Cytogeography of Naturalized Solidago canadensis Populations in Europe
Abstract
1. Introduction
2. Results
2.1. Molecular Markers and Phylogenetic Analysis
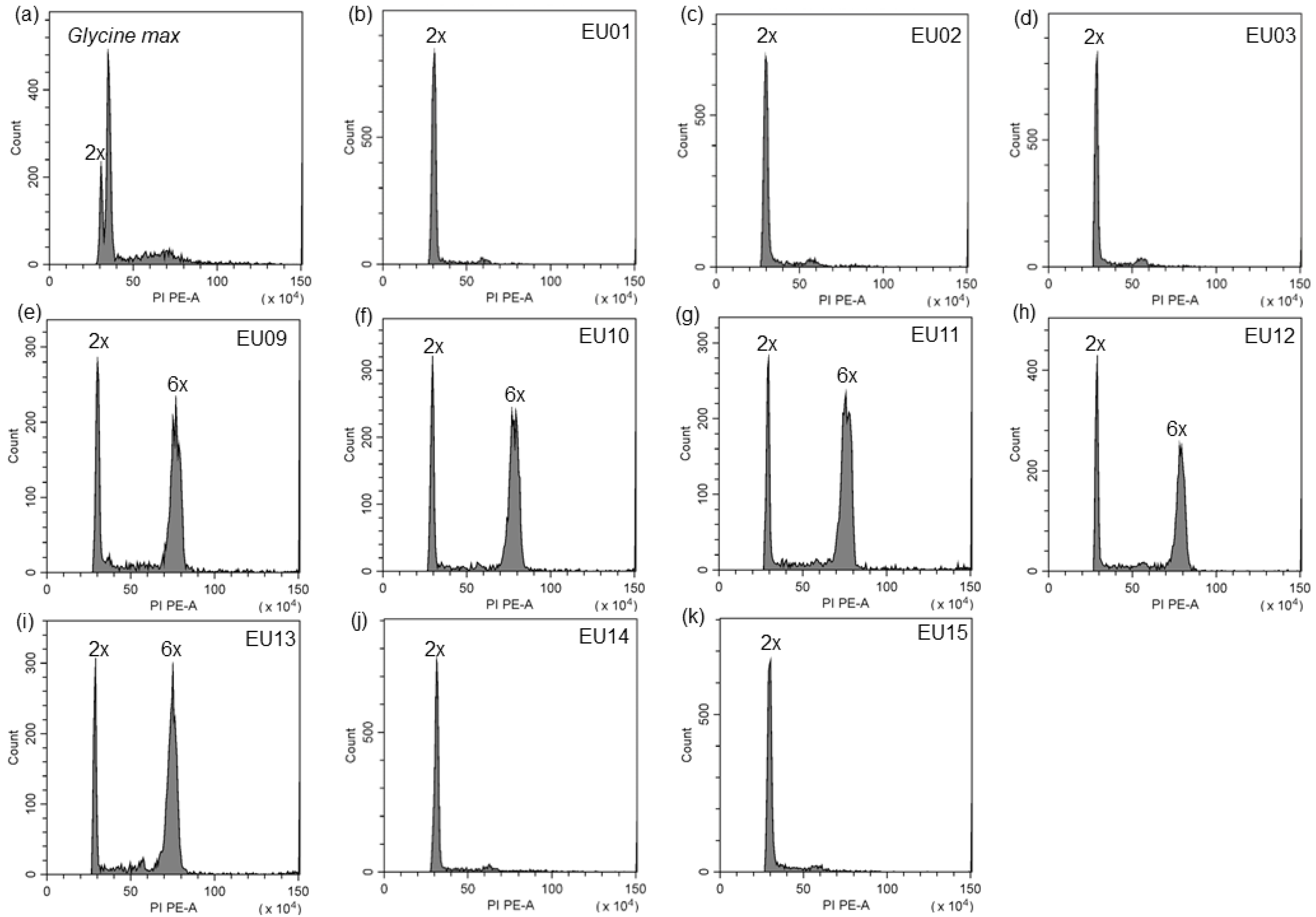
2.2. Ploidy Level of S. canadensis Populations in Europe
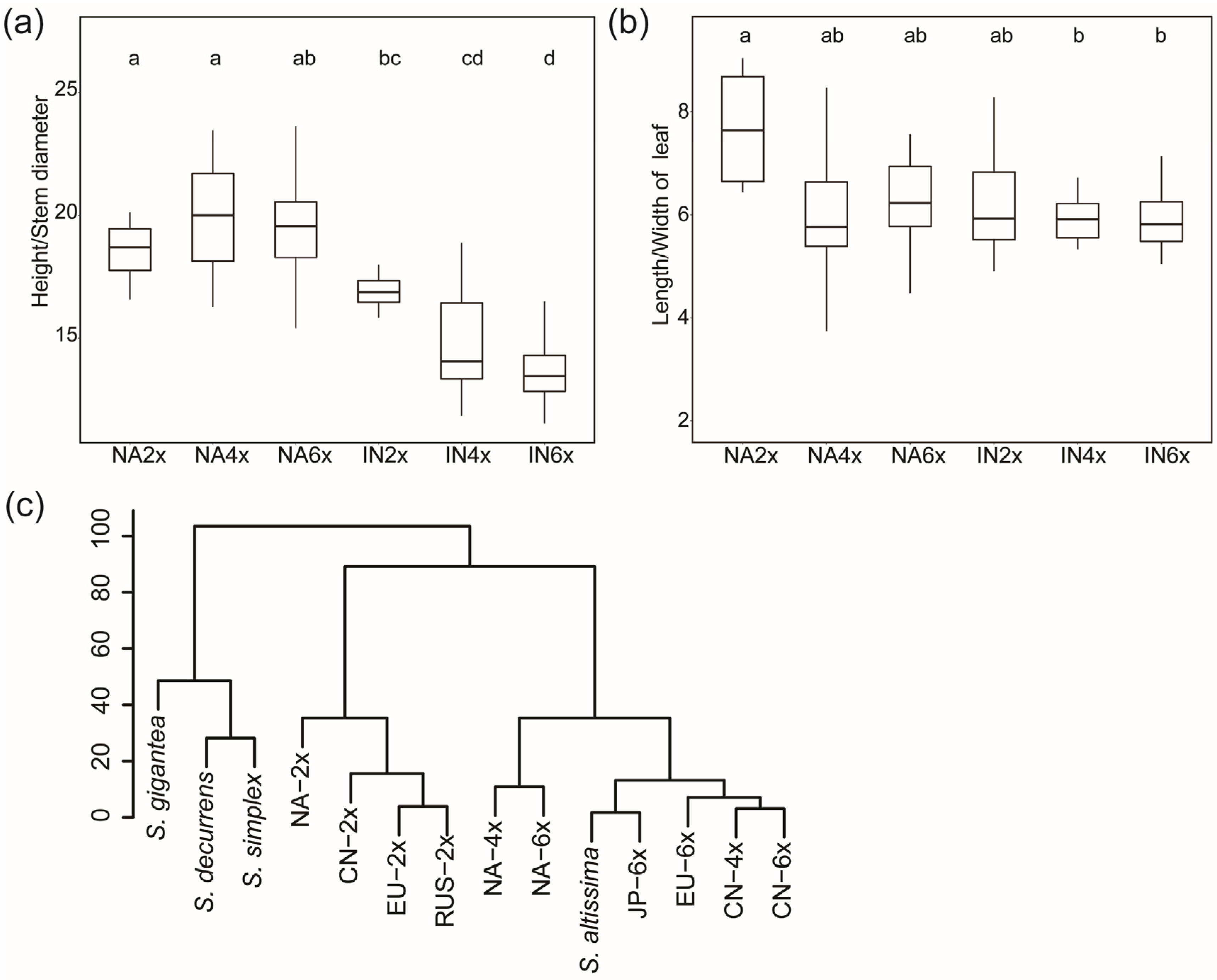
2.3. Comparison of the Morphological Traits of Different Geo-Cytotype Populations
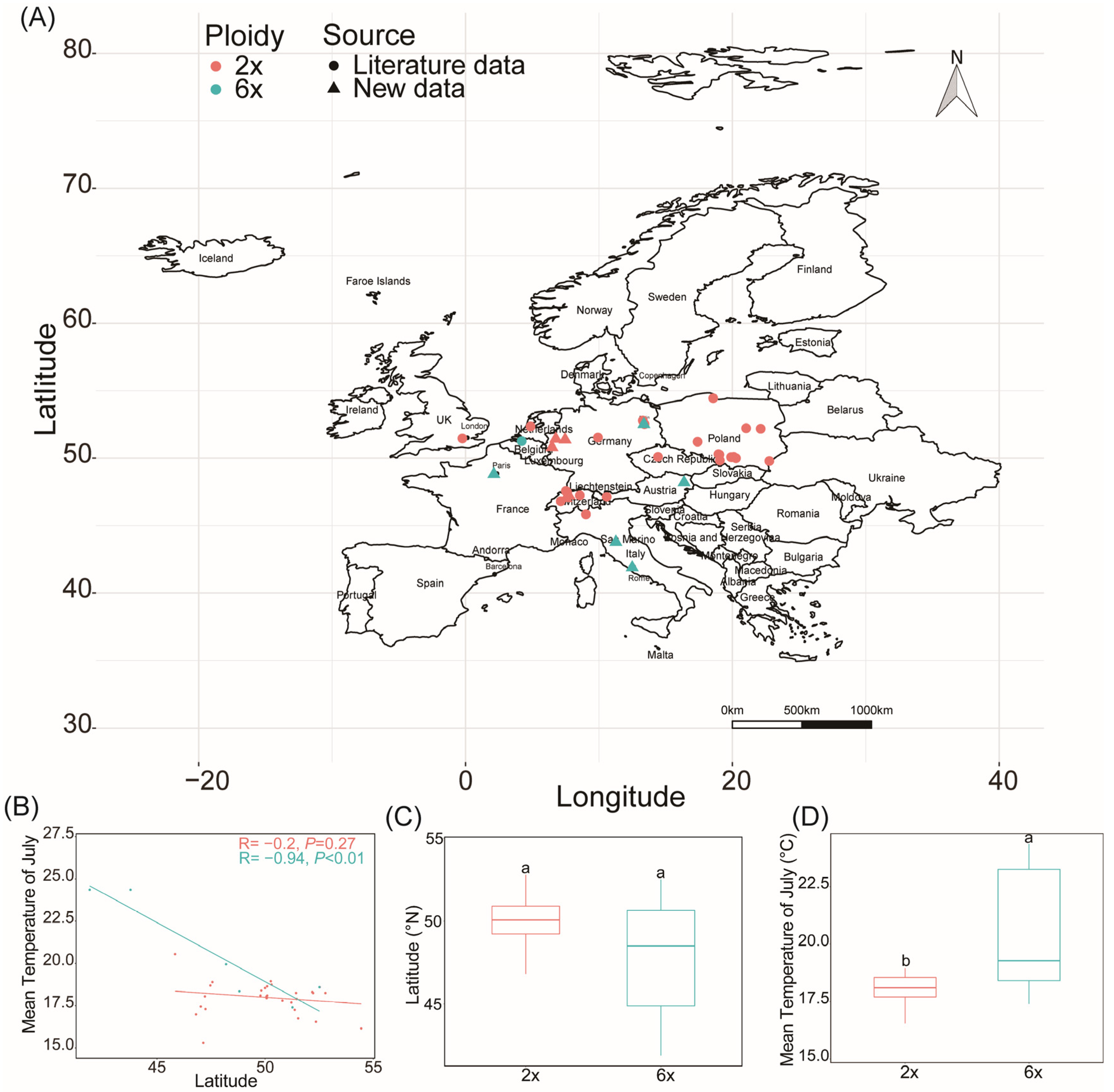
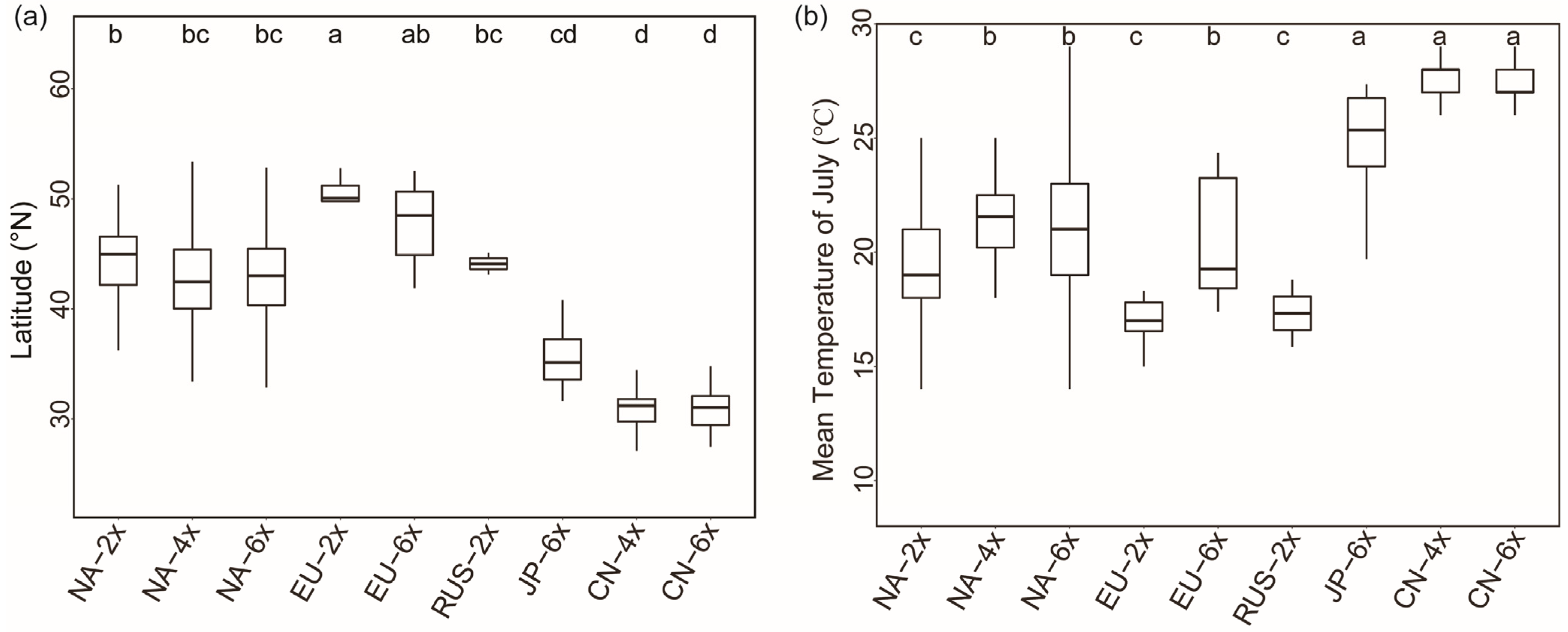
2.4. Geographical Distribution in Europe and Global Differentiation of Ecological Niches of Different Ploidy Populations
3. Discussion
3.1. The Relationship of S. canadensis and S. altissima
3.2. Ploidy and Environment Determine the Morphological Diversity of S. canadensis
3.3. Polyploid S. canadensis Has Been Successfully Invaded and Widely Distributed in Europe
3.4. Ploidy-Driven Ecological Niche Differentiation of S. canadensis in the Northern Hemisphere Depends on Environmental Factors
4. Materials and Methods
4.1. Experimental Materials
4.2. DNA Extraction and Molecular Identification
4.3. Construct Phylogenetic Tree
4.4. Determination of Ploidy Level
4.5. Measurement of Morphological Traits
4.6. Geographical Differentiation Analysis of Different Ploidy Populations
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weber, E. The dynamics of plant invasions: A case study of three exotic goldenrod species (Solidago L.) in Europe. J. Biogeogr. 1998, 25, 147–154. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H. Interactions between alien goldenrods (Solidago and Euthamia species) and comparison with native species in Central Europe. Flora-Morphol. Distrib. Funct. Ecol. Plants 2016, 218, 51–61. [Google Scholar] [CrossRef]
- Melville, M.R.; Morton, J.K. A biosystematic study of the Solidago canadensis (Compositae) complex. I. The Ontario populations. Can. J. Bot. 1982, 60, 976–997. [Google Scholar] [CrossRef]
- Semple, J.C.; Ringius, G.S.; Leeder, C.; Morton, G. Chromosome numbers of goldenrods, Euthamia and Solidago (Compositae: Astereae). II. Additional counts with comments on cytogeography. Brittonia 1984, 36, 280–292. [Google Scholar] [CrossRef]
- Van Kleunen, M.; Schmid, B. No evidence for an evolutionary increased competitive ability in an invasive plant. Ecology 2003, 84, 2816–2823. [Google Scholar] [CrossRef]
- Cheng, J.; Li, J.; Zhang, Z.; Lu, H.; Chen, G.; Yao, B.; Dong, Y.; Ma, L.; Yuan, X.; Xu, J. Autopolyploidy-driven range expansion of a temperate-originated plant to pan-tropic under global change. Ecol. Monogr. 2020, 91, e01445. [Google Scholar] [CrossRef]
- Voser-Huber, M.L. Studien an Eingebürgerten Arten der Gattung solidago L.: Probleme mit S. gigantea, S. canadensis und S. graminifolia in Naturschutzgebieten des Schweizerischen Mittellandes; Lubrecht & Cramer Limited: Port Jervis, NY, USA, 1983. [Google Scholar]
- Schlaepfer, D.R.; Edwards, P.J.; Billeter, R. Why only tetraploid Solidago gigantea (Asteraceae) became invasive: A common garden comparison of ploidy levels. Oecologia 2010, 163, 661–673. [Google Scholar] [CrossRef]
