Genome-Wide Association Study on Seedling Phenotypic Traits of Wheat under Different Nitrogen Conditions
Abstract
:1. Introduction
2. Results
2.1. Trait Phenotyping
2.2. SNP Distribution, Linkage Disequilibrium, and Population Structure
2.3. GWAS of Root-Related Traits
2.4. GWAS of Shoot-Related Traits
2.5. GWAS of Biomass and Root–Shoot Ratio
2.6. Haplotype Analysis
2.7. Candidate Genes for Pleiotropic SNPs
3. Discussion
3.1. Phenotypic Analysis of Wheat under Different Conditions
3.2. Analysis Related Loci of Wheat Seedling Traits
3.3. Prediction of Relevant Candidate Genes
4. Materials and Methods
4.1. Plant Material
4.2. Experimental Design
4.3. Phenotype Measurement and Data Analysis
4.4. Population Structure and Linkage Disequilibrium Analysis
4.5. Genome-Wide Association Analysis and Candidate Gene Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, J.; Wang, M.; Li, W.; He, X.; Teng, W.; Ma, W.; Zhao, X.; Hu, M.; Li, H.; Zhang, Y.; et al. Reducing expression of a nitrate-responsive bZIP transcription factor increases grain yield and N use in wheat. Plant Biotechnol. J. 2019, 17, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Mueller, N.D.; Gerber, J.S.; Johnston, M.; Ray, D.K.; Ramankutty, N.; Foley, J.A. Closing yield gaps through nutrient and water management. Nature 2012, 490, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Rabieyan, E.; Bihamta, M.R.; Moghaddam, M.E.; Mohammadi, V.; Alipour, H. Genome-wide association mapping for wheat morphometric seed traits in Iranian landraces and cultivars under rain-fed and well-watered conditions. Sci. Rep. 2022, 12, 17839. [Google Scholar] [CrossRef]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty Years from Transport to Signaling Networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef]
- Comadira, G.; Rasool, B.; Karpinska, B.; Morris, J.; Verrall, S.R.; Hedley, P.E.; Foyer, C.H.; Hancock, R.D. Nitrogen deficiency in barley (Hordeum vulgare) seedlings induces molecular and metabolic adjustments that trigger aphid resistance. J. Exp. Bot. 2015, 66, 3639–3655. [Google Scholar] [CrossRef]
- Duan, Y.H.; Zhang, Y.L.; Ye, L.T.; Fan, X.R.; Xu, G.H.; Shen, Q.R. Responses of Rice Cultivars with Different Nitrogen Use Efficiency to Partial Nitrate Nutrition. Ann. Bot. 2007, 99, 1153–1160. [Google Scholar] [CrossRef]
- Van der Werf, A.; van Nuenen, M. Contribution of physiological and morphological plant traits to a species competitive ability at high and low nitrogen supply. Oecologia 1993, 94, 434–440. [Google Scholar] [CrossRef]
- Forde, B.; Lorenzo, H. The nutritional control of root development. Plant Soil 2001, 232, 51–68. [Google Scholar] [CrossRef]
- Peng, Y.; Li, X.; Li, C. Temporal and Spatial Profiling of Root Growth Revealed Novel Response of Maize Roots under Various Nitrogen Supplies in the Field. PLoS ONE 2012, 7, e37726. [Google Scholar] [CrossRef]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef]
