Unlocking Nature’s Clock: CRISPR Technology in Flowering Time Engineering
Abstract
:1. Flowering Time Matters
2. Molecular Mechanisms Regulating Flowering Time in Arabidopsis
2.1. Photoperiod Pathway
2.2. Autonomous Pathway
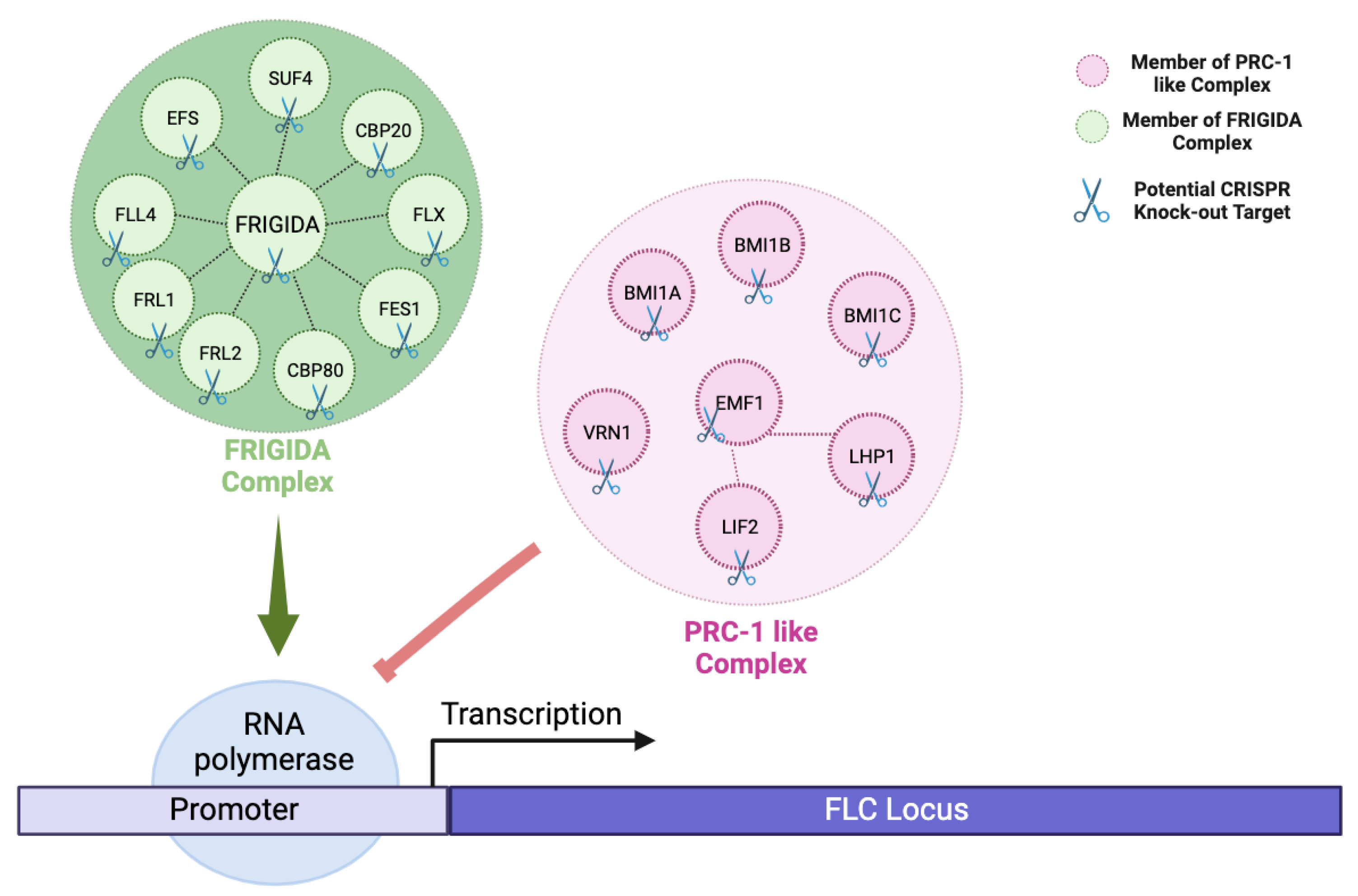
2.3. Vernalization Pathway
2.4. Hormonal Pathway
2.5. Sugar Pathway
2.6. Aging Pathway
2.7. Temperature Pathway
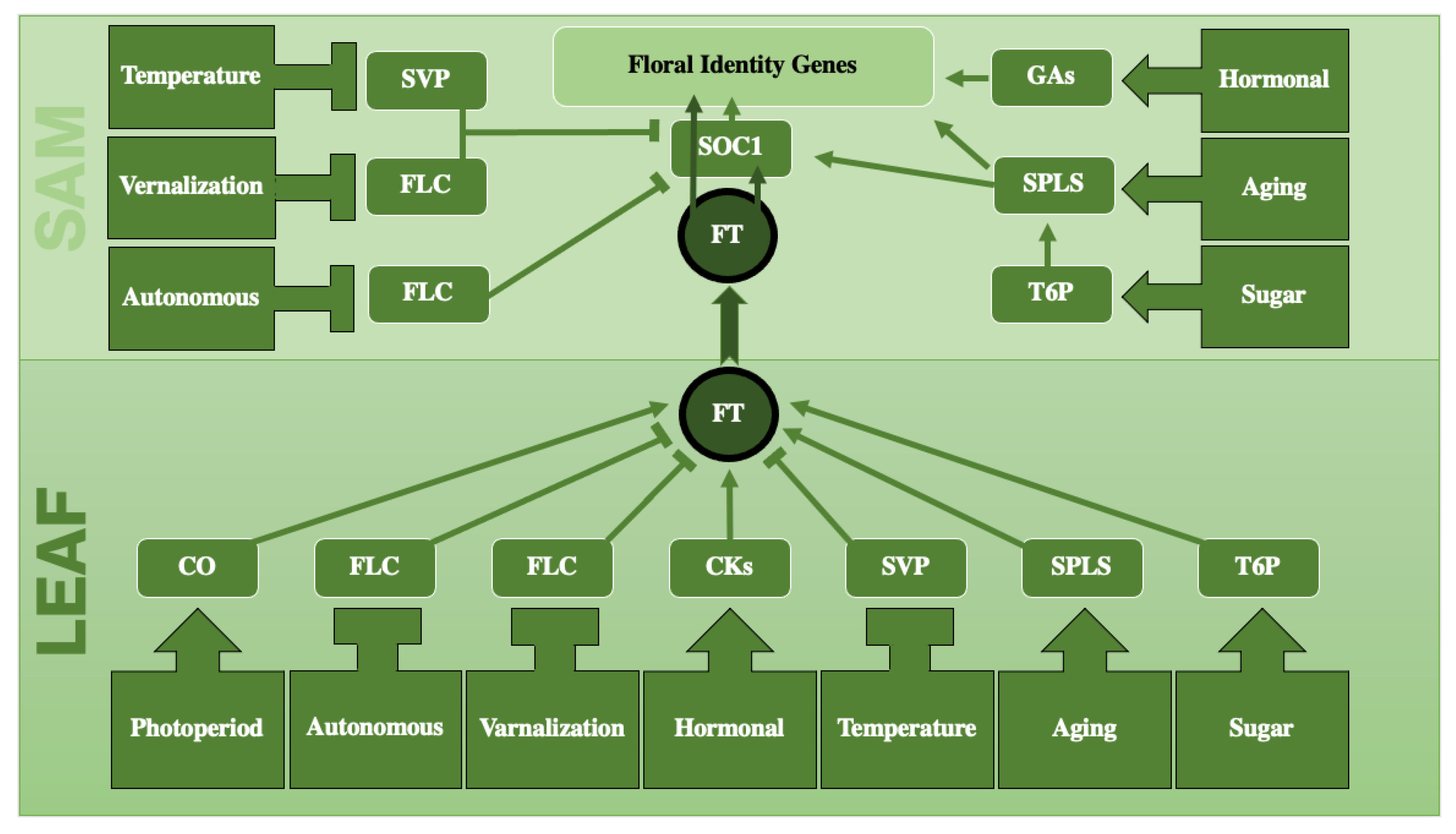
2.8. Interconnectedness between Pathways
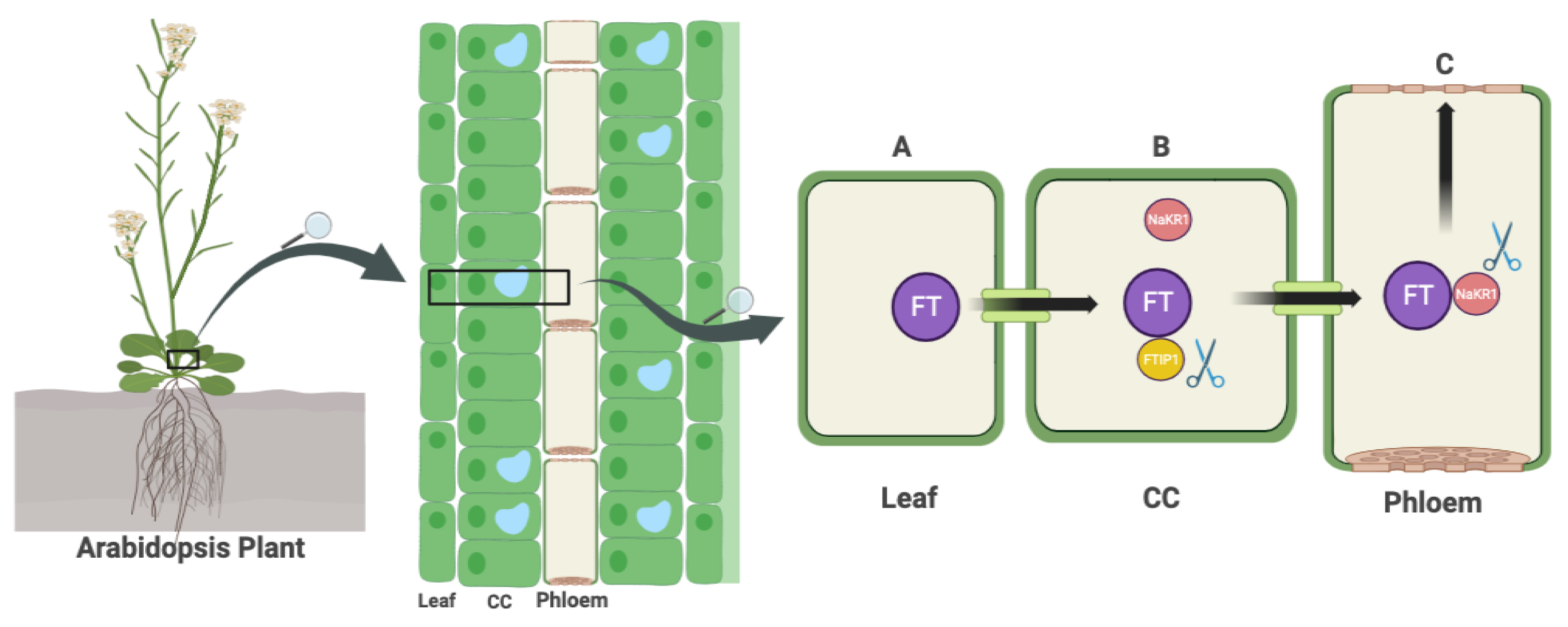
2.9. Transportation of FT
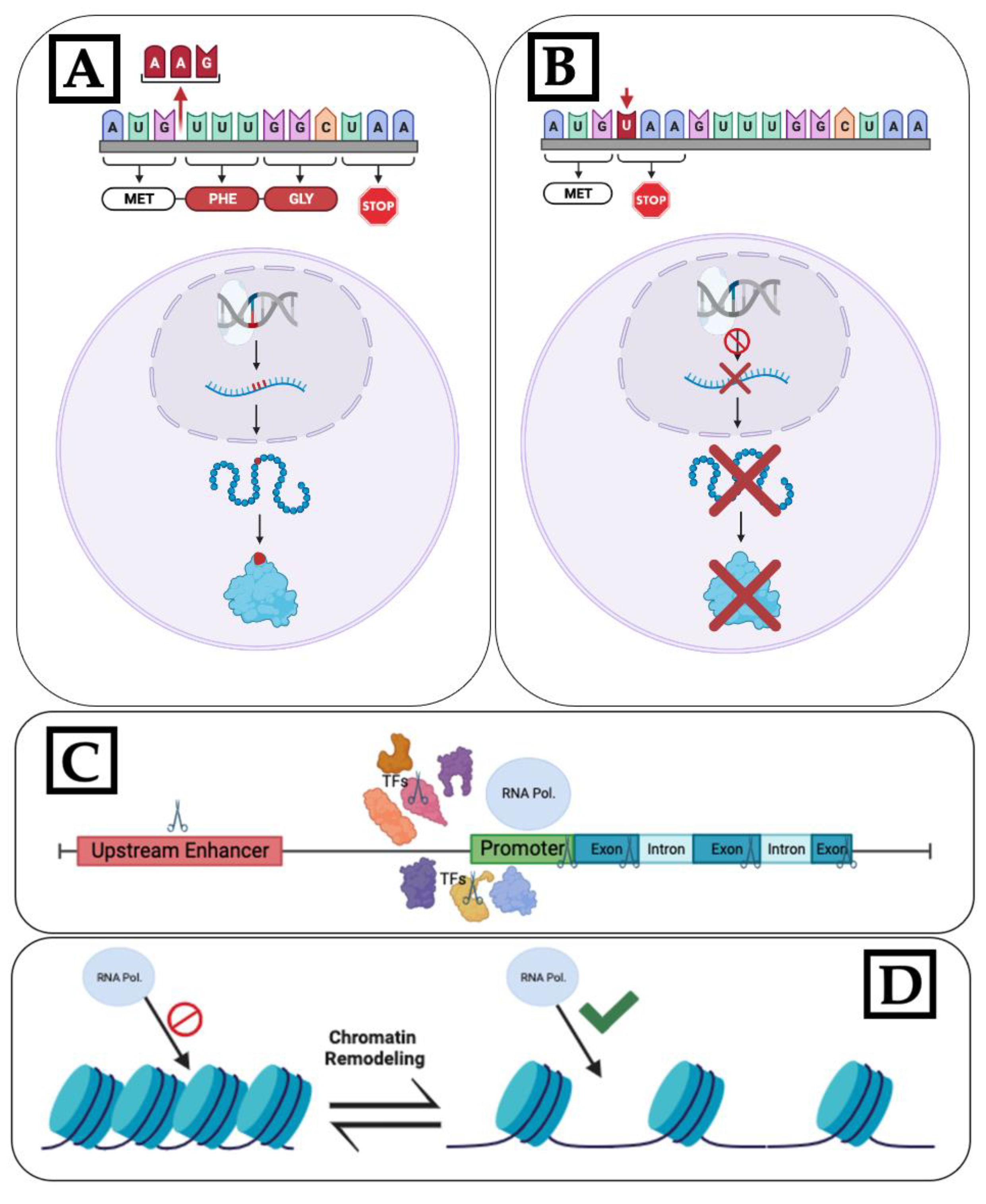
3. Optimal Gene Targeting Strategy for Flowering Time Engineering
3.1. Altering Flowering Time
3.2. Stop Flowering
4. CRISPR-Mediated Modulation of Flowering Time in Literature
| Author(s) | Year | Directed Gene(s) | Mechanism | Plant | Results in Flowering |
|---|---|---|---|---|---|
| Torre et al. [110] | 2022 | AaFRAT1 | KO | Alpine cress | Early |
| Charrier et al. [111] | 2019 | MdTFL1.1, PcTFL1.1 | KO | Apple, Pear | Early |
| Liu et al. [112] | 2019 | TFL1, AP1, SVP | KO | Arabidopsis | Abnormal flower development |
| Ning et al. [113] | 2015 | NACs | KO | Arabidopsis | Early |
| Nobusawa et al. [114] | 2022 | AMP1 | KO | Arabidopsis | Early |
| Capovilla et al. [115] | 2017 | FLM-β | KO | Arabidopsis | Early |
| Branchat et al. [116] | 2020 | FDP, fd | KO | Arabidopsis | Early, late |
| Lian et al. [117] | 2021 | MIR172s | KO and OE | Arabidopsis | Early, late, no effect |
| Yan et al. [118] | 2017 | KHZ1 and KHZ2 | KO and OE | Arabidopsis | Late and early |
| Hyun et al. [119] | 2015 | FT and SPL | KO | Arabidopsis | Late |
| Hou et al. [120] | 2019 | AtMIR396 | KO | Arabidopsis | Late |
| Yao et al. [121] | 2019 | mir167a | KO | Arabidopsis | Late |
| Huang et al. [122] | 2019 | OsNCED5 | OE | Arabidopsis | Late |
| Wang et al. [123] | 2021 | RBP45D | KO | Arabidopsis | Late |
| Zhao et al. [124] | 2022 | CIS1 | KO | Arabidopsis | Late |
| Yang et al. [125] | 2023 | AtAGL79 | KO | Arabidopsis | Late |
| Pyott et al. [126] | 2016 | eIF(iso)4E | KO | Arabidopsis | No effect |
| Soto et al. [127] | 2022 | FT2 | KO | Aspen | No report |
| Qin et al. [128] | 2019 | FTL9 | KO | Brachypodium | Late |
| Jeong et al. [92] | 2019 | BraFLC2, BraFLC3 | KO | Cabbage | Early |
| Jung et al. [129] | 2021 | BrSOC1 | KO | Cabbage | Early |
| Hong et al. [130] | 2021 | BrVRN1 | KO | Cabbage | Late |
| Shin et al. [131] | 2023 | BrLFY | KO | Cabbage | Late |
| Lee et al. [132] | 2023 | BrFT1 and BrFT2 | KO | Cabbage | Late |
| Shin et al. [91] | 2022 | AGL19s, AGL24s | KO | Cabbage | Late |
| Park et al. [133] | 2019 | GI | KO | Cabbage | No report |
| Bellec et al. [134] | 2022 | 15 genes | KO | Camelina | Early |
| Jiang et al. [135] | 2018 | BnaSDG8.A, BnaSDG8.C | KO | Canola | Early |
| Sriboon et al. [99] | 2020 | BnaC03.TFL1 | KO | Canola | Early |
| Guo et al. [136] | 2022 | BnaCOL9 | KO | Canola | Early |
| Ahmar et al. [137] | 2022 | BnaSVPs | KO | Canola | Early |
| Zhou et al. [138] | 2022 | BnaSVP, BnaSEP1 | KO | Canola | Early, no effect |
| Odipio et al. [139] | 2018 | TFL1-like | KO | Cassava | Early |
| Bull et al. [140] | 2018 | AtFT | Ectopic expression | Cassava | Early |
| Liu et al. [141] | 2023 | CiTFL1a, CiTFL1b | KO | Chrysantemum | Early |
| Huang et al. [142] | 2017 | ZmCCT9 | KO | Corn | Early |
| Li et al. [143] | 2020 | ZmPHYC1, ZmPHYC2 | KO | Corn | Early |
| Takahashi et al. [144] | 2022 | GtFT2 | KO | Gentian | Late |
| Ying et al. [145] | 2022 | BdRFS | KO and OE | Brachypodium | Early, late |
| Sheng et al. [146] | 2021 | YSL3 | KO | Brachypodium | Late |
| Herath et al. [147] | 2022 | AcBFT2 | KO | Kiwi | No effect |
| Gasic et al. [148] | 2019 | AcCEN4, AcCEN | KO | Kiwi | Early |
| Varkonyi et al. [149] | 2021 | CEN, CEN4, SyGl | KO | Kiwi | Early |
| Choi et al. [10] | 2022 | SOC1 | KO | Lettuce | Late |
| Singer et al. [150] | 2021 | MsSPL8 | KO | Lucerne | Early |
| Galindo-Sotomonte et al. [151] | 2023 | MSAD_264347 | KO | Lucerne | Late |
| Wolabu et al. [152] | 2023 | MsFTa1 | KO | Lucerne | Late |
| Shibuya et al. [153] | 2018 | EPHEMERAL1 | KO | Morning Glory | Delay in petal aging |
| Andre et al. [154] | 2022 | FT2b | OE | Populus | Early |
| Elorriaga et al. [155] | 2018 | PLFY, PAG1, PAG2 | KO | Populus | No report |
| Lebedeva et al. [156] | 2022 | StLFY | KO | Potato | Late, non-flowering |
| Li et al. [157] | 2017 | Hd2, Hd4, Hd5 | KO | Rice | Early |
| Brambilla et al. [158] | 2017 | hbf1, hbf2 | KO | Rice | Early |
| Zhou et al. [159] | 2018 | Ghd8 | KO | Rice | Early |
| Cui et al. [160] | 2019 | se14 | KO | Rice | Early |
| Wang et al. [161] | 2020 | OsGhd7 | KO | Rice | Early |
| Karthika et al. [162] | 2021 | MSH2 | KO | Rice | Early |
| Leon et al. [163] | 2021 | OsGA20ox2 | KO | Rice | Early |
| Sun et al. [164] | 2022 | qHD5 | KO | Rice | Early |
| Yin et al. [165] | 2023 | HBP1 | KO | Rice | Early |
| Sedeek et al. [166] | 2023 | Hd2, Hd4, Hd5 | KO | Rice | Early |
| Guo et al. [167] | 2022 | OsFTL4 | KO | Rice | Early |
| Sun et al. [168] | 2021 | OsLHY | KO | Rice | Early, late |
| Zhang et al. [169] | 2020 | OsCCTs | KO | Rice | Early, late, no effect |
| Cui Y et al. [170] | 2021 | 14 genes | KO | Rice | Early, late, no effect |
| Yasui et al. [171] | 2017 | MADS3 | KO | Rice | Early/Late |
| Wu et al. [172] | 2020 | Ehd1 | KO | Rice | Late |
| Li et al. [173] | 2021 | OsLHY | KO | Rice | Late |
| Liu et al. [174] | 2021 | OsHd2 | KO | Rice | Late |
| Zhang et al. [175] | 2022 | ga3ox-2 | KO | Rice | Late |
| Xu et al. [176] | 2023 | OsLUX | KO | Rice | Late |
| Zhang et al. [177] | 2022 | ospil12-1 and ospil12-2 | KO | Rice | Late |
| Andrade et al. [178] | 2022 | LUX, ELF3 | KO | Rice | Non-flowering |
| Dai et al. [179] | 2021 | HbFT1-2, HbTFL1-3 | KO | Rubber Tree | No report |
| Wang et al. [180] | 2022 | SiPHYC | KO | Setaria | Early |