- Semple, J.C.; Rahman, H.; Bzovsky, S.; Sorour, M.K.; Kornobis, K.; Laphitz, R.L.; Tong, L. A multivariate morphometric study of the Solidago altissima complex and S. canadensis (Asteraceae: Astereae). Phytoneuron 2015, 10, 1–31. [Google Scholar]
- Croat, T. Solidago canadensis complex of the Great Plains. Brittonia 1972, 24, 317–326. [Google Scholar] [CrossRef]
- Dong, M.; Lu, J.Z.; Zhang, W.J.; Chen, J.K.; Li, B. Canada goldenrod (Solidago canadensis): An invasive alien weed rapidly spreading in China. J. Syst. Evol. 2006, 44, 72. [Google Scholar] [CrossRef]
- Weber, E. Biological flora of central Europe: Solidago altissima L. Flora 2000, 195, 123–134. [Google Scholar] [CrossRef]
- Verloove, F.; Zonneveld, B.J.; Semple, J.C. First evidence for the presence of invasive Solidago altissima (Asteraceae) in Europe. Willdenowia 2017, 47, 69–75. [Google Scholar] [CrossRef]
- Kato-Noguchi, H.; Kato, M. Allelopathy and Allelochemicals of Solidago canadensis L. and S. altissima L. for Their Naturalization. Plants 2022, 11, 3235. [Google Scholar] [CrossRef] [PubMed]
- Szymura, M.; Szymura, T.H. Historical contingency and spatial processes rather than ecological niche differentiation explain the distribution of invasive goldenrods (Solidago and Euthamia). Plant Ecol. 2016, 217, 565–582. [Google Scholar] [CrossRef]
- Musiał, K.; Pagitz, K.; Gudžinskas, Z.; Łazarski, G.; Pliszko, A. Chromosome numbers in hybrids between invasive and native Solidago (Asteraceae) species in Europe. Phytotaxa 2020, 471, 267–275. [Google Scholar] [CrossRef]
- Pliszko, A.; Zalewska-Gałosz, J. Molecular evidence for hybridization between invasive Solidago canadensis and native S. virgaurea. Biol. Invasions 2016, 18, 3103–3108. [Google Scholar] [CrossRef]
- Hayward, A.C.; Tollenaere, R.; Dalton-Morgan, J.; Batley, J. Molecular marker applications in plants. Plant Genotyping 2015, 1245, 13–27. [Google Scholar]
- Guo, X.; Ren, M.; Ding, J. Do the introductions by botanical gardens facilitate the invasion of Solidago canadensis (Asterceae) in China? Weed Res. 2016, 56, 442–451. [Google Scholar] [CrossRef]
- Prentis, P.J.; Wilson, J.R.; Dormontt, E.E.; Richardson, D.M.; Lowe, A.J. Adaptive evolution in invasive species. Trends Plant Sci. 2008, 13, 288–294. [Google Scholar] [CrossRef]
- Tsutsui, N.D.; Suarez, A.V.; Holway, D.A.; Case, T.J. Reduced genetic variation and the success of an invasive species. Proc. Natl. Acad. Sci. USA 2000, 97, 5948–5953. [Google Scholar] [CrossRef]
- Te Beest, M.; Le Roux, J.J.; Richardson, D.M.; Brysting, A.K.; Suda, J.; Kubešová, M.; Pyšek, P. The more the better? The role of polyploidy in facilitating plant invasions. Ann. Bot. 2012, 109, 19–45. [Google Scholar] [CrossRef] [PubMed]
- Pandit, M.K.; White, S.M.; Pocock, M.J. The contrasting effects of genome size, chromosome number and ploidy level on plant invasiveness: A global analysis. New Phytol. 2014, 203, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Suda, J.; Meyerson, L.A.; Leitch, I.J.; Pyšek, P. The hidden side of plant invasions: The role of genome size. New Phytol. 2015, 205, 994–1007. [Google Scholar] [CrossRef] [PubMed]