- Xia, J.; Zhu, D.; Chang, H.; Yan, X.; Yan, Y. Effects of water-deficit and high-nitrogen treatments on wheat resistant starch crystalline structure and physicochemical properties. Carbohydr. Polym. 2020, 234, 115905. [Google Scholar] [CrossRef] [PubMed]
- Gaudin, A.C.; McClymont, S.A. Novel temporal, fine-scale and growth variation phenotypes in roots of adult-stage maize (Zea mays L.) in response to low nitrogen stress. Plant Cell Environ. 2011, 34, 2122–2137. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhuang, Z.; Cai, H.; Cheng, S.; Soomro, A.A.; Liu, Z.; Gu, R.; Mi, G.; Yuan, L.; Chen, F. Use of genotype-environment interactions to elucidate the pattern of maize root plasticity to nitrogen deficiency. J. Integr. Plant Biol. 2016, 58, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; Giehl, R.F.H.; von Wirén, N. The Root Foraging Response under Low Nitrogen Depends on DWARF1-Mediated Brassinosteroid Biosynthesis. Plant Physiol. 2020, 183, 998–1010. [Google Scholar] [CrossRef]
- Ju, X.-T.; Xing, G.-X.; Chen, X.-P.; Zhang, S.-L.; Zhang, L.-J.; Liu, X.-J.; Cui, Z.-L.; Yin, B.; Christie, P.; Zhu, Z.-L.; et al. Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc. Natl. Acad. Sci. USA 2009, 106, 3041–3046. [Google Scholar] [CrossRef]
- Wuebbles, D.J. Nitrous Oxide: No Laughing Matter. Science 2009, 326, 56–57. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, X.; Vitousek, P. Chinese agriculture: An experiment for the world. Nature 2013, 497, 33–35. [Google Scholar] [CrossRef]
- Dimkpa, C.O.; Fugice, J.; Singh, U.; Lewis, T.D. Development of fertilizers for enhanced nitrogen use efficiency—Trends and perspectives. Sci. Total. Environ. 2020, 731, 139113. [Google Scholar] [CrossRef]
- Wan, X.; Wu, W.; Shah, F. Nitrogen fertilizer management for mitigating ammonia emission and increasing nitrogen use efficiencies by 15N stable isotopes in winter wheat. Sci. Total. Environ. 2021, 790, 147587. [Google Scholar] [CrossRef]
- Guo, J.H.; Liu, X.J.; Zhang, Y.; Shen, J.L.; Han, W.X.; Zhang, W.F.; Christie, P.; Goulding, K.W.T.; Vitousek, P.M.; Zhang, F.S. Significant Acidification in Major Chinese Croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, K.; Chen, H.; Yang, H.; Zheng, T.; Huang, X.; Fan, G. Simultaneously genetic selection of wheat yield and grain protein quality in rice–wheat and soybean–wheat cropping systems through critical nitrogen efficiency-related traits. Front. Plant Sci. 2022, 13, 899387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Yang, X.-B.; Li, T.-X.; Yu, H.-Y. Genotype difference in nitrogen utilization efficiency of wheat. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2011, 22, 369–375. (In Chinese) [Google Scholar]
- Zhang, Z.; Xiong, S.; Wei, Y.; Meng, X.; Wang, X.; Ma, X. The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci. Rep. 2017, 7, 1000. [Google Scholar] [CrossRef] [PubMed]
