Insights into the Bioinformatics and Transcriptional Analysis of the Elongator Complexes (ELPs) Gene Family of Wheat: TaELPs Contribute to Wheat Abiotic Stress Tolerance and Leaf Senescence
Abstract
1. Introduction
2. Results
2.1. Identification and Annotation of Wheat ELP Family Members
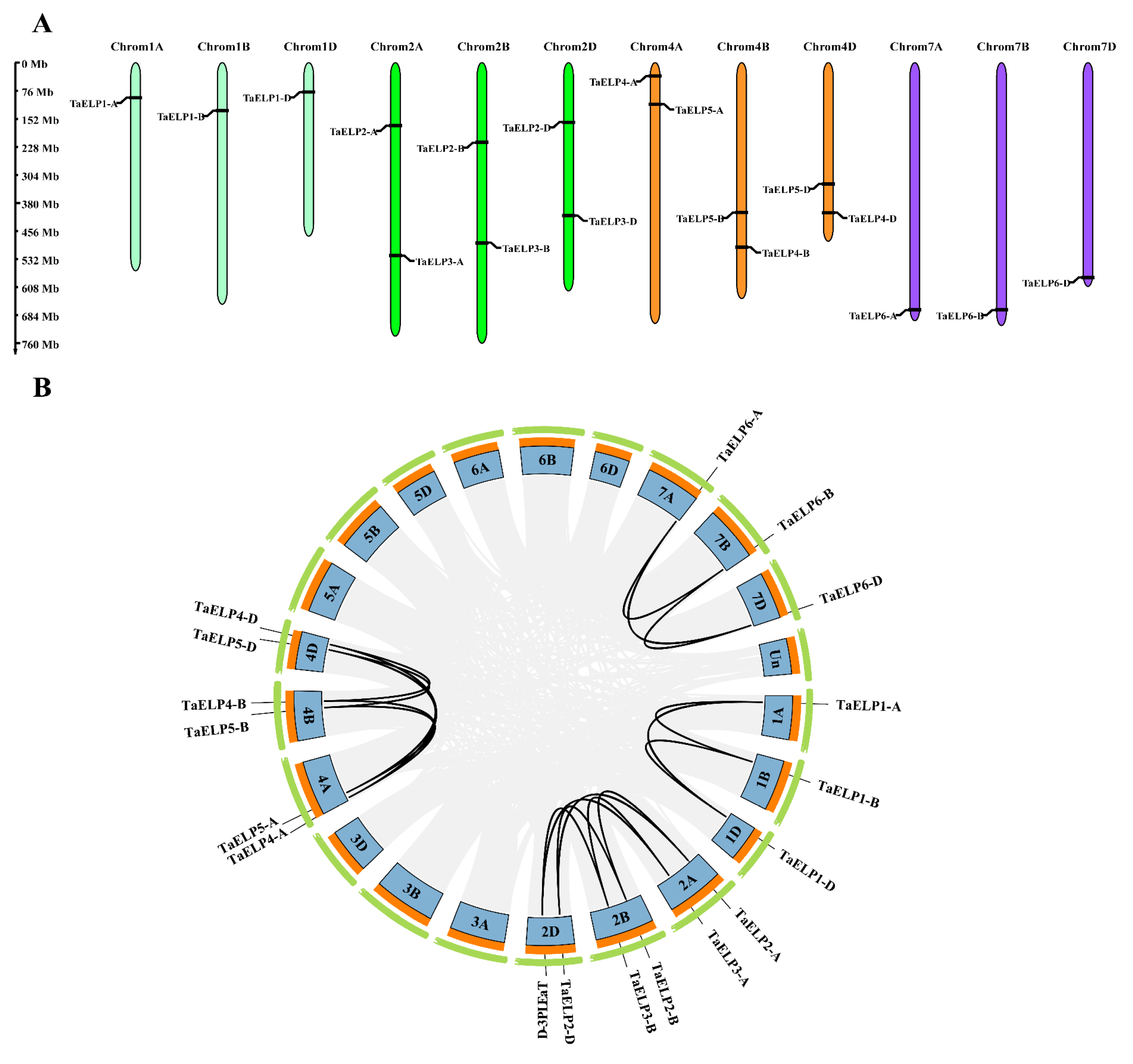
2.2. Chromosomal Distribution and Gene Duplication of TaELPs
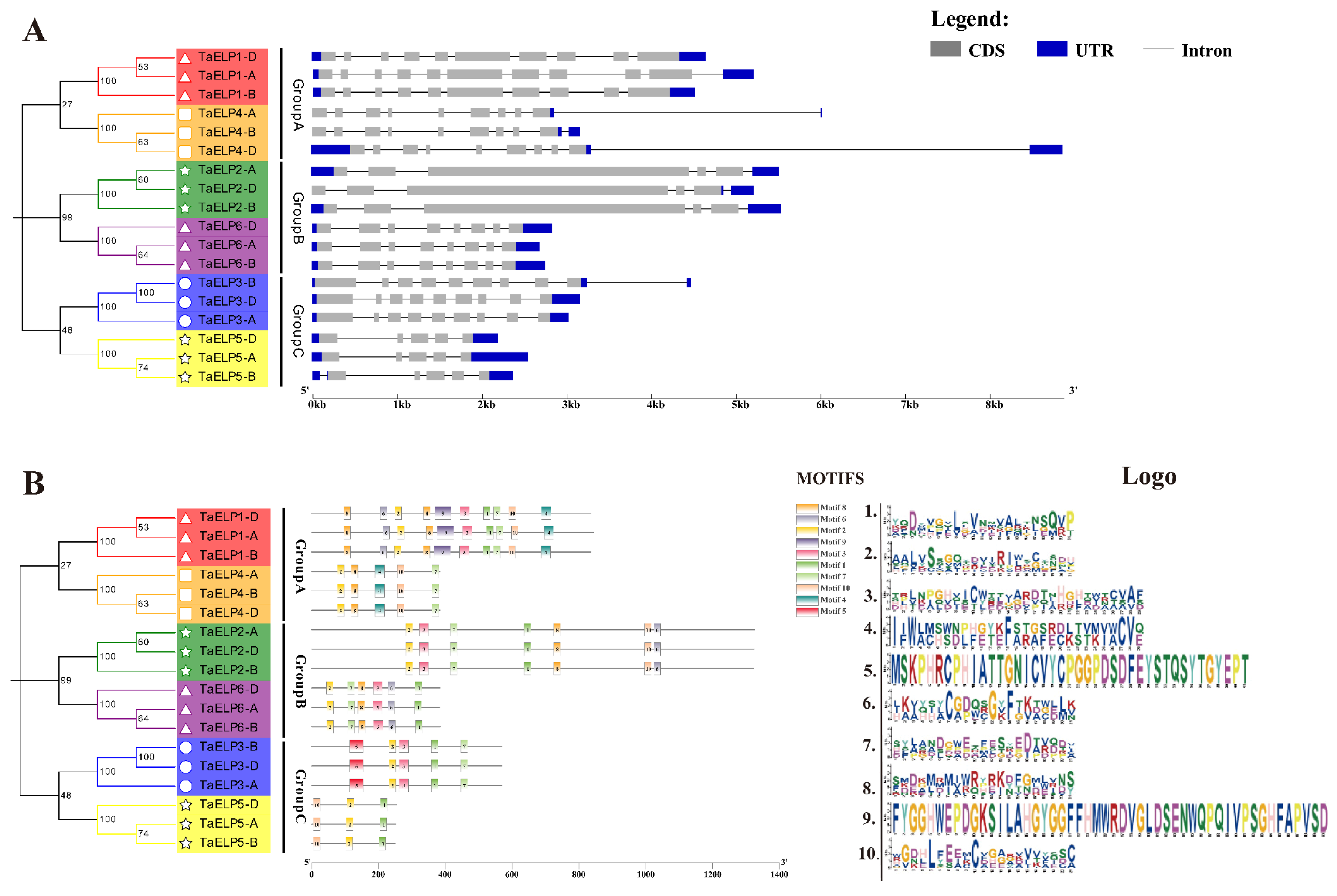
2.3. Phylogenetic and Cluster Analysis of Wheat TaELPs
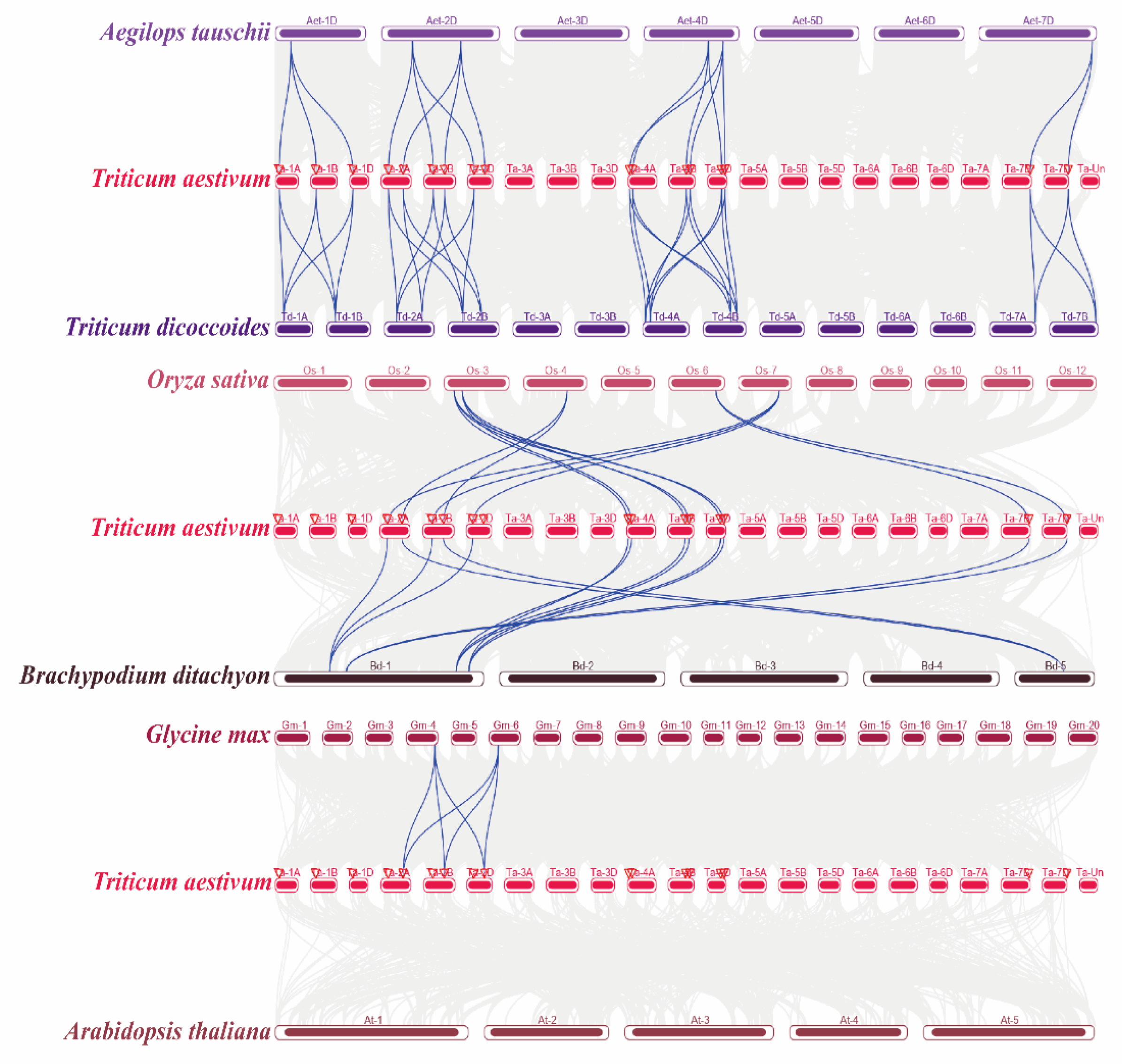
2.4. Orthologous Analysis of Wheat TaELPs Genes
2.5. Gene Structure and Conserved Motif Analysis of TaELPs Genes
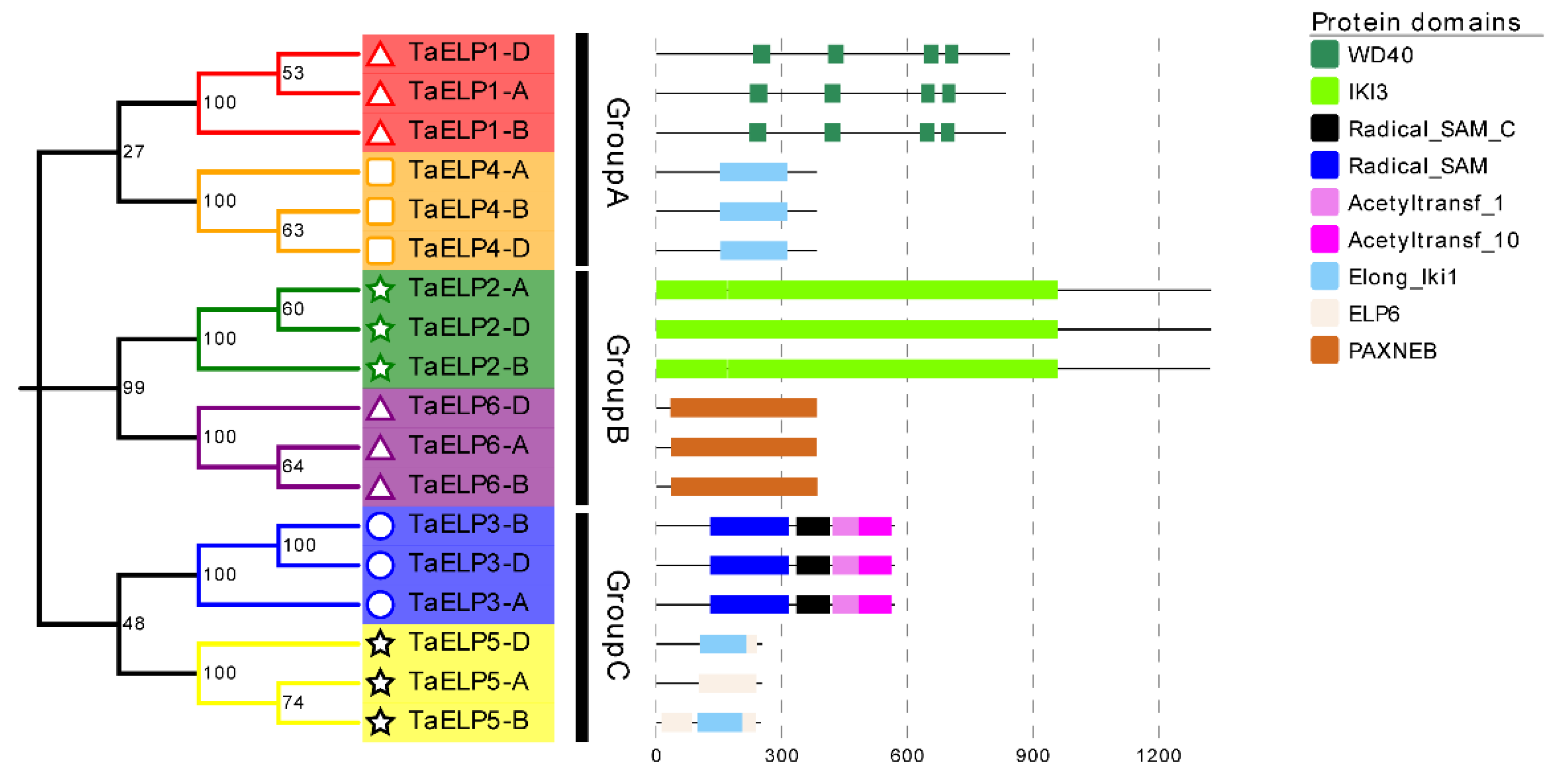
2.6. Protein Conservation Domain and 3-D Protein Structure Analysis of TaELPs Gene
2.7. Cis-Acting Element Regulation (CARE) Analysis of Wheat TaELPs Genes
2.8. TaELP Expression Pattern Prediction Analysis
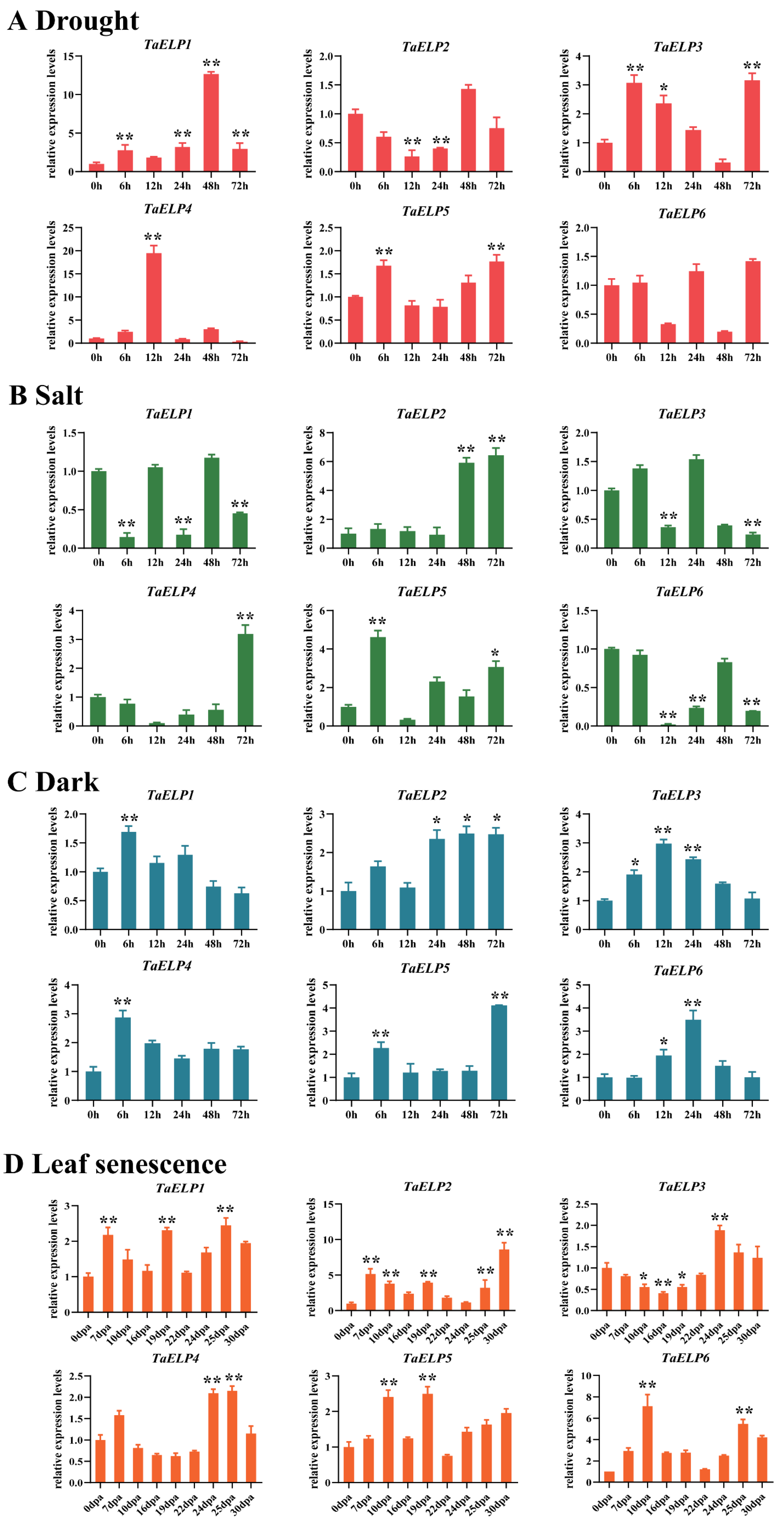
2.9. Expression Pattern Validation Analysis of TaELPs
2.10. Phenotypes of TaELP2-Silenced Wheat Seedlings in Dark-Induced Leaf Senescence
2.11. Prediction of Protein–Protein Interactions of Wheat ELPs
3. Discussion
3.1. Evolution and Genetic Relationship of TaELPs
3.2. Structural Diversity of Wheat TaELPs
3.3. Transcription Analysis of TaELP Genes Reveals Its Role in Wheat Growth, Development, and Abiotic Stress Tolerance
4. Materials and Methods
4.1. Identification of Wheat ELPs Family Gene Members
4.2. Sequence Alignment and Phylogenetic Tree Construction
4.3. Chromosomal Localization, Gene Duplication, and Collinearity Analysis
4.4. Subcellular Localization and 3D Structure Modeling
4.5. Structure, Domain, and Motif Analysis of TaELP Genes
4.6. Analysis of Cis-Acting Regulatory Elements (CAREs) and Protein Interaction Networks
4.7. Tissue-Specific Expression and Stress Analysis of Wheat TaELPs
4.8. Plant Materials and Treatments
4.9. Virus-Induced Gene Silencing (VIGS) and Chlorophyll Content Determination
4.10. RNA Extraction, cDNA First-Strand Synthesis, and Real-Time PCR Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bradsher, J.; Jackson, K.; Conaway, R.; Conaway, J. RNA polymerase II transcription factor SIII. I. Identification, purification, and properties. J. Biol. Chem. 1993, 268, 25587–25593. [Google Scholar] [CrossRef] [PubMed]
- Price, D.; Sluder, A.; Greenleaf, A. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 1989, 9, 1465–1475. [Google Scholar] [PubMed]
- Otero, G.; Fellows, J.; Li, Y.; de Bizemont, T.; Dirac, A.; Gustafsson, C.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 1999, 3, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Winkler, G.S.; Petrakis, T.G.; Ethelberg, S.; Tokunaga, M.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J. Biol. Chem. 2001, 276, 32743–32749. [Google Scholar] [CrossRef]