- Husband, B.C.; Baldwin, S.J.; Suda, J. The incidence of polyploidy in natural plant populations: Major patterns and evolutionary processes. In Plant Genome Diversity Volume 2; Springer: Vienna, Austria, 2013; pp. 255–276. [Google Scholar]
- Glennon, K.; Ritchie, M.; Segraves, K. Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecol. Lett. 2014, 17, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Broz, A.K.; Manter, D.K.; Bowman, G.; Müller-Schärer, H.; Vivanco, J.M. Plant origin and ploidy influence gene expression and life cycle characteristics in an invasive weed. BMC Plant Biol. 2009, 9, 33. [Google Scholar] [CrossRef]
- Hamilton, J.A.; Miller, J.M. Adaptive introgression as a resource for management and genetic conservation in a changing climate. Conserv. Biol. 2016, 30, 33–41. [Google Scholar] [CrossRef]
- Pfennig, K.S.; Kelly, A.L.; Pierce, A.A. Hybridization as a facilitator of species range expansion. Proc. R. Soc. B Biol. Sci. 2016, 283, 20161329. [Google Scholar] [CrossRef]
- Liu, S.; Chen, S.; Chen, Y.; Guan, Z.; Yin, D.; Chen, F. In vitro induced tetraploid of Dendranthema nankingense (Nakai) Tzvel. shows an improved level of abiotic stress tolerance. Sci. Hortic. 2011, 127, 411–419. [Google Scholar] [CrossRef]
- Soliman, W.S.; Fujimori, M.; Tase, K.; Sugiyama, S.-i. Heat tolerance and suppression of oxidative stress: Comparative analysis of 25 cultivars of the C3 grass Lolium perenne. Environ. Exp. Bot. 2012, 78, 10–17. [Google Scholar] [CrossRef]
- Tu, Y.; Jiang, A.; Gan, L.; Hossain, M.; Zhang, J.; Peng, B.; Xiong, Y.; Song, Z.; Cai, D.; Xu, W. Genome duplication improves rice root resistance to salt stress. Rice 2014, 7, 15. [Google Scholar] [CrossRef]
- Moura, R.F.; Queiroga, D.; Vilela, E.; Moraes, A.P. Polyploidy and high environmental tolerance increase the invasive success of plants. J. Plant Res. 2021, 134, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Wan, J.Z.; Qu, H.; Zhang, Z.X. Climatic niche shift of aquatic plant invaders between native and invasive ranges: A test using 10 species across different biomes on a global scale. Knowl. Manag. Aquat. Ecosyst. 2017, 418, 27. [Google Scholar] [CrossRef]
- Atwater, D.Z.; Ervine, C.; Barney, J.N. Climatic niche shifts are common in introduced plants. Nat. Ecol. Evol. 2018, 2, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Deane, D.C.; Li, S.; Wu, Y.; Sui, X.; Xu, H.; Chu, C.; He, F.; Fang, S. Invasion success and impacts depend on different characteristics in non-native plants. Divers. Distrib. 2021, 27, 1194–1207. [Google Scholar] [CrossRef]
- Padilla-García, N.; Šrámková, G.; Záveská, E.; Šlenker, M.; Clo, J.; Zeisek, V.; Lučanová, M.; Rurane, I.; Kolář, F.; Marhold, K. Niche differentiation following whole-genome duplication? The importance of considering the intricate evolutionary history of polyploids when assessing climatic niche evolution. bioRxiv 2022. [Google Scholar] [CrossRef]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Španiel, S.; Marhold, K.; Hodálová, I.; Lihová, J. Diploid and tetraploid cytotypes of Centaurea stoebe (Asteraceae) in Central Europe: Morphological differentiation and cytotype distribution patterns. Folia Geobot. 2008, 43, 131–158. [Google Scholar] [CrossRef]