- Kasemsap, P.; Bloom, A.J. Breeding for Higher Yields of Wheat and Rice through Modifying Nitrogen Metabolism. Plants 2022, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Li, G.; Zhang, H.; Powers, C.; Fang, T.; Chen, Y.; Wang, S.; Zhu, X.; Carver, B.F.; Yan, L. Nitrogen use efficiency is regulated by interacting proteins relevant to development in wheat. Plant Biotechnol. J. 2018, 16, 1214–1226. [Google Scholar] [CrossRef]
- Hawkesford, M.J. Genetic variation in traits for nitrogen use efficiency in wheat. J. Exp. Bot. 2017, 68, 2627–2632. [Google Scholar] [CrossRef]
- Yan, H.; Shi, H.; Hu, C.; Luo, M.; Xu, C.; Wang, S.; Li, N.; Tang, W.; Zhou, Y.; Wang, C.; et al. Transcriptome Differences in Response Mechanisms to Low-Nitrogen Stress in Two Wheat Varieties. Int. J. Mol. Sci. 2021, 22, 12278. [Google Scholar] [CrossRef]
- Hasan, N.; Choudhary, S.; Naaz, N.; Sharma, N.; Laskar, R.A. Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J. Genet. Eng. Biotechnol. 2021, 19, 128. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.-C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y.; Zhao, B.; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Vikas, V.K.; Pradhan, A.K.; Budhlakoti, N.; Mishra, D.C.; Chandra, T.; Bhardwaj, S.C.; Kumar, S.; Sivasamy, M.; Jayaprakash, P.; Nisha, R.; et al. Multi-locus genome-wide association studies (ML-GWAS) reveal novel genomic regions associated with seedling and adult plant stage leaf rust resistance in bread wheat (Triticum aestivum L.). Heredity 2022, 128, 434–449. [Google Scholar] [CrossRef]
- Wang, W.; Guo, W.; Le, L.; Yu, J.; Wu, Y.; Li, D.; Wang, Y.; Wang, H.; Lu, X.; Qiao, H.; et al. Integration of high-throughput phenotyping, GWAS, and predictive models reveals the genetic architecture of plant height in maize. Mol. Plant 2023, 16, 354–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, T.; Yang, J.; Wang, H.; Ji, W.; Xu, Y.; Yang, Z.; Xu, C.; Li, P. GWAS and Transcriptome Analysis Reveal Key Genes Affecting Root Growth under Low Nitrogen Supply in Maize. Genes 2022, 13, 1632. [Google Scholar] [CrossRef]
- Schmidt, L.; Nagel, K.; Galinski, A.; Sannemann, W.; Pillen, K.; Maurer, A. Unraveling Genomic Regions Controlling Root Traits as a Function of Nitrogen Availability in the MAGIC Wheat Population WM-800. Plants 2022, 11, 3520. [Google Scholar] [CrossRef]
- Fu, Y.; Liu, J.; Xia, Z.; Wang, Q.; Zhang, S.; Zhang, G.; Lu, H. Genome-Wide Association Studies of Maize Seedling Root Traits under Different Nitrogen Levels. Plants 2022, 11, 1417. [Google Scholar] [CrossRef]
- Xiong, H.; Guo, H.; Zhou, C.; Guo, X.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. A combined association mapping and t-test analysis of SNP loci and candidate genes involving in resistance to low nitrogen traits by a wheat mutant population. PLoS ONE 2019, 14, e0211492. [Google Scholar] [CrossRef]