- Krogan, N.J.; Gr Ee Nblatt, J.F. Characterization of a Six-Subunit Holo-Elongator Complex Required for the Regulated Expression of a Group of Genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002, 21, 8203–8212. [Google Scholar] [CrossRef]
- Mehlgarten, C.; Jablonowski, D.; Wrackmeyer, U.; Tschitschmann, S.; Sondermann, D.; Jäger, G.; Gong, Z.; Byström, A.; Schaffrath, R.; Breunig, K. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol. Microbiol. 2010, 76, 1082–1094. [Google Scholar] [CrossRef]
- Glatt, S.; Létoquart, J.; Faux, C.; Taylor, N.; Séraphin, B.; Müller, C. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. [Google Scholar] [CrossRef]
- Versées, W.; De Groeve, S.; Van Lijsebettens, M. Elongator, a conserved multitasking complex? Mol. Microbiol. 2010, 76, 1065–1069. [Google Scholar] [CrossRef]
- Dalwadi, U.; Yip, C.K. Structural insights into the function of Elongator. Cell. Mol. Life Sci. 2018, 75, 1613–1622. [Google Scholar] [CrossRef]
- Abdel-Fattah, W.; Jablonowski, D.; Santo, R.D.; Ring, K.L.T.; Stark, M.J.R. Phosphorylation of Elp1 by Hrr25 Is Required for Elongator-Dependent tRNA Modification in Yeast. PLoS Genet. 2015, 11, e1004931. [Google Scholar] [CrossRef]
- Wittschieben, B.; Otero, G.; de Bizemont, T.; Fellows, J.; Erdjument-Bromage, H.; Ohba, R.; Li, Y.; Allis, C.; Tempst, P.; Svejstrup, J. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol. Cell 1999, 4, 123–128. [Google Scholar] [CrossRef]
- Chinenov, Y. A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem. Sci 2002, 27, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Zhang, X.; Zhai, H.; Liu, J.; Wu, F.; Li, C.; Chen, Q. Shortrootelongator Is Required for Root Stem Cell Maintenance by Regulating Transcription. Plant Physiol. 2019, 179, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Zhao, W.; Diao, W.; Xie, X.; Wang, Z.; Zhang, J.; Shen, Y.; Long, J. Crystal structure of elongator subcomplex Elp4-6. J. Biol. Chem. 2012, 287, 21501–21508. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Mou, Z. Elongator and its epigenetic role in plant development and responses to abiotic and biotic stresses. Front. Plant Sci. 2015, 6, 296. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Jin, X.-H.; Wang, Y.-M.; Zheng, B.; Chen, P. Recent Advances in the Role of the Elongator Complex in Plant Physiology and tRNA Modification: A Review. J. Integr. Agr. 2014, 13, 1640–1650. [Google Scholar] [CrossRef]
- Jones, J.; Dangl, J. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Yanfei, W.; Qin, H.; Qi, Z.; Xuezhu, D.; Feng, S. Genome-wide identification and expression analysis of Elongator complex family genes in response to abiotic stresses in rice. Acta Agron. Sin. 2021, 48, 644–655. [Google Scholar]
- Zhou, X.; Hua, D.; Chen, Z.; Zhou, Z.; Gong, Z. Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J. 2009, 60, 79–90. [Google Scholar] [CrossRef]