- Treier, U.A.; Broennimann, O.; Normand, S.; Guisan, A.; Schaffner, U.; Steinger, T.; Müller-Schärer, H. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology 2009, 90, 1366–1377. [Google Scholar] [CrossRef]
- Schlaepfer, D.R.; Edwards, P.J.; Semple, J.C.; Billeter, R. Cytogeography of Solidago gigantea (Asteraceae) and its invasive ploidy level. J. Biogeogr. 2008, 35, 2119–2127. [Google Scholar] [CrossRef]
- Schlaepfer, D.R.; Edwards, P.J.; Widmer, A.; Billeter, R. Phylogeography of native ploidy levels and invasive tetraploids of Solidago gigantea. Mol. Ecol. 2008, 17, 5245–5256. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, X.; Xue, L.; Yao, B.; Lu, H.; Tian, Z.; Li, J.; Zhou, X.; Zhang, Y.; Zia Ul Haq, M. Polyploidization contributes to evolution of competitive ability: A long term common garden study on the invasive Solidago canadensis in China. Oikos 2020, 129, 700–713. [Google Scholar] [CrossRef]
- Xu, J.; Xue, L.; Qiang, S. Comparison on morphological characteristics of different cellular types of Solidago canadensis from native and introduced ranges at seedling stage. J. Plant Resour. Environ. 2018, 27, 44–51. [Google Scholar]
- Yang, X.; Cheng, J.; Yao, B.; Lu, H.; Zhang, Y.; Xu, J.; Song, X.; Sowndhararajan, K.; Qiang, S. Polyploidy-promoted phenolic metabolism confers the increased competitive ability of Solidago canadensis. Oikos 2021, 130, 1014–1025. [Google Scholar] [CrossRef]
- Weber, E. Current and potential ranges of three exotic goldenrods (Solidago) in Europe. Conserv. Biol. 2001, 15, 122–128. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H.; Kreitschitz, A. Morphological and cytological diversity of goldenrods (Solidago L. and Euthamia Nutt.) from south-western Poland. Biodivers. Res. Conserv. 2015, 38, 41–49. [Google Scholar] [CrossRef]
- Weber, E.; Schmid, B. Das neophytenproblem. Diss. Bot. 1993, 196, 209–227. [Google Scholar]
- Follak, S.; Eberius, M.; Essl, F.; Fürdös, A.; Sedlacek, N.; Trognitz, F. Invasive alien plants along roadsides in Europe. EPPO Bull. 2018, 48, 256–265. [Google Scholar] [CrossRef]
- Abhilasha, D.; Quintana, N.; Vivanco, J.; Joshi, J. Do allelopathic compounds in invasive Solidago canadensis s.l. restrain the native European flora? J. Ecol. 2008, 96, 993–1001. [Google Scholar] [CrossRef]
- Weber, E. Morphological variation of the introduced perennial Solidago canadensis L. sensu lato (Asteraceae) in Europe. Bot. J. Linn. Soc. 1997, 123, 197–210. [Google Scholar] [CrossRef]
- Eckert, S.; Herden, J.; Stift, M.; Joshi, J.; van Kleunen, M. Manipulation of cytosine methylation does not remove latitudinal clines in two invasive goldenrod species in Central Europe. Mol. Ecol. 2021, 30, 222–236. [Google Scholar] [CrossRef]
- Suda, J.; Krahulcová, A.; Trávníek, P.; Krahulec, F. Ploidy level versus DNA ploidy level: An appeal for consistent terminology. Taxon 2006, 55, 447–450. [Google Scholar] [CrossRef]
- Doležel, J.; Greilhuber, J.; Suda, J. Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2007, 2, 2233–2244. [Google Scholar] [CrossRef] [PubMed]