- Li, J.; Xin, W.; Wang, W.; Zhao, S.; Xu, L.; Jiang, X.; Duan, Y.; Zheng, H.; Yang, L.; Liu, H.; et al. Mapping of Candidate Genes in Response to Low Nitrogen in Rice Seedlings. Rice 2022, 15, 51. [Google Scholar] [CrossRef]
- Pflugfelder, D.; Kochs, J.; Koller, R.; Jahnke, S.; Mohl, C.; Pariyar, S.; Fassbender, H.; A Nagel, K.; Watt, M.; van Dusschoten, D. The root system architecture of wheat establishing in soil is associated with varying elongation rates of seminal roots: Quantification using 4D magnetic resonance imaging. J. Exp. Bot. 2022, 73, 2050–2060. [Google Scholar] [CrossRef] [PubMed]
- Erenoglu, E.B.; Kutman, U.B. Improved nitrogen nutrition enhances root uptake, root-to-shoot translocation and remobilization of zinc (65Zn) in wheat. New Phytol. 2011, 189, 438–448. [Google Scholar] [CrossRef]
- Lynch, J.P. Steep, cheap and deep: An ideotype to optimize water and N acquisition by maize root systems. Ann. Bot. 2013, 112, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, J.; Wang, C.; Ma, G.; Wei, Q.; Lu, H.; Xie, Y.; Ma, D.; Kang, G. Root Growth, Water and Nitrogen Use Efficiencies in Winter Wheat Under Different Irrigation and Nitrogen Regimes in North China Plain. Front. Plant Sci. 2018, 9, 1798. [Google Scholar] [CrossRef] [PubMed]
- Forde, B.G. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 2014, 21, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.; Palta, J.A.; Rebetzke, G.J.; Milroy, S.P. Wheat genotypes with high early vigour accumulate more nitrogen and have higher photosynthetic nitrogen use efficiency during early growth. Funct. Plant Biol. 2014, 41, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Wang, J.; Li, J.; Zhao, H.; Liu, H.; Zheng, H.; Yang, L.; Wang, C.; Yang, F.; Chen, J.; et al. Candidate Gene Analysis for Nitrogen Absorption and Utilization in Japonica Rice at the Seedling Stage Based on a Genome-Wide Association Study. Front. Plant Sci. 2021, 12, 670861. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yi, X.; Tang, S.; Chen, W.; Wu, F.; Yang, X.; Jiang, X.; Shi, H.; Ma, J.; Chen, G.; et al. Dissection of Phenotypic and Genetic Variation of Drought-Related Traits in Diverse Chinese Wheat Landraces. Plant Genome 2019, 12, 190025. [Google Scholar] [CrossRef]
- Rathan, N.D.; Krishna, H.; Ellur, R.K.; Sehgal, D.; Govindan, V.; Ahlawat, A.K.; Krishnappa, G.; Jaiswal, J.P.; Singh, J.B.; Sv, S.; et al. Genome-wide association study identifies loci and candidate genes for grain micronutrients and quality traits in wheat (Triticum aestivum L.). Sci. Rep. 2022, 12, 7037. [Google Scholar] [CrossRef]
- Lynch, J.P. Roots of the Second Green Revolution. Aust. J. Bot. 2007, 55, 493–512. [Google Scholar] [CrossRef]
- Fan, Y.; Miguez-Macho, G.; Jobbágy, E.G.; Jackson, R.B.; Otero-Casal, C. Hydrologic regulation of plant rooting depth. Proc. Natl. Acad. Sci. USA 2017, 114, 10572–10577. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, Q.; Meng, D.; Ren, X.; Li, H.; Su, Z.; Zhang, N.; Zhi, L.; Ji, J.; Li, J.; et al. QMrl-7B Enhances Root System, Biomass, Nitrogen Accumulation and Yield in Bread Wheat. Plants 2021, 10, 764. [Google Scholar] [CrossRef]