- Nelissen, H.; Fleury, D.; Bruno, L.; Robles, P.; De Veylder, L.; Traas, J.; Micol, J.; Van Montagu, M.; Inzé, D.; Van Lijsebettens, M. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc. Natl. Acad. Sci. USA 2005, 102, 7754–7759. [Google Scholar] [CrossRef]
- Berná, G.; Robles, P.; Micol, J. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics 1999, 152, 729–742. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, H. DRL1, a Homolog of the Yeast TOT4/KTI12 Protein, Has a Function in Meristem Activity and Organ Growth in Plants. Plant Cell 2003, 15, 639–654. [Google Scholar] [CrossRef] [PubMed]
- DeFraia, C.; Mou, Z. The role of the Elongator complex in plants. Plant Signal. Behav. 2011, 6, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhang, H.; Jablonowski, D.; Zhou, X.; Ren, X.; Hong, X.; Schaffrath, R.; Zhu, J.; Gong, Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol. Cell. Biol. 2006, 26, 6902–6912. [Google Scholar] [CrossRef] [PubMed]
- DeFraia, C.T.; Zhang, X.; Mou, Z. Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J. 2010, 64, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Defraia, C.; Wang, Y.; Yao, J.; Mou, Z. Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol. 2013, 13, 102. [Google Scholar] [CrossRef]
- Zhu, M.; Li, Y.; Chen, G.; Ren, L.; Xie, Q.; Zhao, Z.; Hu, Z. Silencing SlELP2L, a tomato Elongator complex protein 2-like gene, inhibits leaf growth, accelerates leaf, sepal senescence and produces dark-green fruit. Sci. Rep. 2015, 5, 7693. [Google Scholar] [CrossRef]
- Wang, K.; Rong, W.; Liu, Y.; Li, H.; Zhang, Z. Wheat Elongator subunit 4 is required for epigenetic regulation of host immune response to Rhizoctonia cerealis. Crop J. 2020, 8, 565–576. [Google Scholar] [CrossRef]
- Pfeifer, M.; Kugler, K.G.; Sandve, S.R.; Zhan, B.; Rudi, H.; Hvidsten, T.R.; Mayer, K.F.X.; Olsen, O.-A. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 2014, 345, 1250091. [Google Scholar] [CrossRef]
- Ray, D.; Mueller, N.; West, P.; Foley, J. Yield Trends Are Insufficient to Double Global Crop Production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef]
- Borrill, P.; Adamski, N.; Uauy, C. Genomics as the key to unlocking the polyploid potential of wheat. New Phytol. 2015, 208, 1008–1022. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, X.; Wang, Q.; Sun, J.; He, D. SorghumGenome-wide identification and analysis of cystatin family genes in Sorghum (Sorghum bicolor (L.) Moench). PeerJ 2021, 9, e10617. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, S.; Ding, L.; Cheng, X.; Kang, Z.; Mao, H. Genome-wide analysis of trehalose-6-phosphate phosphatases (TPP) gene family in wheat indicates their roles in plant development and stress response. BMC Plant Biol. 2022, 22, 120. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Gruszka, D. In-Silico Study of Brassinosteroid Signaling Genes in Rice Provides Insight Into Mechanisms Which Regulate Their Expression. Front. Genet. 2022, 13, 953458. [Google Scholar] [CrossRef]