- Semple, J.C.; Uesugi, A. Solidago altissima var. pluricephala (Asteraceae: Astereae) in Australia, Tonga, and Hawaii. Phytoneuron 2017, 40, 1–16. [Google Scholar]
- Szymura, M.; Szymura, T.H. Soil preferences and morphological diversity of goldenrods (Solidago L.) from south-western Poland. Acta Soc. Bot. Pol. 2013, 82, 107–115. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H. Growth, phenology, and biomass allocation of alien Solidago species in central Europe. Plant Species Biol. 2015, 30, 245–256. [Google Scholar] [CrossRef]
- Navas, M.-L.; Moreau-Richard, J. Can traits predict the competitive response of herbaceous Mediterranean species? Acta Oecologica 2005, 27, 107–114. [Google Scholar] [CrossRef]
- Schnitzler, A.; Muller, S. Ecology and biogeography of highly invasive plants in Europe: Giant knotweeds from Japan (Fallopia japonica and F. sachalinensis). Rev. d’Ecologie 1998, 53, 3–38. [Google Scholar]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef]
- Thébault, A.; Gillet, F.; Müller-Schärer, H.; Buttler, A. Polyploidy and invasion success: Trait trade-offs in native and introduced cytotypes of two Asteraceae species. Plant Ecol. 2011, 212, 315–325. [Google Scholar] [CrossRef]
- Vichiato, M.R.d.M.; Vichiato, M.; Pasqual, M.; Rodrigues, F.A.; Castro, D.M.D. Morphological effects of induced polyploidy in Dendrobium nobile Lindl.(Orchidaceae). Crop Breed. Appl. Biotechnol. 2014, 14, 154–159. [Google Scholar] [CrossRef]
- Hülskamp, M. Plant trichomes: A model for cell differentiation. Nat. Rev. Mol. Cell Biol. 2004, 5, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Hartnett, D.C.; Abrahamson, W.G. The effects of stem gall insects on life history patterns in Solidago canadensis. Ecology 1979, 60, 910–917. [Google Scholar] [CrossRef]
- Bicknell, R.; Boase, M.; Morgan, E. Developing polyploid genotypes of Leptospermum scoparium. N. Z. J. Crop Hortic. Sci. 2019, 47, 273–281. [Google Scholar] [CrossRef]
- Denaeghel, H.E.; Van Laere, K.; Leus, L.; Lootens, P.; Van Huylenbroeck, J.; Van Labeke, M.-C. The variable effect of polyploidization on the phenotype in Escallonia. Front. Plant Sci. 2018, 9, 354. [Google Scholar] [CrossRef] [PubMed]
- Kameyama, Y.; Kudo, G. Flowering phenology influences seed production and outcrossing rate in populations of an alpine snowbed shrub, Phyllodoce aleutica: Effects of pollinators and self-incompatibility. Ann. Bot. 2009, 103, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Sax, D.F.; Brown, J.H. The paradox of invasion. Glob. Ecol. Biogeogr. 2000, 9, 363–371. [Google Scholar] [CrossRef]
- Bradshaw, A.D. Evolutionary significance of phenotypic plasticity in plants. Adv. Genet. 1965, 13, 115–155. [Google Scholar]
- Schlichting, C.D. The evolution of phenotypic plasticity in plants. Annu. Rev. Ecol. Syst. 1986, 17, 667–693. [Google Scholar] [CrossRef]
- Szymura, M.; Szymura, T.H.; Rajsz, A.; Świerszcz, S. Is phenotypic plasticity an explanation for the invasiveness of goldenrods (Solidago and Euthamia) in Europe? Plant Species Biol. 2019, 34, 73–84. [Google Scholar] [CrossRef]