- Ehdaie, B.; Merhaut, D.J.; Ahmadian, S.; Hoops, A.C.; Khuong, T.; Layne, A.P.; Waines, J.G. Root System Size Influences Water-Nutrient Uptake and Nitrate Leaching Potential in Wheat. J. Agron. Crop Sci. 2010, 196, 455–466. [Google Scholar] [CrossRef]
- Cui, F.; Zhao, C.; Ding, A.; Li, J.; Wang, L.; Li, X.; Bao, Y.; Li, J.; Wang, H. Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor. Appl. Genet. 2014, 127, 659–675. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Kong, F.M.; Xu, Y.-F.; Zhao, Y.; Liang, X.; Wang, Y.-Y.; An, D.-G.; Li, S.-S. QTL mapping for seedling traits in wheat grown under varying concentrations of N, P and K nutrients. Theor. Appl. Genet. 2012, 124, 851–865. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, R.; Tong, Y.; Zhao, H.; Xie, Q.; Liu, D.; Zhang, A.; Li, B.; Xu, H.; An, D. Mapping QTLs for yield and nitrogen-related traits in wheat: Influence of nitrogen and phosphorus fertilization on QTL expression. Theor. Appl. Genet. 2014, 127, 59–72. [Google Scholar] [CrossRef]
- Gahlaut, V.; Jaiswal, V.; Singh, S.; Balyan, H.S.; Gupta, P.K. Multi-Locus Genome Wide Association Mapping for Yield and Its Contributing Traits in Hexaploid Wheat under Different Water Regimes. Sci. Rep. 2019, 9, 19486. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Mullan, D.; Zhang, C.; Zhao, S.; Li, X.; Zhang, A.; Lu, Z.; Wang, Y.; Yan, G. Major genomic regions responsible for wheat yield and its components as revealed by meta-QTL and genotype–phenotype association analyses. Planta 2020, 252, 65. [Google Scholar] [CrossRef] [PubMed]
- Fatima, I.; Gao, Y.; Xu, X.; Jin, J.; Duan, S.; Zhen, W.; Xie, C.; Ma, J. Genome-Wide Association Mapping of Seedling Biomass and Root Traits Under Different Water Conditions in Wheat. Front. Genet. 2021, 12, 663557. [Google Scholar] [CrossRef]
- Fang, X.Z.; Fang, S.Q.; Ye, Z.Q.; Liu, D.; Zhao, K.L.; Jin, C.W. NRT1.1 Dual-Affinity Nitrate Transport/Signalling and its Roles in Plant Abiotic Stress Resistance. Front. Plant Sci. 2021, 12, 715694. [Google Scholar] [CrossRef]
- Li, J.; Zhao, C. Arabidopsis NRT1.2 interacts with the PHOSPHOLIPASE Dα1 (PLDα1) to positively regulate seed germination and seedling development in response to ABA treatment. Biochem. Biophys. Res. Commun. 2020, 533, 104–109. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Tsay, Y.-F. Arabidopsis Nitrate Transporter NRT1.9 Is Important in Phloem Nitrate Transport. Plant Cell 2011, 23, 1945–1957. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Lv, X.-F.; Li, J.-Y.; Yi, H.-Y.; Gong, J.-M. Arabidopsis NRT1.5 Is Another Essential Component in the Regulation of Nitrate Reallocation and Stress Tolerance. Plant Physiol. 2012, 159, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-Y.; Fu, Y.