- Islam, M.A.; Rahman, M.M.; Rahman, M.M.; Jin, X.; Sun, L.; Zhao, K.; Wang, S.; Sikdar, A.; Noor, H.; Jeon, J.-S.; et al. In Silico and Transcription Analysis of Trehalose-6-phosphate Phosphatase Gene Family of Wheat: Trehalose Synthesis Genes Contribute to Salinity, Drought Stress and Leaf Senescence. Genes 2021, 12, 1652. [Google Scholar] [CrossRef]
- Sivakumar, K.; Balaji, S.; Gangaradhakrishnan. In silico characterization of antifreeze proteins using computational tools and servers. J. Chem. Sci. 2008, 119, 571–579. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama; et al. Genome-Wide Identification and Characterization of the Brassinazole-resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Afzal, F.; Chaudhari, S.K.; Gul, A.; Farooq, A.; Ali, H.; Nisar, S.; Sarfraz, B.; Shehzadi, K.J.; Mujeeb-Kazi, A. Bread Wheat (Triticum aestivum L.) Under Biotic and Abiotic Stresses: An Overview. In Crop Production and Global Environmental Issues; Hakeem, K.R., Ed.; Springer International Publishing: Cham, Switzerland, 2015; pp. 293–317. [Google Scholar]
- Gu, Z.; Cavalcanti, A.; Chen, F.; Bouman, P.; Li, W. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef]
- Li, H.; Nie, F.; Zhu, L.; Mu, M.; Fan, R.; Li, J.; Shaheen, A.; Liu, Y.; Li, C.; Liu, W.; et al. New insights into the dispersion history and adaptive evolution of taxon Aegilops tauschii in China. J. Genet. Genom. 2022, 49, 185–194. [Google Scholar] [CrossRef]
- Cannon, S.; Mitra, A.; Baumgarten, A.; Young, N.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Finn, R.; Tate, J.; Mistry, J.; Coggill, P.; Sammut, S.; Hotz, H.; Ceric, G.; Forslund, K.; Eddy, S.; Sonnhammer, E.; et al. The Pfam protein families database. Nucleic Acids Res. 2008, 36, D281–D288. [Google Scholar] [CrossRef] [PubMed]
- Bordoli, L.; Schwede, T. Automated Protein Structure Modeling with SWISS-MODEL Workspace and the Protein Model Portal. In Homology Modeling: Methods and Protocols; Orry, A.J.W., Abagyan, R., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 107–136. [Google Scholar]
- Jain, B.; Pandey, S. WD40 Repeat Proteins: Signalling Scaffold with Diverse Functions. Protein J. 2018, 37, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Petrakis, T.; Søgaard, T.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J. Physical and functional interaction between Elongator and the chromatin-associated Kti12 protein. J. Biol. Chem. 2005, 280, 19454–19460. [Google Scholar] [CrossRef]
- Abhinandan, K.; Skori, L.; Stanic, M.; Hickerson, N.M.N.; Jamshed, M.; Samuel, M.A. Abiotic Stress Signaling in Wheat—An Inclusive Overview of Hormonal Interactions During Abiotic Stress Responses in Wheat. Front. Plant Sci. 2018, 9, 734. [Google Scholar] [CrossRef] [PubMed]