| Zhu et al. [181] | 2022 | spp1 | KO | Setaria | No effect |
| Char et al. [182] | 2019 | SbFT | KO | Sorghum | Late |
| Han et al. [183] | 2019 | E1 | KO | Soy | Early |
| Wang et al. [184] | 2020 | Gmprr37 | KO | Soy | Early |
| Wang et al. [185] | 2020 | GmNMHC5 | OE | Soy | Early |
| Zhaobo Li et al. [186] | 2021 | LNK2 | KO | Soy | Early |
| Wan et al. [187] | 2022 | E1 | KO | Soy | Early |
| Zhai et al. [188] | 2022 | E1 | KO and OE | Soy | Early, late |
| Cai et al. [189] | 2018 | GmFT2a | KO | Soy | Late |
| Wang et al. [190] | 2019 | GmLCLa1-4 | KO | Soy | Late |
| Cong Li et al. [191] | 2020 | GmPRR3bH6 | KO | Soy | Late |
| Chen et al. [192] | 2020 | GmAP1 | KO | Soy | Late |
| Zhao et al. A [193] | 2022 | GmPHYAs | KO | Soy | Late |
| Schmidt et al. [194] | 2020 | NtFT5 | KO | Tobacco | Non-flowering |
| Soyk et al. [195] | 2017 | SP5G | KO | Tomato | Early |
| Lemmon et al. [196] | 2018 | SP5G | KO | Tomato | Early |
| Li et al. [197] | 2018 | SP and SP5G | KO | Tomato | Early |
| Hu et al. [198] | 2022 | SlDOF9s | KO | Tomato | Early |
| Moreira et al. [199] | 2022 | SP3C | KO and OE | Tomato | Early, late |
| Xu et al. [200] | 2016 | S1BOP | KO | Tomato | No effect |
| Lin et al. [201] | 2021 | SlMIR172c, SlMIR172d | KO | Tomato | No report |
| Kwon et al. [202] | 2019 | SP5G, SP, SlER | KO | Tomato | Early |
| Gupta et al. [203] | 2022 | TaSPL13 | KO | Wheat | Early |
| Sun et al. [204] | 2023 | TaTFL1-5 | KO | Wheat | Early |
| Chen et al. [205] | 2022 | FT-D1 | KO | Wheat | Late |
| Errum et al. [206] | 2023 | TaPpd | KO | Wheat | Late |
| Huiyun Liu et al. [207] | 2020 | TaAQ and TaDq | KO | Wheat | No report |
5. Perspectives and Challenges of CRISPR-Mediated Flowering Time Engineering
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaudinier, A.; Blackman, B.K. Evolutionary processes from the perspective of flowering time diversity. New Phytol. 2020, 225, 1883–1898. [Google Scholar] [CrossRef]
- Anderson, J.T.; Willis, J.H.; Mitchell-Olds, T. Evolutionary genetics of plant adaptation. Trends Genet. 2011, 27, 258–266. [Google Scholar] [CrossRef]
- Thomson, J.D. Flowering phenology, fruiting success and progressive deterioration of pollination in an early-flowering geophyte. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 3187–3199. [Google Scholar] [CrossRef]
- Kehrberger, S.; Holzschuh, A. How does timing of flowering affect competition for pollinators, flower visitation and seed set in an early spring grassland plant? Sci. Rep. 2019, 9, 15593. [Google Scholar] [CrossRef]
- Osnato, M. Evolution of flowering time genes in rice: From the paleolithic to the anthropocene. Plant Cell Environ. 2023, 46, 1046–1059. [Google Scholar] [CrossRef]
- Cockram, J.; Jones, H.; Leigh, F.J.; O’Sullivan, D.; Powell, W.; Laurie, D.A.; Greenland, A.J. Control of flowering time in temperate cereals: Genes, domestication, and sustainable productivity. J. Exp. Bot. 2007, 58, 1231–1244. [Google Scholar] [CrossRef]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Wang, J.; Wu, F.; Zhu, S.; Xu, Y.; Cheng, Z.; Wang, J.; Li, C.; Sheng, P.; Zhang, H.; Cai, M.; et al. Overexpression of OsMYB1R1–VP64 fusion protein increases grain yield in rice by delaying flowering time. FEBS Lett. 2016, 590, 3385–3396. [Google Scholar] [CrossRef]
- Chen, C.; Huang, W.; Hou, K.; Wu, W. Bolting, an Important Process in Plant Development, Two Types in Plants. J. Plant Biol. 2019, 62, 161–169. [Google Scholar] [CrossRef]
- Choi, S.H.; Ahn, W.S.; Jie, E.Y.; Cho, H.-S.; Kim, S.W. Development of late-bolting plants by CRISPR/Cas9-mediated genome editing from mesophyll protoplasts of lettuce. Plant Cell Rep. 2022, 41, 1627–1630. [Google Scholar] [CrossRef]
- Tominaga, A.; Ito, A.; Sugiura, T.; Yamane, H. How Is Global Warming Affecting Fruit Tree Blooming? “Flowering (Dormancy) Disorder” in Japanese Pear (Pyrus pyrifolia) as a Case Study. Front. Plant Sci. 2021, 12, 787638. [Google Scholar] [CrossRef] [PubMed]
- Fadón, E.; Herrera, S.; Guerrero, B.I.; Guerra, M.E.; Rodrigo, J. Chilling and Heat Requirements of Temperate Stone Fruit Trees (Prunus sp.). Agronomy 2020, 10, 409. [Google Scholar] [CrossRef]
- Dong, X.; Jiang, X.; Kuang, G.; Wang, Q.; Zhong, M.; Jin, D.; Hu, J. Genetic control of flowering time in woody plants: Roses as an emerging model. Plant Divers 2017, 39, 104–110. [Google Scholar] [CrossRef]
- Haberman, A.; Bakhshian, O.; Cerezo-Medina, S.; Paltiel, J.; Adler, C.; Ben-Ari, G.; Mercado, J.A.; Pliego-Alfaro, F.; Lavee, S.; Samach, A. A possible role for flowering locus T-encoding genes in interpreting environmental and internal cues affecting olive (Olea europaea L.) flower induction. Plant Cell Environ. 2017, 40, 1263–1280. [Google Scholar] [CrossRef]
- Ahmar, S.; Gill, R.A.; Jung, K.-H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and Molecular Techniques from Simple Breeding to Speed Breeding in Crop Plants: Recent Advances and Future Outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef]
- Abdul Fiyaz, R.; Ajay, B.C.; Ramya, K.T.; Aravind Kumar, J.; Sundaram, R.M.; Subba Rao, L.V. Speed Breeding: Methods and Applications. In Accelerated Plant Breeding; Gosal, S., Wani, S., Eds.; Springer: Cham, Switzerland, 2020; Volume 1. [Google Scholar] [CrossRef]
- Rafferty, N.E.; Diez, J.M.; Bertelsen, C.D. Changing Climate Drives Divergent and Nonlinear Shifts in Flowering Phenology across Elevations. Curr. Biol. 2020, 30, 432–441.e3. [Google Scholar] [CrossRef]
- Anderson, J.T.; Inouye, D.W.; McKinney, A.M.; Colautti, R.I.; Mitchell-Olds, T. Phenotypic plasticity and adaptive evolution contribute to advancing flowering phenology in response to climate change. Proc. R. Soc. B Biol. Sci. 2012, 279, 3843–3852. [Google Scholar] [CrossRef]