- Du, L.; Liu, H.; Yan, M.; Li, J.; Li, J. Individual plasticity of the shade response of the invasive Solidago canadensis in China. PLoS ONE 2017, 12, e0170049. [Google Scholar] [CrossRef]
- Li, J.; Liu, H.; Yan, M.; Du, L. No evidence for local adaptation to salt stress in the existing populations of invasive Solidago canadensis in China. PLoS ONE 2017, 12, e0175252. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, G.; Weber, E.; Edwards, P.J. Introduced plants of the invasive Solidago gigantea (Asteraceae) are larger and grow denser than conspecifics in the native range. Divers. Distrib. 2004, 10, 11–19. [Google Scholar] [CrossRef]
- van Kleunen, M.; Fischer, M. Adaptive rather than non-adaptive evolution of Mimulus guttatus in its invasive range. Basic Appl. Ecol. 2008, 9, 213–223. [Google Scholar] [CrossRef]
- Halverson, K.; Heard, S.B.; Nason, J.D.; Stireman III, J.O. Origins, distribution, and local co-occurrence of polyploid cytotypes in Solidago altissima (Asteraceae). Am. J. Bot. 2008, 95, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Broennimann, O.; Treier, U.A.; Müller-Schärer, H.; Thuiller, W.; Peterson, A.; Guisan, A. Evidence of climatic niche shift during biological invasion. Ecol. Lett. 2007, 10, 701–709. [Google Scholar] [CrossRef] [PubMed]
- Perera, P.C.D.; Szymura, T.H.; Zając, A.; Chmolowska, D.; Szymura, M. Drivers of Solidago species invasion in Central Europe—Case study in the landscape of the Carpathian Mountains and their foreground. Ecol. Evol. 2021, 11, 12429–12444. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J. Study on the Mechanism of Successful Invasion of Solidago canadensis Driven by Polyploidization. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2020. [Google Scholar]
- Montagnani, C.; Casazza, G.; Gentili, R.; Caronni, S.; Citterio, S. Kudzu in Europe: Niche conservatism for a highly invasive plant. Biol. Invasions 2022, 24, 1017–1032. [Google Scholar] [CrossRef]
- Petitpierre, B.; Kueffer, C.; Broennimann, O.; Randin, C.; Daehler, C.; Guisan, A. Climatic niche shifts are rare among terrestrial plant invaders. Science 2012, 335, 1344–1348. [Google Scholar] [CrossRef]
- Pandit, M.K.; Pocock, M.J.; Kunin, W.E. Ploidy influences rarity and invasiveness in plants. J. Ecol. 2011, 99, 1108–1115. [Google Scholar] [CrossRef]
- Duchoslav, M.; Jandová, M.; Kobrlová, L.; Šafářová, L.; Brus, J.; Vojtěchová, K. Intricate distribution patterns of six cytotypes of Allium oleraceum at a continental scale: Niche expansion and innovation followed by niche contraction with increasing ploidy level. Front. Plant Sci. 2020, 11, 591137. [Google Scholar] [CrossRef]
- Wilfried, T. Biodiversity: Climate change and the ecologist. Nature 2007, 448, 7153. [Google Scholar]
- Wan, J.Z.; Chen, L.X.; Gao, S.; Song, Y.B.; Tang, S.L.; Yu, F.H.; Li, J.M.; Dong, M. Ecological niche shift between diploid and tetraploid plants of Fragaria (Rosaceae) in China. S. Afr. J. Bot. 2019, 121, 68–75. [Google Scholar] [CrossRef]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Arumuganathan, K.; Earle, E. Estimation of nuclear DNA content of plants by flow cytometry. Plant Mol. Biol. Report. 1991, 9, 229–241. [Google Scholar] [CrossRef]