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.-Z.; Zhang, Y.; Li, H.-M.; Huang, J.; et al. The Arabidopsis Nitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.-C.; Lin, C.-S.; Hsu, P.-K.; Lin, S.-H.; Tsay, Y.-F. The Arabidopsis Nitrate Transporter NRT1.7, Expressed in Phloem, Is Responsible for Source-to-Sink Remobilization of Nitrate. Plant Cell 2009, 21, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Buchner, P.; Hawkesford, M.J. Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. J. Exp. Bot. 2014, 65, 5697–5710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Park, J.J.; Yi, J.; Yoon, J.; Cho, L.; Ping, J.; Jeong, H.J.; Cho, S.K.; Kim, W.T.; An, G. OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 2011, 65, 194–205. [Google Scholar] [CrossRef]
- Al-Saharin, R.; Hellmann, H.; Mooney, S. Plant E3 Ligases and Their Role in Abiotic Stress Response. Cells 2022, 11, 890. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y. Plant U-box E3 ligases PUB25 and PUB26 control organ growth in Arabidopsis. New Phytol. 2021, 229, 403–413. [Google Scholar] [CrossRef]
- Beyer, S.; Daba, S.; Tyagi, P.; Bockelman, H.; Brown-Guedira, G.; IWGSC; Mohammadi, M. Loci and candidate genes controlling root traits in wheat seedlings—A wheat root GWAS. Funct. Integr. Genom. 2019, 19, 91–107. [Google Scholar] [CrossRef]
- Min, H.J.; Jung, Y.J.; Kang, B.G.; Kim, W.T. CaPUB1, a Hot Pepper U-box E3 Ubiquitin Ligase, Confers Enhanced Cold Stress Tolerance and Decreased Drought Stress Tolerance in Transgenic Rice (Oryza sativa L.). Mol. Cells 2016, 39, 250–257. [Google Scholar] [CrossRef]
- Zhu, J.; Kaeppler, S.M.; Lynch, J.P. Mapping of QTLs for lateral root branching and length in maize (Zea mays L.) under differential phosphorus supply. Theor. Appl. Genet. 2005, 111, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Manfioletti, G.; Schneider, C. A new and fast method for preparing high quality lambda DNA suitable for sequencing. Nucleic Acids Res. 1988, 16, 2873–2884. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Kang, H.M.; A Zaitlen, N.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient Control of Population Structure in Model Organism Association Mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, F.; Yan, X.; Zhang, X.; Dong, Z.; Cui, D.; Chen, F. Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol. J. 2017, 15, 953–969. [Google Scholar] [CrossRef]
- Jin, J.; Duan, S.; Qi, Y.; Yan, S.; Li, W.; Li, B.; Xie, C.; Zhen, W.; Ma, J. Identification of a novel genomic region associated with resistance to Fusarium crown rot in wheat. Theor. Appl. Genet. 2020, 133, 2063–2073. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, D.; Tang, X.; Yuan, M.; Zhang, D.; Xu, M.; Duan, Y.; Ren, H.; Zeng, Q.; Wu, J.; et al. Genome-Wide Association Study on Root System Architecture and Identification of Candidate Genes in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2022, 23, 1843. [Google Scholar] [CrossRef]