- Siegmund, J.F.; Wiedermann, M.; Donges, J.F.; Donner, R.V. Impact of temperature and precipitation extremes on the flowering dates of four German wildlife shrub species. Biogeosciences 2016, 13, 5541–5555. [Google Scholar] [CrossRef]
- Mohandass, D.; Zhao, J.-L.; Xia, Y.-M.; Campbell, M.J.; Li, Q.-J. Increasing temperature causes flowering onset time changes of alpine ginger Roscoea in the Central Himalayas. J. Asia-Pac. Biodivers 2015, 8, 191–198. [Google Scholar] [CrossRef]
- Lambert, A.M.; Miller-Rushing, A.J.; Inouye, D.W. Changes in snowmelt date and summer precipitation affect the flowering phenology of Erythronium grandiflorum (glacier lily; Liliaceae). Am. J. Bot. 2010, 97, 1431–1437. [Google Scholar] [CrossRef]
- Menzel, A.; Sparks, T.H.; Estrella, N.; Koch, E.; Aasa, A.; Ahas, R.; Alm-Kübler, K.; Bissolli, P.; Braslavská, O.; Briede, A.; et al. European phenological response to climate change matches the warming pattern. Glob. Chang. Biol. 2006, 12, 1969–1976. [Google Scholar] [CrossRef]
- Craufurd, P.Q.; Wheeler, T.R. Climate change and the flowering time of annual crops. J. Exp. Bot. 2009, 60, 2529–2539. [Google Scholar] [CrossRef]
- Hu, Q.; Weiss, A.; Feng, S.; Baenziger, P.S. Earlier winter wheat heading dates and warmer spring in the U.S. Great Plains. Agric. For. Meteorol. 2005, 135, 284–290. [Google Scholar] [CrossRef]
- Drogoudi, P.; Kazantzis, K.; Blanke, M. Climate change effects on cherry flowering in northern Greece. Acta Hortic. 2017, 1162, 45–50. [Google Scholar] [CrossRef]
- Fitter, A.H.; Fitter, R.S.R. Rapid Changes in Flowering Time in British Plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef]
- Garner, W.W.; Allard, H.A. Effect of the relative length of day and night and other environmental factors on the growth and reproduction of plants. Mon. Wea. Rev. 1920, 48, 415. [Google Scholar] [CrossRef]
- Zeevaart, J.A. Florigen Coming of Age after 70 Years. Plant Cell 2006, 18, 1783–1789. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Böhlenius, H.; Eriksson, S.; Parcy, F.; Nilsson, O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 2005, 309, 1694–1696. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Yu, Z.; Chen, W.; Wang, Y.; Zhang, P.; Shi, N.; Hong, Y. Mobile Flowering Locus T RNA—Biological Relevance and Biotechnological Potential. Front. Plant Sci. 2022, 12, 792192. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, Y.; Money, T.; Bradley, D. A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 2005, 102, 7748–7753. [Google Scholar] [CrossRef] [PubMed]
- Corbesier, L.; Coupland, G. Photoperiodic flowering of Arabidopsis: Integrating genetic and physiological approaches to characterization of the floral stimulus. Plant Cell Environ. 2004, 28, 54–66. [Google Scholar] [CrossRef]
- Bouché, F.; Lobet, G.; Tocquin, P.; Périlleux, C. FLOR-ID: An interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 2016, 44, D1167–D1171. [Google Scholar] [CrossRef] [PubMed]
- Takagi, H.; Hempton, A.K.; Imaizumi, T. Photoperiodic flowering in Arabidopsis: Multilayered regulatory mechanisms of CONSTANS and the florigen FLOWERING LOCUS T. Plant Commun. 2023, 4, 100552. [Google Scholar] [CrossRef]
- Wenkel, S.; Turck, F.; Singer, K.; Gissot, L.; Le Gourrierec, J.; Samach, A.; Coupland, G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 2006, 18, 2971–2984. [Google Scholar] [CrossRef]
- Shim, J.S.; Kubota, A.; Imaizumi, T. Circadian Clock and Photoperiodic Flowering in Arabidopsis: CONSTANS Is a Hub for Signal Integration. Plant Physiol. 2017, 173, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef]
- Mockler, T.; Yang, H.; Yu, X.; Parikh, D.; Cheng, Y.-C.; Dolan, S.; Lin, C. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc. Natl. Acad. Sci. USA 2003, 100, 2140–2145. [Google Scholar] [CrossRef] [PubMed]
- Leivar, P.; Monte, E.; Al-Sady, B.; Carle, C.; Storer, A.; Alonso, J.M.; Ecker, J.R.; Quail, P.H. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 2008, 20, 337–352. [Google Scholar] [CrossRef]
- Sawa, M.; Kay, S.A. GIGANTEA directly activates Flowering Locus T in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2011, 108, 11698–11703. [Google Scholar] [CrossRef]
- Brandoli, C.; Petri, C.; Egea-Cortines, M.; Weiss, J. Gigantea: Uncovering New Functions in Flower Development. Genes 2020, 11, 1142. [Google Scholar] [CrossRef]
- Rival, P.; Press, M.O.; Bale, J.; Grancharova, T.; Undurraga, S.F.; Queitsch, C. The conserved PFT1 tandem repeat is crucial for proper flowering in Arabidopsis thaliana. Genetics 2014, 198, 747–754. [Google Scholar] [CrossRef]
- Steinbach, Y.; Hennig, L. Arabidopsis MSI1 functions in photoperiodic flowering time control. Front. Plant Sci. 2014, 5, 77. [Google Scholar] [CrossRef]
- Morris, K.; Jackson, S.P. DAY NEUTRAL FLOWERING does not act through GIGANTEA and FKF1 to regulate CONSTANS expression and flowering time. Plant Signal Behav. 2010, 5, 1105–1107. [Google Scholar] [CrossRef]
- Valverde, F. CONSTANS and the evolutionary origin of photoperiodic timing of flowering. J. Exp. Bot. 2011, 62, 2453–2463. [Google Scholar] [CrossRef]
- Ogawa, K.I.; Tasaka, Y.; Mino, M.; Tanaka, Y.; Iwabuchi, M. Association of Glutathione with Flowering in Arabidopsis thaliana. Plant Cell Physiol. 2001, 42, 524–530. [Google Scholar] [CrossRef]
- Kyung, J.; Jeon, M.; Lee, I. Recent advances in the chromatin-based mechanism of FLOWERING LOCUS C repression through autonomous pathway genes. Front. Plant Sci. 2022, 13, 964931. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.-Z.; Zhou, Y.-P.; Lv, T.-X.; Xie, C.-P.; Tian, C.-E. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol. Mol. Biol. Plants 2017, 23, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H. Current understanding of flowering pathways in plants: Focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants. Hortic. Environ. Biotechnol. 2020, 61, 209–227. [Google Scholar] [CrossRef]
