T.K.; Axtell, J.D. Agrobacterium-mediated transformation of Sorghum: Factors that affect transformation efficiency. Genet Mol. Biol. 2004, 27, 259–269. [Google Scholar] [CrossRef]
- Sheikholeslam, S.N.; Weeks, D.P. Acetosyringone promotes high efficiency transformation of Arabidopsis thaliana explants by Agrobacterium tumefaciens. Plant Mol. Biol. 1987, 8, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Uranbey, S.; Sevimay, C.S.; Kaya, M.D.; İpek, A.; Sancak, C.; Başalma, D.; Er, C.; Özcan, S. Influence of different co-cultivation temperatures, periods and media on Agrobacterium tumefaciens-mediated gene transfer. Biol. Plant 2005, 49, 53–57. [Google Scholar] [CrossRef]
- Stefano, B.; Patrizia, B.; Matteo, C.; Massimo, G. Inverse PCR and quantitative PCR as alternative methods to southern blotting analysis to assess transgene copy number and characterize the integration site in transgenic woody plants. Biochem. Genet. 2016, 54, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Mason, G.; Provero, P.; Vaira, A.M.; Accotto, G.P. Estimating the number of integrations in transformed plants by quantitative real-time PCR. BMC Biotechnol. 2002, 2, 20. [Google Scholar] [CrossRef]
- Faize, M.; Faize, L.; Burgos, L. Using quantitative real-time PCR to detect chimeras in transgenic tobacco and apricot and to monitor their dissociation. BMC Biotechnol. 2010, 10, 53. [Google Scholar] [CrossRef]
- Yang, L.; Ding, J.; Zhang, C.; Jia, J.; Weng, H.; Liu, W.; Zhang, D. Estimating the copy number of transgenes in transformed rice by real-time quantitative PCR. Plant Cell Rep. 2005, 23, 759–763. [Google Scholar] [CrossRef]
- You-wen, Q.; Xue-jun, G.; Bang-ruo, Q.; Lu, L.; Zhen, Z. Establishment of TaqMan real-time quantitative PCR assay for foreign gene copy numbers in transgenic soybean. J. Northeast. Agric. Univ. 2012, 19, 48–52. [Google Scholar] [CrossRef]
- Subramanyam, K.; Subramanyam, K.; Sailaja, K.V.; Srinivasulu, M.; Lakshmidevi, K. Highly efficient Agrobacterium-mediated transformation of banana Cv. Rasthali (AAB) via sonication and vacuum infiltration. Plant Cell Rep. 2011, 30, 425–436. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1972; ISBN 0879691069. [Google Scholar]
- Prakash, D.P.; Deepali, B.S.; Asokan, R.; Ramachandra, Y.L.; Anand, L.; Hanur, V.S. Effect of antibiotics and gelling agents in transformation of brinjal (Solanum melongena L.) Cv. Manjarigota. J. Hortic. Sci. 2007, 2, 19–25. [Google Scholar]
- Li, Z.T.; Dhekney, S.; Dutt, M.; Aman, M.; Tattersall, J.; Kelley, K.T.; Gray, D.J. Optimizing Agrobacterium-mediated transformation of grapevine. In Vitro Cell. Dev. Biol.-Plant 2006, 42, 220–227. [Google Scholar] [CrossRef]
- Cano, V.; Martínez, M.T.; San José, M.C.; Couselo, J.L.; Varas, E.; Bouza-Morcillo, L.; Toribio, M.; Corredoira, E. Regeneration of transgenic plants by Agrobacterium-mediated transformation of Quercus ilex L. somatic embryos with the gene CsTL1. New For. 2020, 51, 1003–1021. [Google Scholar] [CrossRef]
- Ribas, A.F.; Dechamp, E.; Champion, A.; Bertrand, B.; Combes, M.; Verdeil, J.; Lapeyre, F.; Lashermes, P.; Etienne, H. Agrobacterium-mediated genetic transformation of Coffea arabica (L.) is greatly enhanced by using established embryogenic callus cultures. BMC Plant Biol. 2011, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Bull, S.; Owiti, J.; Niklaus, M.; Beeching, J.R.; Gruissem, W.; Vanderschuren, H. Agrobacterium-mediated transformation of friable embryogenic calli and regeneration of transgenic cassava. Nat. Protoc. 2009, 4, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Plant Transformation and Genome Editing-GUS Assay Protocol. Available online: https://web.uri.edu/ctwg/ (accessed on 3 November 2022).
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, E45. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic Concentrations (mg/L) | Antibacterial Effect | |
|---|---|---|
| Carbenicillin | Cefotaxime | |
| 0 | 0 | ● |
| 50 | 0 | ● |
| 0 | 50 | ● |
| 50 | 50 | ○ |
| 100 | 0 | ○ |
| 0 | 100 | ○ |
| 100 | 100 | ○ |
| 200 | 0 | ○ |
| 0 | 200 | ○ |
| 200 | 200 | ○ |
| 250 | 250 | ○ |
| 300 | 300 | ○ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordeiro, D.; Alves, A.; Ferraz, R.; Casimiro, B.; Canhoto, J.; Correia, S. An Efficient Agrobacterium-Mediated Genetic Transformation Method for Solanum betaceum Cav. Embryogenic Callus. Plants 2023, 12, 1202. https://doi.org/10.3390/plants12051202
Cordeiro D, Alves A, Ferraz R, Casimiro B, Canhoto J, Correia S. An Efficient Agrobacterium-Mediated Genetic Transformation Method for Solanum betaceum Cav. Embryogenic Callus. Plants. 2023; 12(5):1202. https://doi.org/10.3390/plants12051202
Chicago/Turabian StyleCordeiro, Daniela, Ana Alves, Ricardo Ferraz, Bruno Casimiro, Jorge Canhoto, and Sandra Correia. 2023. "An Efficient Agrobacterium-Mediated Genetic Transformation Method for Solanum betaceum Cav. Embryogenic Callus" Plants 12, no. 5: 1202. https://doi.org/10.3390/plants12051202
APA StyleCordeiro, D., Alves, A., Ferraz, R., Casimiro, B., Canhoto, J., & Correia, S. (2023). An Efficient Agrobacterium-Mediated Genetic Transformation Method for Solanum betaceum Cav. Embryogenic Callus. Plants, 12(5), 1202. https://doi.org/10.3390/plants12051202