- Suda, J.; Trávníček, P. Reliable DNA ploidy determination in dehydrated tissues of vascular plants by DAPI flow cytometry—New prospects for plant research. Cytom. Part A 2006, 69, 273–280. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, version 3.6.0; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
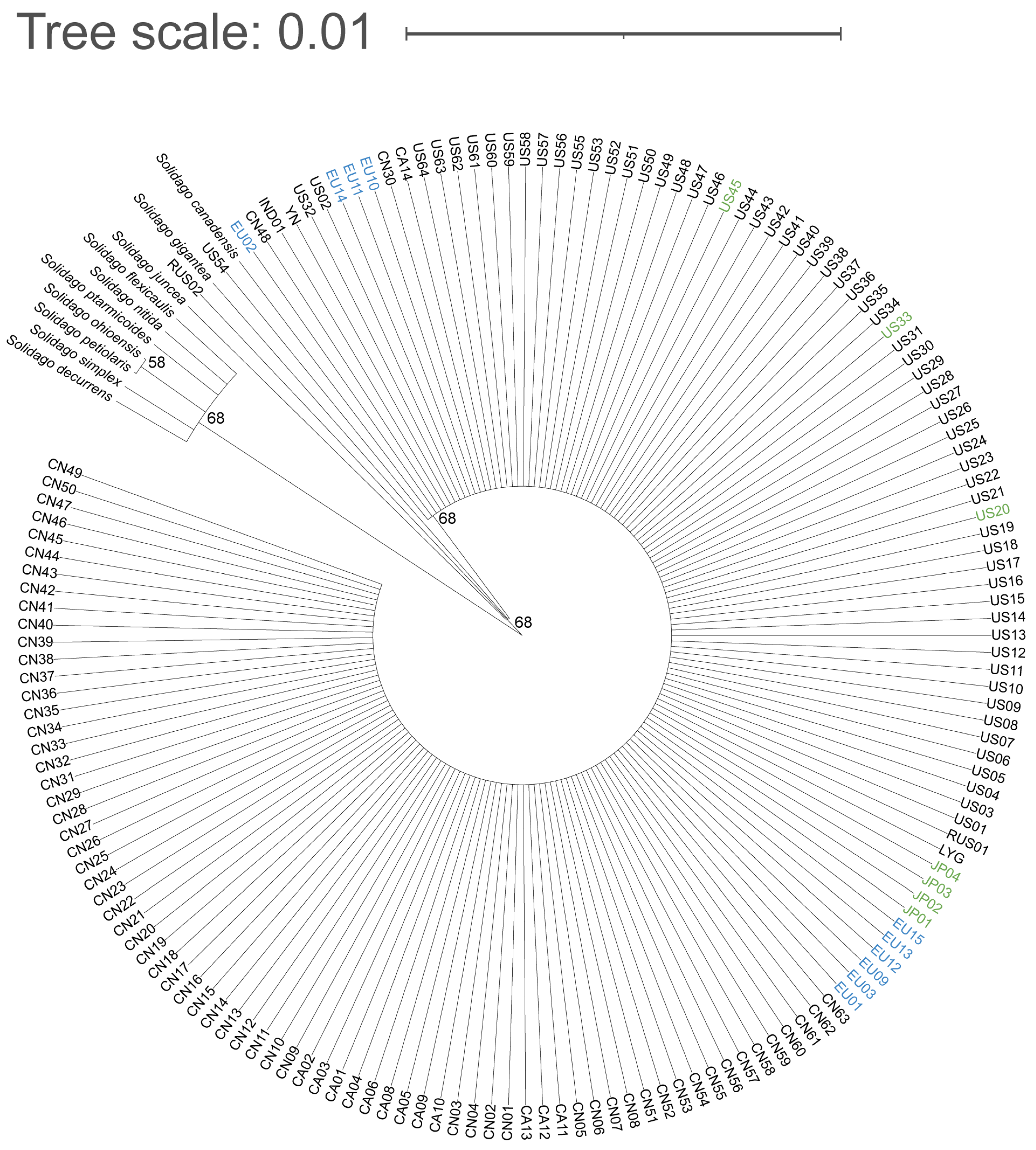
| Population Codes | Accession Number from Genbank | Species | Query Cover | Homology Comparison to S. canadensis | Variable Sites |
|---|---|---|---|---|---|
| EU01 | HQ142590.1 | Solidago canadensis | 100% | 100% | |
| EU02 | HQ142590.1 | Solidago canadensis | 100% | 100% | |
| EU03 | HQ142590.1 | Solidago canadensis | 100% | 100% | |
| EU09 | MT610958.1 | Solidago canadensis | 100% | 100% | |
| EU10 | MT610958.1 | Solidago canadensis | 100% | 99.85% | 492 (A→G) |
| EU11 | MT610958.1 | Solidago canadensis | 100% | 99.85% | 433 (T→C) |
| EU12 | MT610958.1 | Solidago canadensis | 100% | 100% | |
| EU13 | MT610958.1 | Solidago canadensis | 100% | 100% | |
| EU14 | MT610958.1 | Solidago canadensis | 100% | 99.85% | 499 (C→T) |
| EU15 | MT610958.1 | Solidago canadensis | 100% | 100% |
| Population Codes | Accession Number from Genbank | Species | Query Cover | Homology Comparison to S. canadensis | Variable Sites |
|---|---|---|---|---|---|
| EU01 | KX214929.1 | Solidago canadensis | 100% | 100% | |
| EU02 | KX214929.1 | Solidago canadensis | 100% | 99.53% | 12 (C→-) |
| EU03 | KX214929.1 | Solidago canadensis | 100% | 100% | |
| EU09 | KX214780.1 | Solidago canadensis | 100% | 100% | |
| EU10 | KX214780.1 | Solidago canadensis | 100% | 100% | |
| EU11 | KX214780.1 | Solidago canadensis | 100% | 100% | |
| EU12 | KX214780.1 | Solidago canadensis | 100% | 100% | |
| EU13 | KX214780.1 | Solidago canadensis | 100% | 100% | |
| EU14 | KX214780.1 | Solidago canadensis | 100% | 100% | |
| EU15 | KX214780.1 | Solidago canadensis | 100% | 100% |
| Group | Height/cm | Stem Diameter/mm | Leaf Length/cm | Leaf Width/cm | Number of Epidermal Hairs on the Main Vein per 2 mm | Number of Epidermal Hairs on the Lateral Vein per 2 mm | Number of Epidermal Hairs on the Netted Venation per 4 mm2 | Length of the Ray Floret/mm | Height of Involucre/ mm | Number of Ray Florets | Number of Disc Florets |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NA-2x | 62.5 ± 2.89 c | 3.39 ± 0.25 d | 8.98 ± 1.61 g | 1.17 ± 0.37 c | 61.95 ± 10.79 a | 31.22 ± 5.33 ab | 53.71 ± 8.67 a | 6.07 ± 0.82 bcd | 3.84 ± 0.51 ef | 10.5 ± 2.68 b | 3.6 ± 1.28 cd |
| NA-4x | 128.78 ± 34.86 b | 6.45 ± 1.58 cd | 13.12 ± 2.53 de | 2.16 ± 0.46 b | 58.5 ± 10.72 a | 29.39 ± 4.9 b | 50.9 ± 11.52 a | 6.03 ± 1.27 bcd | 4.72 ± 0.52 b | 12.87 ± 3.3 a | 4.96 ± 1.87 b |
| NA-6x | 137.99 ± 30.06 ab | 7.3 ± 1.92 cd | 13.78 ± 2.67 cd | 2.2 ± 0.57 ab | 60.2 ± 12.03 a | 32.34 ± 6.98 a | 50.55 ± 12.5 a | 5.9 ± 0.93 cd | 3.97 ± 0.45 def | 8.57 ± 1.74 cd | 3.53 ± 1.34 cd |
| RUS-2x | 62.4 ± 1.95 c | 4.07 ± 0.25 cd | 12.11 ± 1.4 ef | 2.01 ± 0.28 bc | 53.82 ± 6.76 ab | 23.22 ± 3.38 c | 31.48 ± 3.85 bc | 5.61 ± 0.6 de | 4.17 ± 0.28 cd | 9.71 ± 2.25 bc | 3.92 ± 1.06 c |