- Woloszynska, M.; Le Gall, S.; Van Lijsebettens, M. Plant Elongator-mediated transcriptional control in a chromatin and epigenetic context. Biochim. Biophys. Acta 2016, 1859, 1025–1033. [Google Scholar] [CrossRef]
- Lawton-Rauh, A. Evolutionary dynamics of duplicated genes in plants. Mol. Phylogenet. Evol. 2003, 29, 396–409. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.; Purugganan, M. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef] [PubMed]
- Delehelle, F.; Cussat-Blanc, S.; Alliot, J.-M.; Luga, H.; Balaresque, P. ASGART: Fast and parallel genome scale segmental duplications mapping. Bioinformatics 2018, 34, 2708–2714. [Google Scholar] [CrossRef]
- Dubcovsky, J.; Dvorak, J. Genome Plasticity a Key Factor in the Success of Polyploid Wheat Under Domestication. Science 2007, 316, 1862–1866. [Google Scholar] [CrossRef]
- Close, P.; Gillard, M.; Ladang, A.; Jiang, Z.; Papuga, J.; Hawkes, N.; Nguyen, L.; Chapelle, J.P.; Bouillenne, F.; Svejstrup, J. DERP6 (ELP5) and C3ORF75 (ELP6) regulate tumorigenicity and migration of melanoma cells as subunits of Elongator. J. Biol. Chem. 2012, 287, 32535. [Google Scholar] [CrossRef]
- Kleinjan, D.A.; Seawright, A.; Elgar, G.; Heyningen, V.V. Characterization of a novel gene adjacent to PAX6, revealing synteny conservation with functional significance. Mamm. Genome 2002, 13, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Nelissen, H.; De Groeve, S.; Fleury, D.; Neyt, P.; Bruno, L.; Bitonti, M.; Vandenbussche, F.; Van der Straeten, D.; Yamaguchi, T.; Tsukaya, H.; et al. Plant Elongator regulates auxin-related genes during RNA polymerase II transcription elongation. Proc. Natl. Acad. Sci. USA 2010, 107, 1678–1683. [Google Scholar] [CrossRef]
- Tran, H.T.; Nimick, M.; Uhrig, R.G.; Templeton, G.; Morrice, N.; Gourlay, R.; DeLong, A.; Moorhead, G.B.G. Arabidopsis thaliana histone deacetylase 14 (HDA14) is an α-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J. 2012, 71, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217–218, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.S.; Wilhelm, K.S.; Thomashow, M.F. The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant. Mol. Biol. 1994, 24, 701–713. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.; Fujii, H.; Zheng, X.; Zhu, J. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2006, 281, 37636–37645. [Google Scholar] [CrossRef]
- Yusuf, C.; Abdullah, J.; Shaharuddin, N.; Abu Seman, I.; Abdullah, M. Characterization of promoter of EgPAL1, a novel PAL gene from the oil palm Elaeis guineensis Jacq. Plant Cell Rep. 2018, 37, 265–278. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Jinyan, G.; Xiaoyu, Z.; Cuixia, Z.; Qiuli, L. Research Progress of cis-elements of Abiotic Stress Inducible Promoters and Associated Transcription Factors. Biotechnol. Bull. 2011, 04, 16–20+30. [Google Scholar]