- Duncan, S.; Holm, S.; Questa, J.; Irwin, J.; Grant, A.; Dean, C. Seasonal shift in timing of vernalization as an adaptation to extreme winter. eLife 2015, 4, e06620. [Google Scholar] [CrossRef]
- Surkova, S.Y.; Samsonova, M.G. Mechanisms of Vernalization-Induced Flowering in Legumes. Int. J. Mol. Sci. 2022, 23, 9889. [Google Scholar] [CrossRef]
- Luo, X.; He, Y. Experiencing winter for spring flowering: A molecular epigenetic perspective on vernalization. J. Integr. Plant Biol. 2020, 62, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Fjellheim, S. Flowering time runs hot and cold. Plant Physiol. 2022, 190, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Li, X.; Chen, W.; Xu, Z.; Chen, M.; Wang, H.; Yu, D. The emerging role of jasmonate in the control of flowering time. J. Exp. Bot. 2022, 73, 11–21. [Google Scholar] [CrossRef]
- Izawa, T. What is going on with the hormonal control of flowering in plants? Plant J. 2021, 105, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Davière, J.-M.; Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar] [CrossRef] [PubMed]
- Conti, L. Hormonal control of the floral transition: Can one catch them all? Dev. Biol. 2017, 430, 288–301. [Google Scholar] [CrossRef]
- Cho, L.-H.; Pasriga, R.; Yoon, J.; Jeon, J.-S.; An, G. Roles of Sugars in Controlling Flowering Time. J. Plant Biol. 2018, 61, 121–130. [Google Scholar] [CrossRef]
- Moghaddam, M.R.B.; Ende, W.V.D. Sugars, the clock and transition to flowering. Front. Plant Sci. 2013, 4, 22. [Google Scholar] [CrossRef]
- Roldán, M.; Gómez-Mena, C.; Ruiz-García, L.; Salinas, J.; Martínez-Zapater, J.M. Sucrose availability on the aerial part of the plant promotes morphogenesis and flowering of Arabidopsis in the dark. Plant J. 1999, 20, 581–590. [Google Scholar] [CrossRef]
- El-Lithy, M.E.; Reymond, M.; Stich, B.; Koornneef, M.; Vreugdenhil, D. Relation among plant growth, carbohydrates and flowering time in the Arabidopsis Landsberg erecta × Kondara recombinant inbred line population. Plant Cell Environ. 2010, 33, 1369–1382. [Google Scholar] [CrossRef] [PubMed]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.-A.; Onai, K.; Furukawa, Y.; Aoki, E.; Araki, T.; Nakamura, K. Effects of Sugar on Vegetative Development and Floral Transition in Arabidopsis. Plant Physiol. 2001, 127, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Kinoshita, A.; Kalluri, N.; Fernández, V.; Falavigna, V.S.; Cruz, T.M.D.; Jang, S.; Chiba, Y.; Seo, M.; Mettler-Altmann, T.; et al. The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual Effects of miR156-Targeted SPL Genes and CYP78A5/KLUH on Plastochron Length and Organ Size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef]
- Cui, L.; Shan, J.; Shi, M.; Gao, J.; Lin, H. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Shikata, M.; Koyama, T.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009, 50, 2133–2145. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.-R.; Bäurle, I. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cao, L.; Zhou, C.-M.; Zhang, T.-Q.; Lian, H.; Sun, Y.; Wu, J.; Huang, J.; Wang, G.; Wang, J.-W. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. eLife 2013, 2, e00269. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Franklin, K.A.; Lee, S.H.; Patel, D.; Kumar, S.V.; Spartz, A.K.; Gu, C.; Ye, S.; Yu, P.; Breen, G.; Cohen, J.D.; et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA 2011, 108, 20231–20235. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.-Y.; Wang, Z.-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Thines, B.C.; Youn, Y.; Duarte, M.I.; Harmon, F.G. The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J. Exp. Bot. 2014, 65, 1141–1151. [Google Scholar] [CrossRef]
- Bäurle, I.; Smith, L.; Baulcombe, D.C.; Dean, C. Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 2007, 318, 109–112. [Google Scholar] [CrossRef]
- Sonmez, C.; Bäurle, I.; Magusin, A.; Dreos, R.; Laubinger, S.; Weigel, D.; Dean, C. RNA 3′ processing functions of Arabidopsis FCA and FPA limit intergenic transcription. Proc. Natl. Acad. Sci. USA 2011, 108, 8508–8513. [Google Scholar] [CrossRef]
- Jung, J.-H.; Seo, P.J.; Ahn, J.H.; Park, C.-M. Arabidopsis RNA-binding protein FCA regulates microRNA172 processing in thermosensory flowering. J. Biol. Chem. 2012, 287, 16007–16016. [Google Scholar] [CrossRef]
- Lee, H.-J.; Jung, J.-H.; Llorca, L.C.; Kim, S.-G.; Lee, S.; Baldwin, I.T.; Park, C.-M. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nat. Commun. 2014, 5, 5473. [Google Scholar] [CrossRef]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015, 16, 31. [Google Scholar] [CrossRef] [PubMed]
- Andrés, F.; Porri, A.; Torti, S.; Mateos, J.; Romera-Branchat, M.; García-Martínez, J.L.; Fornara, F.; Gregis, V.; Kater, M.M.; Coupland, G. SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 2014, 111, E2760–E2769. [Google Scholar] [CrossRef]
- Capovilla, G.; Schmid, M.; Posé, D. Control of flowering by ambient temperature. J. Exp. Bot. 2015, 66, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of Flowering by the miR172 Target SMZ. PLoS Biol. 2009, 7, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Posé, D.; Verhage, L.; Ott, F.; Yant, L.; Mathieu, J.; Angenent, G.C.; Immink, R.G.H.; Schmid, M. Temperature-dependent regulation of flowering by antagonistic FLM variants. Nature 2013, 503, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, C.; Hou, X.; Xi, W.; Shen, L.; Tao, Z.; Wang, Y.; Yu, H. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012, 10, e1001313. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, L.; Shen, L.; Yu, H. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nat. Plants 2016, 2, 16075. [Google Scholar] [CrossRef]