| EU-2x | 64.46 ± 2.64 c | 3.88 ± 0.46 d | 11.21 ± 1.39 f | 2.07 ± 0.42 b | 55.67 ± 4.9 ab | 22.29 ± 2.87 cd | 33.41 ± 2.82 b | 5.25 ± 0.36 e | 3.57 ± 0.47 f | 10.96 ± 1.65 b | 3.48 ± 0.59 cd |
| EU-6x | 141.22 ± 14.63 ab | 10.72 ± 1.29 ab | 15.48 ± 1.94 b | 2.82 ± 0.56 a | 53.75 ± 3.68 ab | 20.63 ± 2.32 cd | 31.31 ± 5.08 bc | 5.83 ± 0.34 cde | 4.13 ± 0.31 cde | 10.84 ± 0.8 b | 3.24 ± 0.52 cd |
| JP-6x | 139.16 ± 5.92 ab | 10.18 ± 0.55 b | 14.9 ± 2.19 bcd | 2.62 ± 0.72 ab | 50.42 ± 12.91 b | 22.61 ± 3.95 cd | 19.55 ± 6.81 cd | 5.38 ± 0.68 de | 3.97 ± 0.61 def | 9.89 ± 1.47 bc | 3.45 ± 0.86 cd |
| CN-2x | 66.54 ± 4.27 c | 3.88 ± 0.6 d | 7.28 ± 1.28 h | 1 ± 0.15 c | 42.8 ± 4.24 b | 21.47 ± 4.48 cd | 26.19 ± 4.01 bcd | 4.82 ± 0.42 e | 3.32 ± 0.31 f | 9 ± 1.05 bcd | 4.4 ± 0.84 bc |
| CN-4x | 145.66 ± 21.41 a | 10.05 ± 2.39 b | 15.39 ± 2.23 b | 2.71 ± 1.1 a | 52.1 ± 15.26 b | 21.55 ± 7.44 cd | 26.21 ± 16.43 bcd | 6.44 ± 0.65 b | 4.32 ± 0.62 c | 10.18 ± 1.84 bc | 3.32 ± 1.1 cd |
| CN-6x | 143.11 ± 19.5 ab | 10.2 ± 2.01 b | 15.88 ± 2.01 b | 2.68 ± 0.47 a | 50.44 ± 13.24 b | 22.07 ± 6.57 cd | 26.4 ± 12.18 bcd | 6.08 ± 0.81 bc | 4.29 ± 0.49 c | 9.83 ± 1.35 bc | 3.2 ± 0.87 d |
| S. altissima | 137.82 ± 7.03 ab | 10.45 ± 0.84 ab | 14.91 ± 2.44 bc | 2.62 ± 0.64 ab | 50.8 ± 10.43 b | 22.04 ± 4.8 cd | 19.04 ± 5.89 cd | 5.08 ± 0.67 de | 3.74 ± 0.64 ef | 9.65 ± 1.24 bc | 3.59 ± 0.74 cd |
| S. decurrens | 106.67 ± 4.51 cb | 7.57 ± 0.5 bcd | 8.71 ± 0.76 gh | 2.02 ± 0.11 bc | 0.73 ± 0.12 d | 1.39 ± 0.65 e | 1.83 ± 0.47 e | 12.38 ± 0.76 a | 6.86 ± 0.65 a | 5.85 ± 0.31 d | 13.7 ± 2.19 a |
| S. gigantea | 144.11 ± 11.92 ab | 12.51 ± 0.8 a | 21.74 ± 2.94 a | 2.87 ± 0.33 a | 19.65 ± 2.22 c | 14.02 ± 2.86 de | 12.36 ± 1.15 d | 5.31 ± 0.6 e | 3.7 ± 0.36 ef | 10.2 ± 1.18 bc | 6.01 ± 1.02 b |
| S. simplex | 112.33 ± 5.51 bc | 8.57 ± 0.7 bc | 7.06 ± 2.04 h | 1.99 ± 0.37 bc | 16.73 ± 4.1 c | 10.33 ± 2.12 de | 16.67 ± 7.79 cd | 4.59 ± 0.3 e | 4.01 ± 0.12 ef | 9.18 ± 0.95 bcd | 2.68 ± 0.38 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, Z.; Cheng, J.; Xu, J.; Feng, D.; Zhong, J.; Yuan, X.; Zhang, Z.; Zhang, Y.; Mao, Z.; Qiang, S. Cytogeography of Naturalized Solidago canadensis Populations in Europe. Plants 2023, 12, 1113. https://doi.org/10.3390/plants12051113
Tian Z, Cheng J, Xu J, Feng D, Zhong J, Yuan X, Zhang Z, Zhang Y, Mao Z, Qiang S. Cytogeography of Naturalized Solidago canadensis Populations in Europe. Plants. 2023; 12(5):1113. https://doi.org/10.3390/plants12051113
Chicago/Turabian StyleTian, Zhongsai, Jiliang Cheng, Jingxuan Xu, Dongyan Feng, Jian Zhong, Xiaoxiao Yuan, Zheng Zhang, Yu Zhang, Zhiyuan Mao, and Sheng Qiang. 2023. "Cytogeography of Naturalized Solidago canadensis Populations in Europe" Plants 12, no. 5: 1113. https://doi.org/10.3390/plants12051113
APA StyleTian, Z., Cheng, J., Xu, J., Feng, D., Zhong, J., Yuan, X., Zhang, Z., Zhang, Y., Mao, Z., & Qiang, S. (2023). Cytogeography of Naturalized Solidago canadensis Populations in Europe. Plants, 12(5), 1113. https://doi.org/10.3390/plants12051113