- Xin-Lei, M.; Rui-Qi, X.; Xiao-Man, S.; Jing-Shi, L.; Peng-Peng, G.; Rui, Y.; Xiao-Hu, L.; Hui, G. Genome-wide identification of the Class III PRX gene family in foxtail millet (Setaria italica L.) and expression analysis under drought stress. Acta Agron. Sin. 2022, 48, 2517–2532. [Google Scholar]
- Wang, L.; Ruan, Y. Regulation of cell division and expansion by sugar and auxin signaling. Front. Plant Sci. 2013, 4, 163. [Google Scholar] [CrossRef] [PubMed]
- Valarmathi, P. Salicylic acid, A Versatile Hormone to combat diseases in cotton. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 33–47. [Google Scholar] [CrossRef]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.; Zhang, Q.; Cui, M.; Zhao, M.; Li, N.; Wang, S.; Wu, R.; Zhang, L.; Cao, Y.; et al. The interaction of ABA and ROS in plant growth and stress resistances. Front. Plant Sci. 2022, 13, 1050132. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 2002, 14, S15–S45. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and Drought Stress Signal Transduction in Plants. Annu. Rev. Plant. Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Kwak, J.M.; Schroeder, J.I. An mRNA Cap Binding Protein, ABH1,Modulates Early Abscisic Acid Signal Transduction in Arabidopsis. Cell 2001, 106, 477–487. [Google Scholar] [CrossRef]
- Xiong, L.; Gong, Z.; Rock, C.; Subramanian, S.; Guo, Y.; Xu, W.; Galbraith, D.; Zhu, J. Modulation of Abscisic Acid Signal Transduction and Biosynthesis by an Sm-like Protein in Arabidopsis. Dev. Cell 2001, 1, 771–781. [Google Scholar] [CrossRef]
- Xiong, L.; Schumaker, K.S.; Zhu, J.-K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef]
- Gong, Z.; Dong, C.; Lee, H.; Zhu, J.; Xiong, L.; Gong, D.; Stevenson, B.; Zhu, J. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell 2005, 17, 256–267. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Gong, Z.; Zhu, J.K. Nuclear RNA Export and Its Importance in Abiotic Stress Responses of Plants. Curr. Top. Microbiol. Immunol. 2008, 326, 235–255. [Google Scholar]
- Nocker, S.v.; Ludwig, P. The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genom. 2003, 4, 50. [Google Scholar] [CrossRef] [PubMed]
- Smejda, M.; Kadziolka, D.; Radczuk, N.; Krutyholowa, R.; Chramiec-Glabik, A.; Kedracka-Krok, S.; Jankowska, U.; Biela, A.; Glatt, S. Same but different—Molecular comparison of human KTI12 and PSTK. Biochim. Biophys. Acta Mol. Cell Res. 2021, 1868, 118945. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, C.; Zhang, Y.; Yan, X.; Jin, X.; Yao, X.; Chen, P.; Zheng, B. PtKTI12 genes influence wobble uridine modifications and drought stress tolerance in hybrid poplar. Tree Physiol. 2020, 40, 1778–1791. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Davidson, R.; Martín del Campo, A. Combinatorial and Computational Investigations of Neighbor-Joining Bias. Front. Genet. 