- Tian, H.; Baxter, I.R.; Lahner, B.; Reinders, A.; Salt, D.E.; Ward, J.M. Arabidopsis NPCC6/NaKR1 Is a phloem mobile metal binding protein necessary for phloem function and root meristem maintenance. Plant Cell 2010, 22, 3963–3979. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, F.; Zhou, X.; Poh, T.X.; Xie, L.; Shen, J.; Yang, L.; Song, S.; Yu, H.; Chen, Y. The tetratricopeptide repeat protein OsTPR075 promotes heading by regulating florigen transport in rice. Plant Cell 2022, 34, 3632–3646. [Google Scholar] [CrossRef]
- Shin, Y.-H.; Lee, H.-M.; Park, Y.-D. CRISPR/Cas9-Mediated Editing of AGAMOUS-like Genes Results in a Late-Bolting Phenotype in Chinese Cabbage (Brassica rapa ssp. pekinensis). Int. J. Mol. Sci. 2022, 23, 15009. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Ahn, H.; Ryu, J.; Oh, Y.; Sivanandhan, G.; Won, K.-H.; Park, Y.D.; Kim, J.-S.; Kim, H.; Lim, Y.P.; et al. Generation of early-flowering Chinese cabbage (Brassica rapa spp. pekinensis) through CRISPR/Cas9-mediated genome editing. Plant Biotechnol. Rep. 2019, 13, 491–499. [Google Scholar] [CrossRef]
- Duplat-Bermúdez, L.; Ruiz-Medrano, R.; Landsman, D.; Mariño-Ramírez, L.; Xoconostle-Cázares, B. Transcriptomic analysis of Arabidopsis overexpressing flowering locus T driven by a meristem-specific promoter that induces early flowering. Gene 2016, 587, 120–131. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef]
- Auge, G.A.; Penfield, S.; Donohue, K. Pleiotropy in developmental regulation by flowering-pathway genes: Is it an evolutionary constraint? New Phytol. 2019, 224, 55–70. [Google Scholar] [CrossRef]
- Li, C.; Fu, Q.; Niu, L.; Luo, L.; Chen, J.; Xu, Z.-F. Three TFL1 homologues regulate floral initiation in the biofuel plant Jatropha curcas. Sci. Rep. 2017, 7, srep43090. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, W.; Wang, L.; Lin, C.; Cong, B.; Sun, C.; Luo, D. TFL1/CEN-like genes control intercalary meristem activity and phase transition in rice. Plant Sci. 2005, 168, 1393–1408. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, Y.; Wu, Z.; Bu, X.; Fan, M.; Zhang, Q. Characterization of TEMINAL FLOWER1 homologs CmTFL1c gene from Chrysanthemum morifolium. Plant Mol. Biol. 2019, 99, 587–601. [Google Scholar] [CrossRef]
- Sriboon, S.; Li, H.; Guo, C.; Senkhamwong, T.; Dai, C.; Liu, K. Knock-out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus. BMC Genet. 2020, 21, 52. [Google Scholar] [CrossRef] [PubMed]
- Tapia-López, R.; García-Ponce, B.; Dubrovsky, J.G.; Garay-Arroyo, A.; Pérez-Ruíz, R.V.; Kim, S.-H.; Acevedo, F.; Pelaz, S.; Alvarez-Buylla, E.R. An AGAMOUS-Related MADS-Box Gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol. 2008, 146, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Novillo, F.; Medina, J.; Salinas, J. Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 2007, 104, 21002–21007. [Google Scholar] [CrossRef] [PubMed]
- Gachomo, E.W.; Baptiste, L.J.; Kefela, T.; Saidel, W.M.; Kotchoni, S.O. The Arabidopsis CURVY1 (CVY1) gene encoding a novel receptor-like protein kinase regulates cell morphogenesis, flowering time and seed production. BMC Plant Biol. 2014, 14, 221. [Google Scholar] [CrossRef]
- Schönrock, N.; Bouveret, R.; Leroy, O.; Borghi, L.; Köhler, C.; Gruissem, W.; Hennig, L. Polycomb-group proteins repressthe floral activator AGL19 in the FLC-independent vernalization pathway. Minerva Anestesiol. 2006, 20, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Long, Y.; Raman, H.; Zou, X.; Wang, J.; Dai, S.; Xiao, Q.; Li, C.; Fan, L.; Liu, B.; et al. A Tourist-like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant Biol. 2012, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, Z.; Li, W.; Ku, L.; Wang, C.; Ye, J.; Li, K.; Yang, N.; Li, Y.; Zhong, T.; et al. CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2013, 110, 16969–16974. [Google Scholar] [CrossRef] [PubMed]
- Salvi, S.; Sponza, G.; Morgante, M.; Tomes, D.; Niu, X.; Fengler, K.A.; Meeley, R.; Ananiev, E.V.; Svitashev, S.; Bruggemann, E.; et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 2007, 104, 11376–11381. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Papikian, A.; Liu, W.; Gallego-Bartolomé, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 729. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Neumann, M.; Duro, D.I.; Schmid, M. CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS ONE 2019, 14, e0222778. [Google Scholar] [CrossRef] [PubMed]
- de la Torre, N.V.; Vayssières, A.; Obeng-Hinneh, E.; Neumann, U.; Zhou, Y.; Lázaro, A.; Roggen, A.; Sun, H.; Stolze, S.C.; Nakagami, H.; et al. FLOWERING REPRESSOR AAA+ ATPase 1 is a novel regulator of perennial flowering in Arabis alpina. New Phytol. 2022, 236, 729–744. [Google Scholar] [CrossRef]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient Targeted Mutagenesis in Apple and First Time Edition of Pear Using the CRISPR-Cas9 System. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Y.; Gao, Y.; Zhang, Q. Targeted deletion of floral development genes in Arabidopsis with CRISPR/Cas9 using the RNA endoribonuclease Csy4 processing system. Hortic. Res. 2019, 6, 99. [Google Scholar] [CrossRef]
- Ning, Y.-Q.; Ma, Z.-Y.; Huang, H.-W.; Mo, H.; Zhao, T.-T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X.-J. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 2015, 43, 1469–1484. [Google Scholar] [CrossRef]
- Nobusawa, T.; Yamatani, H.; Kusaba, M. Early flowering phenotype of the Arabidopsis altered meristem program1 mutant is dependent on the FLOWERING LOCUS T-mediated pathway. Plant Biotechnol. 2022, 39, 317–321. [Google Scholar] [CrossRef]
- Capovilla, G.; Symeonidi, E.; Wu, R.; Schmid, M. Contribution of major FLM isoforms to temperature-dependent flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5117–5127. [Google Scholar] [CrossRef]
- Romera-Branchat, M.; Severing, E.; Pocard, C.; Ohr, H.; Vincent, C.; Née, G.; Martinez-Gallegos, R.; Jang, S.; Andrés, F.; Madrigal, P.; et al. Functional Divergence of the Arabidopsis Florigen-Interacting bZIP Transcription Factors FD and FDP. Cell Rep. 2020, 31, 107717. [Google Scholar] [CrossRef]