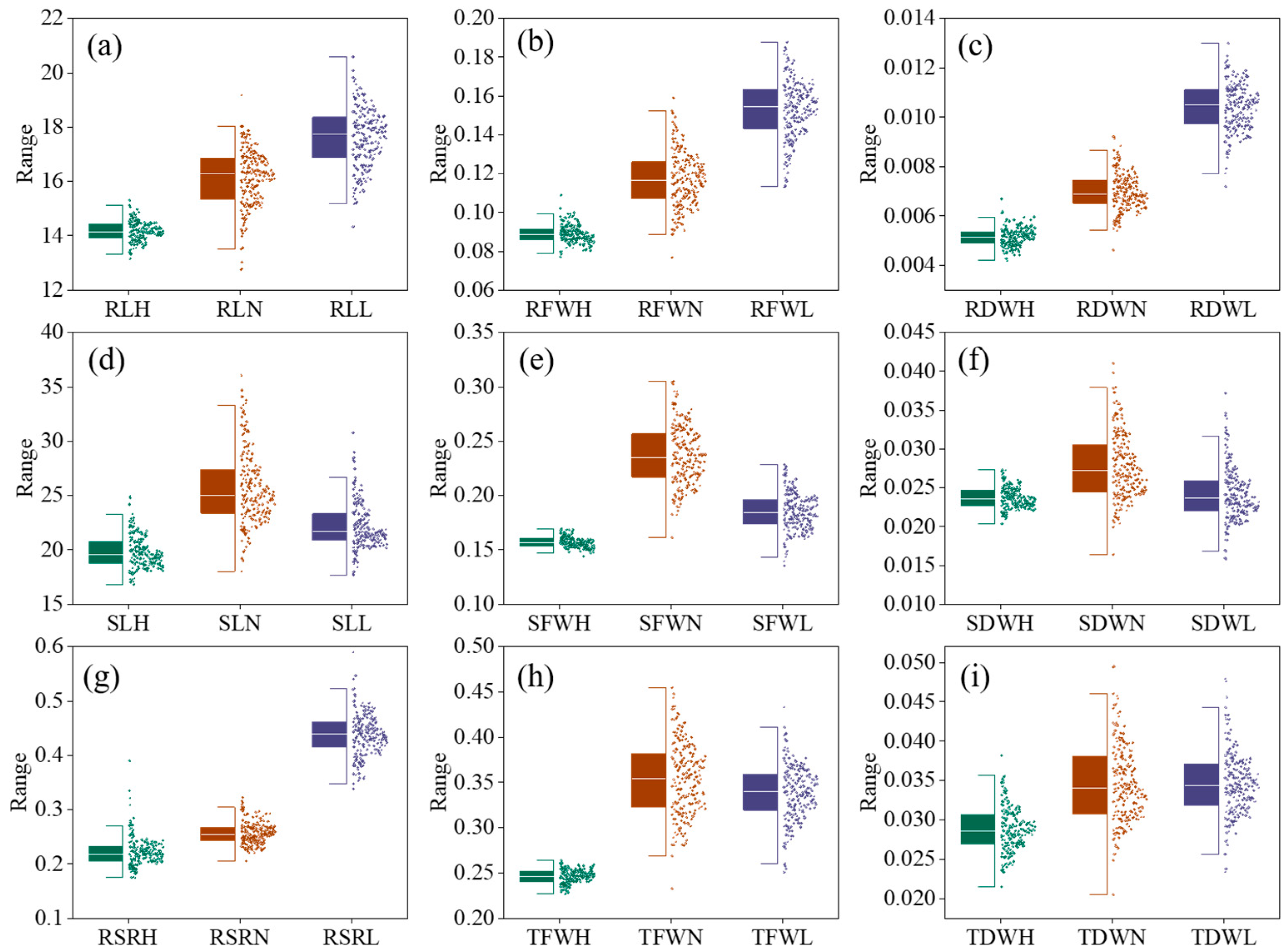

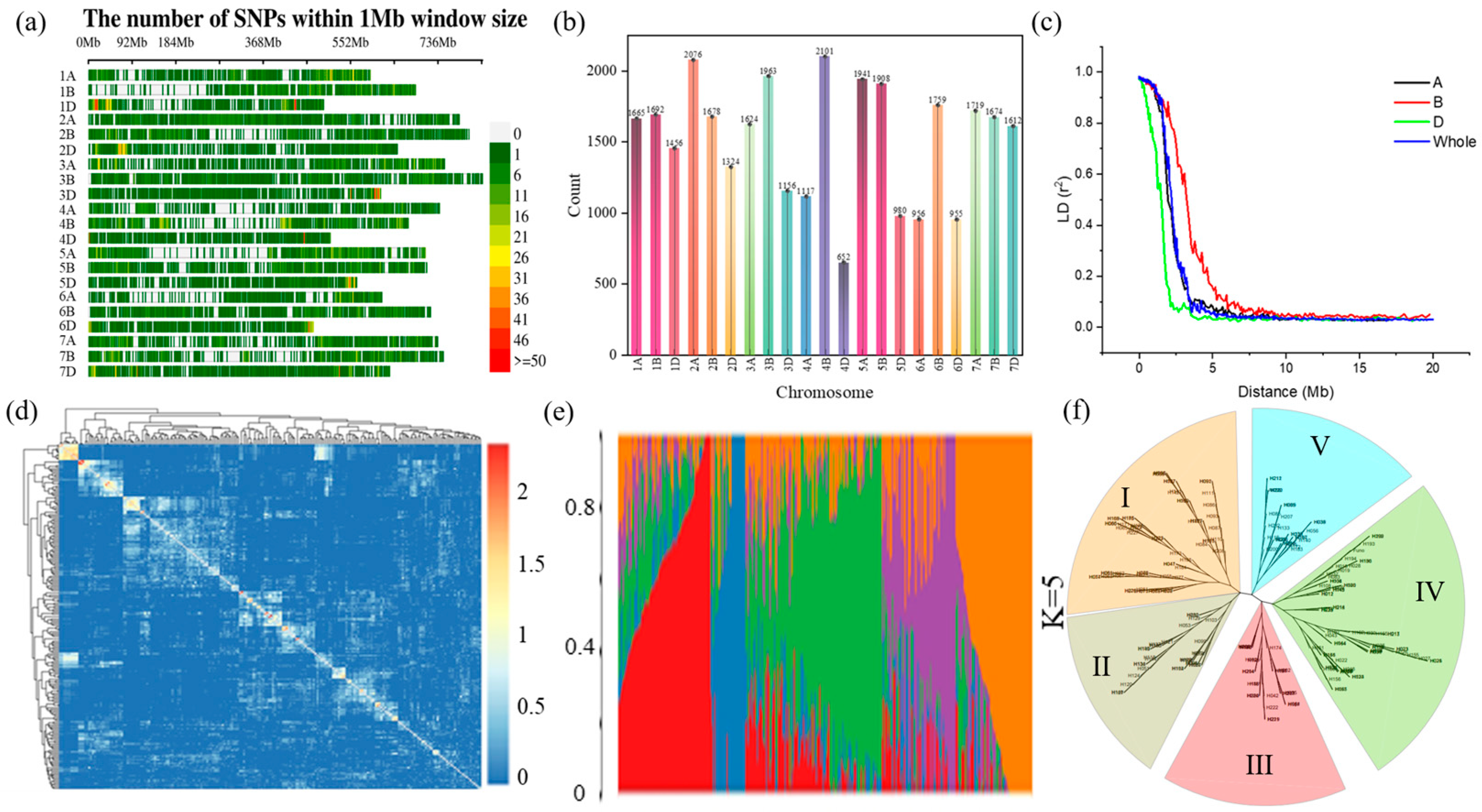
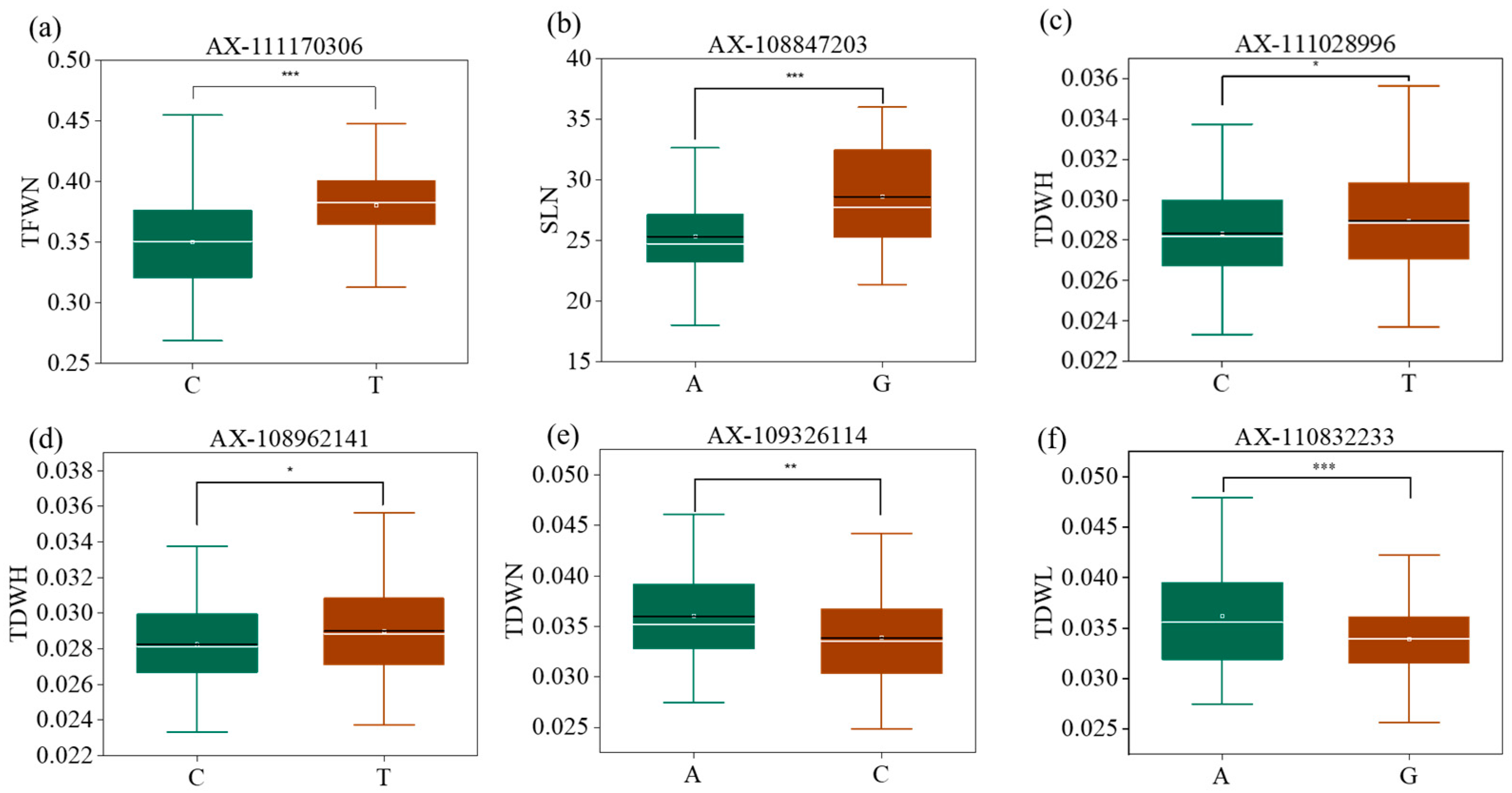
| Conditions | Index | RL | RSR | RDW | RFW | SL | SDW | SFW | TDW | TFW |
|---|---|---|---|---|---|---|---|---|---|---|
| HN | Min | 13.14 | 0.17 | 0.004 | 0.077 | 16.82 | 0.020 | 0.144 | 0.022 | 0.227 |
| Max | 15.30 | 0.39 | 0.007 | 0.109 | 24.97 | 0.027 | 0.170 | 0.038 | 0.264 | |
| Ave | 14.17 | 0.22 | 0.005 | 0.089 | 19.83 | 0.024 | 0.157 | 0.029 | 0.246 | |
| SD | 0.36 | 0.03 | 0.001 | 0.005 | 1.51 | 0.001 | 0.005 | 0.003 | 0.008 | |
| Coef. of var (%) | 2.56 | 12.15 | 7.130 | 5.272 | 7.62 | 5.850 | 2.987 | 9.666 | 3.122 | |
| h2 | 0.92 | 0.47 | 0.470 | 0.774 | 0.57 | 0.381 | 0.860 | 0.398 | 0.618 | |
| NN | Min | 12.76 | 0.21 | 0.005 | 0.077 | 18.02 | 0.021 | 0.161 | 0.015 | 0.233 |
| Max | 19.17 | 0.32 | 0.009 | 0.159 | 36.03 | 0.050 | 0.305 | 0.064 | 0.455 | |
| Ave | 16.02 | 0.26 | 0.007 | 0.117 | 25.74 | 0.035 | 0.237 | 0.035 | 0.354 | |
| SD | 1.09 | 0.02 | 0.001 | 0.014 | 3.43 | 0.005 | 0.027 | 0.009 | 0.041 | |
| Coef. of var (%) | 6.76 | 7.93 | 11.299 | 12.161 | 13.34 | 14.268 | 11.519 | 24.837 | 11.44 | |
| h2 | 0.79 | 0.72 | 0.684 | 0.597 | 0.40 | 0.777 | 0.673 | 0.327 | 0.620 | |
| LN | Min | 14.33 | 0.34 | 0.007 | 0.113 | 17.69 | 0.016 | 0.135 | 0.023 | 0.251 |
| Max | 20.59 | 0.66 | 0.013 | 0.206 | 30.78 | 0.037 | 0.229 | 0.048 | 0.434 | |
| Ave | 17.58 | 0.44 | 0.010 | 0.154 | 22.27 | 0.024 | 0.185 | 0.035 | 0.339 | |
| SD | 1.12 | 0.04 | 0.001 | 0.016 | 2.16 | 0.004 | 0.017 | 0.004 | 0.031 | |
| Coef. of var (%) | 6.39 | 9.49 | 9.097 | 10.065 | 9.70 | 14.362 | 9.341 | 11.671 | 9.044 | |
| h2 | 0.92 | 0.62 | 0.698 | 0.473 | 0.59 | 0.135 | 0.450 | 0.288 | 0.467 |
| Marker | Chromosome | Position | Pleiotropic Effect | SNP | p-Value |
|---|---|---|---|---|---|
| AX-108847203 | 1A | 567714120 | RDWL, RDWN, SLL, SLN, SDWL, SDWN, TDWL, TDWN | G | <0.001 |
| AX-111170306 | 1A | 567967912 | RDWN, SLL, SLN, SDWL, SDWN, SFWN, TDWL, TDWN, TFWN | T | <0.001 |
| AX-108737478 | 1B | 375098459 | TDWL, TFWN | C | <0.001 |
| AX-111695833 | 1B | 374522868 | RDWL, TDWH | A | <0.001 |
| AX-108962141 | 1B | 403813869 | RDWN, RFWN, SLL, SDWL, SDWN, TDWH, TDWL, TDWN | T | <0.001 |
| AX-110392069 | 1B | 639638582 | SDWN, SFWN, TDWH, TDWN, TFWN | G | <0.001 |
| AX-109326114 | 1D | 21743856 | RDWN, SLL, SDWL, SDWN, SFWN, TDWN | A | <0.001 |
| AX-111161220 | 1B | 414806318 | RDWN, TDWH | T | <0.001 |
| AX-108903381 | 1B | 415260486 | RSRH, SFWN | A | <0.001 |
| AX-108786044 | 2A | 698009743 | RDWL, SFWH, TDWH, TDWL, TFWH | A | <0.001 |
| AX-110362294 | 1B | 317448975 | RSRH, SFWN, TFWN | T | <0.001 |
| AX-111575379 | 2B | 461886409 | RDWN, TDWH, TDWL | A | <0.001 |
| AX-111672733 | 2B | 572628142 | SDWL, TDWH, TDWL | C | <0.001 |
| AX-110010230 | 1B | 325283703 | RSRH, SFWN, TFWN | C | <0.001 |