2020, 11, 584785. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Howe, E.; Holton, K.; Nair, S.; Schlauch, D.; Sinha, R.; Quackenbush, J. MeV: MultiExperiment Viewer. In Biomedical Informatics for Cancer Research; Ochs, M.F., Casagrande, J.T., Davuluri, R.V., Eds.; Springer: Boston, MA, USA, 2010; pp. 267–277. [Google Scholar]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Paolacci, A.; Tanzarella, O.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Chen, W.; Yang, J. Another formula for calculating the gene change rate in real-time RT-PCR. Mol. Biol. Rep. 2009, 36, 2165–2168. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | Splice | PC | ORF | Chromosome Location | Introns | Exons | Length | M.W. | PI | Instability | Aliphatic Index | GRAVY | SL Prediction | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | Chr | Strand | Start | End | Chr Length | (aa) | (KDa) | Index | ||||||||||
| TaELP1-A | TraesCS1A02G104700.1 | 1 | II | 2511 | 1A | reverse | 100,291,939 | 100,297,147 | 594,102,056 | 10 | 11 | 836 | 91.40 | 6.26 | 44.07 | 84.67 | −0.093 | cytosol |
| TaELP1-B | TraesCS1B02G116100.1 | 1 | II | 2508 | 1B | reverse | 136,647,720 | 136,652,235 | 689,851,870 | 9 | 10 | 835 | 91.36 | 6.23 | 42.10 | 86.05 | −0.090 | cytosol |
| TaELP1-D | TraesCS1D02G096900.1 | 2 | II | 2535 | 1D | reverse | 83,724,571 | 83,729,227 | 495,453,186 | 9 | 10 | 844 | 92.51 | 6.25 | 43.44 | 83.98 | −0.085 | nucleus |
| TaELP2-A | TraesCS2A02G203700.1 | 1 | III | 3978 | 2A | reverse | 179,670,464 | 179,675,987 | 780,798,557 | 5 | 6 | 1325 | 147.31 | 5.60 | 44.21 | 90.59 | −0.163 | cytosol, nucleus, plasma membrane |
| TaELP2-B | TraesCS2B02G231000.1 | 1 | III | 3975 | 2B | reverse | 227,082,327 | 227,087,872 | 801,256,715 | 5 | 6 | 1324 | 147.14 | 5.52 | 44.05 | 90.95 | −0.161 | cytosol, nucleus, plasma membrane |
| TaELP2-D | TraesCS2D02G212000.1 | 1 | III | 3978 | 2D | forward | 170,443,244 | 170,448,462 | 651,852,609 | 5 | 6 | 1325 | 147.17 | 5.57 | 42.54 | 90.60 | −0.158 | cytosol, nucleus, plasma membrane |
| TaELP3-A | TraesCS2A02G320900.1 | 1 | I | 1710 | 2A | forward | 550,539,215 | 550,542,666 | 780,798,557 | 8 | 9 | 569 | 63.57 | 8.88 | 35.59 | 85.55 | −0.310 | cytosol |
| TaELP3-B | TraesCS2B02G361800.1 | 1 | I | 1710 | 2B | reverse | 514,861,001 | 514,865,480 | 801,256,715 | 9 | 10 | 569 | 63.61 | 8.88 | 35.55 | 85.89 | −0.307 | cytosol |
| TaELP3-D | TraesCS2D02G341600.1 | 1 | I | 1710 | 2D | forward | 436,369,113 | 436,372,717 | 651,852,609 | 8 | 9 | 569 | 63.58 | 8.88 | 35.55 | 85.55 | −0.312 | cytosol |
| TaELP4-A | TraesCS4A02G045700.1 | 1 | V | 1155 | 4A | forward | 37,776,004 | 37,782,022 | 744,588,157 | 9 | 10 | 384 | 42.72 | 5.36 | 51.44 | 84.32 | −0.397 | cytosol |