- Lian, H.; Wang, L.; Ma, N.; Zhou, C.-M.; Han, L.; Zhang, T.-Q.; Wang, J.-W. Redundant and specific roles of individual MIR172 genes in plant development. PLoS Biol. 2021, 19, e3001044. [Google Scholar] [CrossRef]
- Yan, Z.; Jia, J.; Yan, X.; Shi, H.; Han, Y. Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol. Biol. 2017, 95, 549–565. [Google Scholar] [CrossRef]
- Hyun, Y.; Kim, J.; Cho, S.W.; Choi, Y.; Kim, J.-S.; Coupland, G. Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 2015, 241, 271–284. [Google Scholar] [CrossRef]
- Hou, N.; Cao, Y.; Li, F.; Yuan, W.; Bian, H.; Wang, J.; Zhu, M.; Han, N. Epigenetic regulation of miR396 expression by SWR1-C and the effect of miR396 on leaf growth and developmental phase transition in Arabidopsis. J. Exp. Bot. 2019, 70, 5217–5229. [Google Scholar] [CrossRef]
- Yao, X.; Chen, J.; Zhou, J.; Yu, H.; Ge, C.; Zhang, M.; Gao, X.; Dai, X.; Yang, Z.-N.; Zhao, Y. An Essential Role for miRNA167 in Maternal Control of Embryonic and Seed Development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef]
- Huang, Y.; Jiao, Y.; Xie, N.; Guo, Y.; Zhang, F.; Xiang, Z.; Wang, R.; Wang, F.; Gao, Q.; Tian, L.; et al. OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019, 287, 110188. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, D.; Scharf, K.; Frank, W.; Leister, D.; Kleine, T. The RNA-binding protein RBP45D of Arabidopsis promotes transgene silencing and flowering time. Plant J. 2022, 109, 1397–1415. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Dent, C.; Liang, H.; Lv, J.; Shang, G.; Liu, Y.; Feng, F.; Wang, F.; Pang, J.; Li, X.; et al. CRY2 interacts with CIS1 to regulate thermosensory flowering via FLM alternative splicing. Nat. Commun. 2022, 13, 7045. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhang, P.; Guo, D.; Wang, N.; Lin, H.; Wang, X.; Niu, L. Transcriptional repressor AGL79 positively regulates flowering time in Arabidopsis. J. Plant Physiol. 2023, 285, 153985. [Google Scholar] [CrossRef] [PubMed]
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 2016, 17, 1276–1288. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Soto, D.; Allona, I.; Perales, M. FLOWERING LOCUS T2 Promotes Shoot Apex Development and Restricts Internode Elongation via the 13-Hydroxylation Gibberellin Biosynthesis Pathway in Poplar. Front. Plant Sci. 2022, 12, 814195. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Bai, Y.; Muhammad, S.; Wu, X.; Deng, P.; Wu, J.; An, H.; Wu, L. Divergent roles of FT-like 9 in flowering transition under different day lengths in Brachypodium distachyon. Nat. Commun. 2019, 10, 812. [Google Scholar] [CrossRef]
- Jung, H.; Lee, A.; Jo, S.H.; Park, H.J.; Jung, W.Y.; Kim, H.-S.; Lee, H.-J.; Jeong, S.-G.; Kim, Y.-S.; Cho, H.S. Nitrogen Signaling Genes and SOC1 Determine the Flowering Time in a Reciprocal Negative Feedback Loop in Chinese Cabbage (Brassica rapa L.) Based on CRISPR/Cas9-Mediated Mutagenesis of Multiple BrSOC1 Homologs. Int. J. Mol. Sci. 2021, 22, 4631. [Google Scholar] [CrossRef]
- Hong, J.K.; Suh, E.J.; Park, S.R.; Park, J.; Lee, Y.-H. Multiplex CRISPR/Cas9 Mutagenesis of BrVRN1 Delays Flowering Time in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Agriculture 2021, 11, 1286. [Google Scholar] [CrossRef]
- Shin, Y.-H.; Park, Y.-D. CRISPR/Cas9-Mediated Mutagenesis of BrLEAFY Delays the Bolting Time in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Int. J. Mol. Sci. 2022, 24, 541. [Google Scholar] [CrossRef]
- Lee, A.; Jung, H.; Park, H.J.; Jo, S.H.; Jung, M.; Kim, Y.-S.; Cho, H.S. Their C-termini divide Brassica rapa FT-like proteins into FD-interacting and FD-independent proteins that have different effects on the floral transition. Front. Plant Sci. 2023, 13, 1091563. [Google Scholar] [CrossRef]
- Park, S.-C.; Park, S.; Jeong, Y.J.; Lee, S.B.; Pyun, J.W.; Kim, S.; Kim, T.H.; Kim, S.W.; Jeong, J.C.; Kim, C.Y. DNA-free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol. Rep. 2019, 13, 483–489. [Google Scholar] [CrossRef]
- Bellec, Y.; Guyon-Debast, A.; François, T.; Gissot, L.; Biot, E.; Nogué, F.; Faure, J.-D.; Tepfer, M. New Flowering and Architecture Traits Mediated by Multiplex CRISPR-Cas9 Gene Editing in Hexaploid Camelina sativa. Agronomy 2022, 12, 1873. [Google Scholar] [CrossRef]
- Jiang, L.; Li, D.; Jin, L.; Ruan, Y.; Shen, W.-H.; Liu, C. Histone lysine methyltransferases BnaSDG8.A and BnaSDG8.C are involved in the floral transition in Brassica napus. Plant J. 2018, 95, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zeng, L.; Chen, H.; Ma, C.; Tu, J.; Shen, J.; Wen, J.; Fu, T.; Yi, B. CRISPR/Cas9-Mediated Targeted Mutagenesis of BnaCOL9 Advances the Flowering Time of Brassica napus L. Int. J. Mol. Sci. 2022, 23, 14944. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Zhai, Y.; Huang, H.; Yu, K.; Khan, M.H.U.; Shahid, M.; Samad, R.A.; Khan, S.U.; Amoo, O.; Fan, C.; et al. Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus L. Crop J. 2022, 10, 67–74. [Google Scholar] [CrossRef]
- Zhou, E.; Zhang, Y.; Wang, H.; Jia, Z.; Wang, X.; Wen, J.; Shen, J.; Fu, T.; Yi, B. Identification and Characterization of the MIKC-Type MADS-Box Gene Family in Brassica napus and Its Role in Floral Transition. Int. J. Mol. Sci. 2022, 23, 4289. [Google Scholar] [CrossRef]
- Odipio, J.; Alicai, T.; Nusinow, D.; Bart, R.; Taylor, N. CRISPR/Cas9-mediated disruption of multiple TFL1-like floral repressors activates flowering in cassava. In Vitro Cellular & Developmental Biology-Animal; Springer: New York, NY, USA, 2018; Volume 54, p. S47. [Google Scholar]