| AX-111405488 | 2D | 59943039 | SDWL, SFWL, TDWH, TDWL | C | <0.001 |
| AX-110926323 | 1B | 325256496 | RSRH, SFWN | T | <0.001 |
| AX-110438187 | 2A | 95830292 | RSRH, SLN | A | <0.001 |
| AX-109815802 | 4B | 10916305 | RFWN, SFWH, SFWN, TFWH, TFWN | A | <0.001 |
| AX-109327593 | 4B | 10944746 | RFWN, SFWH, SFWN, TDWH, TFWN | A | <0.001 |
| AX-109365636 | 2B | 758808487 | SDWN, SFWN, TDWH, TFWN | T | <0.001 |
| AX-110832233 | 4D | 507128999 | SLL, SLN, SDWL, SDWN, TDWH, TDWL, TDWN | A | <0.001 |
| AX-111051748 | 5B | 44358312 | SDWL, TDWH, TDWL | A | <0.001 |
| AX-89696347 | 2D | 647533340 | RLH, SDWL, TDWL | C | <0.001 |
| AX-109325100 | 6A | 61431458 | SDWN, SFWN, TDWH, TDWN, TFWN | A | <0.001 |
| AX-111342539 | 2D | 100544989 | TDWH, TFWN | A | <0.001 |
| AX-109651481 | 6A | 310569613 | SDWL, SFWL, TDWH, TDWL, TFWL | C | <0.001 |
| AX-108871687 | 6B | 553420734 | RDWL, RDWN, TDWH, TDWL | G | <0.001 |
| AX-111605442 | 2D | 220630228 | TDWH, TFWN | C | <0.001 |
| AX-110525380 | 3B | 25322853 | RFWH, TFWH | T | <0.001 |
| AX-111123066 | 7A | 55431597 | SDWN, SFWN, TDWH, TDWN, TFWN | T | <0.001 |
| AX-111526214 | 4D | 310458135 | SFWN, TDWH, TFWN | C | <0.001 |
| AX-110483224 | 7B | 611776655 | SLL, SLN, SDWN | C | <0.001 |
| AX-109327847 | 7B | 612256762 | SLL, SLN, SDWN, SFWN | A | <0.001 |
| AX-111485326 | 7B | 627697145 | SDWN, TDWL, TDWN | A | <0.001 |
| AX-109401644 | 5B | 50655258 | SDWN, TDWN | A | <0.001 |
| AX-111721212 | 5D | 553775393 | RDWL, RFWN, TDWL | C | <0.001 |
| AX-111762061 | 6B | 192866992 | RFWN, SDWN, SFWN, TDWH, TFWN | T | <0.001 |
| AX-109624261 | 7A | 113627131 | SFWN, TDWH, TFWN | G | <0.001 |
| AX-110595073 | 7B | 627636405 | SLL, SDWN, TDWN | A | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, C.; Li, J.; Liu, J.; Zhang, D.; Jin, L.; Yang, N.; Bai, B.; Wang, Z.; Feng, S.; Ru, Z.; et al. Genome-Wide Association Study on Seedling Phenotypic Traits of Wheat under Different Nitrogen Conditions. Plants 2023, 12, 4050. https://doi.org/10.3390/plants12234050
Hu C, Li J, Liu J, Zhang D, Jin L, Yang N, Bai B, Wang Z, Feng S, Ru Z, et al. Genome-Wide Association Study on Seedling Phenotypic Traits of Wheat under Different Nitrogen Conditions. Plants. 2023; 12(23):4050. https://doi.org/10.3390/plants12234050
Chicago/Turabian StyleHu, Chenchen, Jinghui Li, Jiajia Liu, Dazhong Zhang, Liqiao Jin, Nian Yang, Bipo Bai, Zenghao Wang, Suwei Feng, Zhengang Ru, and et al. 2023. "Genome-Wide Association Study on Seedling Phenotypic Traits of Wheat under Different Nitrogen Conditions" Plants 12, no. 23: 4050. https://doi.org/10.3390/plants12234050
APA StyleHu, C., Li, J., Liu, J., Zhang, D., Jin, L., Yang, N., Bai, B., Wang, Z., Feng, S., Ru, Z., & Hu, T. (2023). Genome-Wide Association Study on Seedling Phenotypic Traits of Wheat under Different Nitrogen Conditions. Plants, 12(23), 4050. https://doi.org/10.3390/plants12234050