| TaELP4-B | TraesCS4B02G259300.1 | 1 | V | 1155 | 4B | forward | 526,726,516 | 526,729,679 | 673,617,499 | 9 | 10 | 384 | 42.71 | 5.38 | 53.49 | 83.05 | −0.413 | cytosol |
| TaELP4-D | TraesCS4D02G259200.1 | 1 | V | 1155 | 4D | reverse | 428,719,059 | 428,727,934 | 509,857,067 | 9 | 10 | 384 | 42.71 | 5.30 | 52.06 | 85.34 | −0.390 | cytosol |
| TaELP5-A | TraesCS4A02G105200.1 | 1 | VI | 759 | 4A | forward | 119,080,643 | 119,083,199 | 744,588,157 | 4 | 5 | 252 | 27.05 | 5.97 | 31.37 | 101.83 | 0.193 | cytosol |
| TaELP5-B | TraesCS4B02G198800.1 | 1 | VI | 753 | 4B | reverse | 427,496,766 | 427,499,136 | 673,617,499 | 5 | 6 | 250 | 27.08 | 5.98 | 33.83 | 99.52 | 0.159 | cytosol |
| TaELP5-D | TraesCS4D02G199700.1 | 1 | VI | 765 | 4D | reverse | 346,452,075 | 346,454,270 | 509,857,067 | 4 | 5 | 254 | 27.24 | 5.91 | 35.06 | 97.56 | 0.163 | cytosol |
| TaELP6-A | TraesCS7A02G522900.2 | 2 | IV | 1152 | 7A | forward | 705,684,956 | 705,687,644 | 736,706,236 | 7 | 8 | 383 | 41.20 | 8.69 | 53.26 | 80.84 | −0.252 | plastid |
| TaELP6-B | TraesCS7B02G439900.1 | 1 | IV | 1158 | 7B | forward | 705,251,306 | 705,254,066 | 750,620,385 | 7 | 8 | 385 | 41.29 | 8.97 | 50.04 | 78.88 | −0.250 | plastid |
| TaELP6-D | TraesCS7D02G512100.1 | 2 | IV | 1155 | 7D | forward | 613,867,768 | 613,870,602 | 638,686,055 | 7 | 8 | 384 | 41.18 | 8.69 | 47.94 | 79.35 | −0.252 | plastid |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, F.; Islam, M.A.; Lv, C.; Jin, X.; Sun, L.; Zhao, K.; Lu, J.; Yan, R.; Zhang, W.; Shi, Y.; et al. Insights into the Bioinformatics and Transcriptional Analysis of the Elongator Complexes (ELPs) Gene Family of Wheat: TaELPs Contribute to Wheat Abiotic Stress Tolerance and Leaf Senescence. Plants 2023, 12, 952. https://doi.org/10.3390/plants12040952
Guo F, Islam MA, Lv C, Jin X, Sun L, Zhao K, Lu J, Yan R, Zhang W, Shi Y, et al. Insights into the Bioinformatics and Transcriptional Analysis of the Elongator Complexes (ELPs) Gene Family of Wheat: TaELPs Contribute to Wheat Abiotic Stress Tolerance and Leaf Senescence. Plants. 2023; 12(4):952. https://doi.org/10.3390/plants12040952
Chicago/Turabian StyleGuo, Feng, Md Ashraful Islam, Chenxu Lv, Xiujuan Jin, Lili Sun, Kai Zhao, Juan Lu, Rongyue Yan, Wenjun Zhang, Yugang Shi, and et al. 2023. "Insights into the Bioinformatics and Transcriptional Analysis of the Elongator Complexes (ELPs) Gene Family of Wheat: TaELPs Contribute to Wheat Abiotic Stress Tolerance and Leaf Senescence" Plants 12, no. 4: 952. https://doi.org/10.3390/plants12040952
APA StyleGuo, F., Islam, M. A., Lv, C., Jin, X., Sun, L., Zhao, K., Lu, J., Yan, R., Zhang, W., Shi, Y., Li, N., & Sun, D. (2023). Insights into the Bioinformatics and Transcriptional Analysis of the Elongator Complexes (ELPs) Gene Family of Wheat: TaELPs Contribute to Wheat Abiotic Stress Tolerance and Leaf Senescence. Plants, 12(4), 952. https://doi.org/10.3390/plants12040952