- Bull, S.E.; Seung, D.; Chanez, C.; Mehta, D.; Kuon, J.-E.; Truernit, E.; Hochmuth, A.; Zurkirchen, I.; Zeeman, S.C.; Gruissem, W.; et al. Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Sci. Adv. 2018, 4, eaat6086. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xue, Y.; Luo, J.; Han, M.; Liu, X.; Jiang, T.; Zhao, Y.; Xu, Y.; Ma, C. Developing a UV–visible reporter-assisted CRISPR/Cas9 gene editing system to alter flowering time in Chrysanthemum indicum. Plant Biotechnol. J. 2023, 21, 1519–1521. [Google Scholar] [CrossRef]
- Huang, C.; Sun, H.; Xu, D.; Chen, Q.; Liang, Y.; Wang, X.; Xu, G.; Tian, J.; Wang, C.; Li, D.; et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 2018, 115, E334–E341. [Google Scholar] [CrossRef]
- Li, Q.; Wu, G.; Zhao, Y.; Wang, B.; Zhao, B.; Kong, D.; Wei, H.; Chen, C.; Wang, H. CRISPR/Cas9-mediated knockout and overexpression studies reveal a role of maize phytochrome C in regulating flowering time and plant height. Plant Biotechnol. J. 2020, 18, 2520–2532. [Google Scholar] [CrossRef]
- Takahashi, H.; Nishihara, M.; Yoshida, C.; Itoh, K. Gentian FLOWERING LOCUS T orthologs regulate phase transitions: Floral induction and endodormancy release. Plant Physiol. 2022, 188, 1887–1899. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Scheible, W.-R.; Lundquist, P.K. A stress-inducible protein regulates drought tolerance and flowering time in Brachypodium and Arabidopsis. Plant Physiol. 2022, 191, 643–659. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Jiang, Y.; Rahmati, M.; Chia, J.-C.; Dokuchayeva, T.; Kavulych, Y.; Zavodna, T.-O.; Mendoza, P.N.; Huang, R.; Smieshka, L.M.; et al. YSL3-mediated copper distribution is required for fertility, seed size and protein accumulation in Brachypodium. Plant Physiol. 2021, 186, 655–676. [Google Scholar] [CrossRef] [PubMed]
- Herath, D.; Voogd, C.; Mayo-Smith, M.; Yang, B.; Allan, A.C.; Putterill, J.; Varkonyi-Gasic, E. CRISPR-Cas9 -mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype. Plant Biotechnol. J. 2022, 20, 2064–2076. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wang, T.; Voogd, C.; Jeon, S.; Drummond, R.S.M.; Gleave, A.P.; Allan, A.C. Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol. J. 2019, 17, 869–880. [Google Scholar] [CrossRef]
- Varkonyi-Gasic, E.; Wang, T.; Cooney, J.; Jeon, S.; Voogd, C.; Douglas, M.J.; Pilkington, S.M.; Akagi, T.; Allan, A.C. Shy Girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signalling. New Phytol. 2021, 230, 1461–1475. [Google Scholar] [CrossRef]
- Singer, S.D.; Hughes, K.B.; Subedi, U.; Dhariwal, G.K.; Kader, K.; Acharya, S.; Chen, G.; Hannoufa, A. The CRISPR/Cas9-Mediated Modulation of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 8 in Alfalfa Leads to Distinct Phenotypic Outcomes. Front. Plant Sci. 2022, 12, 774146. [Google Scholar] [CrossRef]
- Galindo-Sotomonte, L.; Jozefkowicz, C.; Gómez, C.; Stritzler, M.; Frare, R.; Bottero, E.; Tajima, H.; Blumwald, E.; Ayub, N.; Soto, G. CRISPR/Cas9-mediated knockout of a polyester synthase-like gene delays flowering time in alfalfa. Plant Cell Rep. 2023, 42, 953–956. [Google Scholar] [CrossRef]
- Wolabu, T.W.; Mahmood, K.; Jerez, I.T.; Cong, L.; Yun, J.; Udvardi, M.; Tadege, M.; Wang, Z.; Wen, J. Multiplex CRISPR/Cas9-mediated mutagenesis of alfalfa FLOWERING LOCUS Ta1 (MsFTa1) leads to delayed flowering time with improved forage biomass yield and quality. Plant Biotechnol. J. 2023, 21, 1383–1392. [Google Scholar] [CrossRef]
- Shibuya, K.; Watanabe, K.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. Biochem. 2018, 131, 53–57. [Google Scholar] [CrossRef]
- André, D.; Marcon, A.; Lee, K.C.; Goretti, D.; Zhang, B.; Delhomme, N.; Schmid, M.; Nilsson, O. FLOWERING LOCUS T paralogs control the annual growth cycle in Populus trees. Curr. Biol. 2022, 32, 2988–2996.e4. [Google Scholar] [CrossRef] [PubMed]
- Elorriaga, E.; Klocko, A.L.; Ma, C.; Strauss, S.H. Variation in Mutation Spectra Among CRISPR/Cas9 Mutagenized Poplars. Front. Plant Sci. 2018, 9, 594. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.; Komakhin, R.; Konovalova, L.; Ivanova, L.; Taranov, V.; Monakhova, Y.; Babakov, A.; Klepikova, A.; Zlobin, N. Development of potato (Solanum tuberosum L.) plants with StLEAFY knockout. Planta 2022, 256, 116. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, W.; Ren, Y.; Tian, X.; Lv, T.; Wang, Z.; Fang, J.; Chu, C.; Yang, J.; Bu, Q. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genom. 2017, 44, 175–178. [Google Scholar] [CrossRef]
- Brambilla, V.; Martignago, D.; Goretti, D.; Cerise, M.; Somssich, M.; de Rosa, M.; Galbiati, F.; Shrestha, R.; Lazzaro, F.; Simon, R.; et al. Antagonistic Transcription Factor Complexes Modulate the Floral Transition in Rice. Plant Cell 2017, 29, 2801–2816. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, G.; Yang, L.; Qiu, L.; He, P.; Nong, C.; Wang, Y.; He, Y.; Xing, Y. Gene diagnosis and targeted breeding for blast-resistant Kongyu 131 without changing regional adaptability. J. Genet. Genom. 2018, 45, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhu, M.; Xu, Z.; Xu, Q. Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor. Appl. Genet. 2019, 132, 1887–1896. [Google Scholar] [CrossRef]
- Wang, G.; Wang, C.; Lu, G.; Wang, W.; Mao, G.; Habben, J.E.; Song, C.; Wang, J.; Chen, J.; Gao, Y.; et al. Knockouts of a late flowering gene via CRISPR–Cas9 confer early maturity in rice at multiple field locations. Plant Mol. Biol. 2020, 104, 137–150. [Google Scholar] [CrossRef]
- Chandrashekar, B.K.; Ag, S.; Makarla, U.; Ramu, V.S. Disruption in the DNA Mismatch Repair Gene MSH2 by CRISPR-Cas9 in Indica Rice Can Create Genetic Variability. J. Agric. Food Chem. 2021, 69, 4144–4152. [Google Scholar] [CrossRef]
- Andrew-Peter-Leon, M.T.; Selvaraj, R.; Kumar, K.K.; Muthamilarasan, M.; Yasin, J.K.; Pillai, M.A. Loss of Function of OsFBX267 and OsGA20ox2 in Rice Promotes Early Maturing and Semi-Dwarfism in γ-Irradiated IWP and Genome-Edited Pusa Basmati-1. Front. Plant Sci. 2021, 12, 714066. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Xue, P.; Wen, X.-X.; Gong, K.; Wang, B.-F.; Xu, P.; Lin, Z.-C.; Peng, Z.-Q.; Fu, J.-L.; Yu, P.; et al. qHD5 encodes an AP2 factor that suppresses rice heading by down-regulating Ehd2 expression. Plant Sci. 2022, 324, 111446. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Yan, Z.; Guan, J.; Huo, Y.; Wang, T.; Li, T.; Cui, Z.; Ma, W.; Wang, X.; Chen, W. Two interacting basic helix-loop-helix transcription factors control flowering time in rice. Plant Physiol. 2023, 192, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Sedeek, K.; Zuccolo, A.; Fornasiero, A.; Weber, A.M.; Sanikommu, K.; Sampathkumar, S.; Rivera, L.F.; Butt, H.; Mussurova, S.; Alhabsi, A.; et al. Multi-omics resources for targeted agronomic improvement of pigmented rice. Nat. Food 2023, 4, 366–371. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Zhang, K.; Chen, J.; Gull, S.; Chen, C.; Hou, Y.; Li, X.; Miao, J.; Zhou, Y.; Liang, G. OsFTL4, an FT-like Gene, Regulates Flowering Time and Drought Tolerance in Rice (Oryza sativa L.). Rice 2022, 15, 47. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, K.; Zhou, Y.; Xiang, L.; He, C.; Zhong, C.; Li, K.; Wang, Q.; Yang, C.; Wang, Q.; et al. Dual function of clock component OsLHY sets critical day length for photoperiodic flowering in rice. Plant Biotechnol. J. 2021, 19, 1644–1657. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, X.; Hu, Y.; Zhou, X.; He, Q.; Liang, L.; Xing, Y. Global analysis of CCT family knockout mutants identifies four genes involved in regulating heading date in rice. J. Integr. Plant Biol. 2021, 63, 913–923. [Google Scholar] [CrossRef]
- Cui, Y.; Xu, Z.; Xu, Q. Elucidation of the relationship between yield and heading date using CRISPR/Cas9 system-induced mutation in the flowering pathway across a large latitudinal gradient. Mol. Breed. 2021, 41, 23. [Google Scholar] [CrossRef]
- Yasui, Y.; Tanaka, W.; Sakamoto, T.; Kurata, T.; Hirano, H.-Y. Genetic Enhancer Analysis Reveals that FLORAL ORGAN NUMBER2 and OsMADS3 Co-operatively Regulate Maintenance and Determinacy of the Flower Meristem in Rice. Plant Cell Physiol. 2017, 58, 893–903. [Google Scholar] [CrossRef]
- Wu, M.; Liu, H.; Lin, Y.; Chen, J.; Fu, Y.; Luo, J.; Zhang, Z.; Liang, K.; Chen, S.; Wang, F. In-Frame and Frame-Shift Editing of the Ehd1 Gene to Develop Japonica Rice with Prolonged Basic Vegetative Growth Periods. Front. Plant Sci. 2020, 11, 307. [Google Scholar] [CrossRef]
- Li, C.; Liu, X.-J.; Yan, Y.; Shah, A.; Liu, Z.; Tao, R.-F.; Yue, E.-K.; Duan, M.-H.; Xu, J. OsLHY regulates photoperiodic flowering through the unique pathways under long-day conditions in rice (Oryza sativa L.). Authorea 2021. [Google Scholar] [CrossRef]
- Liu, X.; Liu, H.; Zhang, Y.; He, M.; Li, R.; Meng, W.; Wang, Z.; Li, X.; Bu, Q. Fine-tuning Flowering Time via Genome Editing of Upstream Open Reading Frames of Heading Date 2 in Rice. Rice 2021, 14, 69. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Deng, L.; Cheng, R.; Hu, J.; Wu, C. RID1 sets rice heading date by balancing its binding with SLR1 and SDG722. J. Integr. Plant Biol. 2022, 64, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhang, Y.; Wen, X.; Yang, Q.; Liu, L.; Hao, S.; Li, J.; Wu, Z.; Shah, L.; Sohail, A.; et al. The clock component OsLUX regulates rice heading through recruiting OsELF3-1 and OsELF4s to repress Hd1 and Ghd7. J. Adv. Res. 2023, 48, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Deng, L.; Zhao, L.; Wu, C. Genome-wide binding analysis of transcription factor Rice Indeterminate 1 reveals a complex network controlling rice floral transition. J. Integr. Plant Biol. 2022, 64, 1690–1705. [Google Scholar] [CrossRef]
- Andrade, L.; Lu, Y.; Cordeiro, A.; Costa, J.M.F.; Wigge, P.A.; Saibo, N.J.M.; Jaeger, K.E. The evening complex integrates photoperiod signals to control flowering in rice. Proc. Natl. Acad. Sci. USA 2022, 119, e2122582119. [Google Scholar] [CrossRef]
- Dai, X.; Yang, X.; Wang, C.; Fan, Y.; Xin, S.; Hua, Y.; Wang, K.; Huang, H. CRISPR/Cas9-mediated genome editing in Hevea brasiliensis. Ind. Crops Prod. 2021, 164, 113418. [Google Scholar] [CrossRef]
- Wang, H.; Jia, G.; Zhang, N.; Zhi, H.; Xing, L.; Zhang, H.; Sui, Y.; Tang, S.; Li, M.; Zhang, H.; et al. Domestication-associated PHYTOCHROME C is a flowering time repressor and a key factor determining Setaria as a short-day plant. New Phytol. 2022, 236, 1809–1823. [Google Scholar] [CrossRef]
- Zhu, C.; Box, M.S.; Thiruppathi, D.; Hu, H.; Yu, Y.; Martin, C.; Doust, A.N.; McSteen, P.; Kellogg, E.A. Pleiotropic and nonredundant effects of an auxin importer in Setaria and maize. Plant Physiol. 2022, 189, 715–734. [Google Scholar] [CrossRef]
- Char, S.N.; Wei, J.; Mu, Q.; Li, X.; Zhang, Z.J.; Yu, J.; Yang, B. An Agrobacterium-delivered CRISPR/Cas9 system for targeted mutagenesis in sorghum. Plant Biotechnol. J. 2020, 18, 319–321. [Google Scholar] [CrossRef]
- Han, J.; Guo, B.; Guo, Y.; Zhang, B.; Wang, X.; Qiu, L.-J. Creation of Early Flowering Germplasm of Soybean by CRISPR/Cas9 Technology. Front. Plant Sci. 2019, 10, 1446. [Google Scholar] [CrossRef]
- Wang, L.; Sun, S.; Wu, T.; Liu, L.; Sun, X.; Cai, Y.; Li, J.; Jia, H.; Yuan, S.; Chen, L.; et al. Natural variation and CRISPR/Cas9-mediated mutation in GmPRR37 affect photoperiodic flowering and contribute to regional adaptation of soybean. Plant Biotechnol. J. 2020, 18, 1869–1881. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Z.; Hou, W.; Chen, L.; Jiang, B.; Liu, W.; Feng, Y.; Wu, C. GmNMHC5, A Neoteric Positive Transcription Factor of Flowering and Maturity in Soybean. Plants 2020, 9, 792. [Google Scholar] [CrossRef]
- Li, Z.; Cheng, Q.; Gan, Z.; Hou, Z.; Zhang, Y.; Li, Y.; Li, H.; Nan, H.; Yang, C.; Chen, L.; et al. Multiplex CRISPR/Cas9-mediated knockout of soybean LNK2 advances flowering time. Crop J. 2021, 9, 767–776. [Google Scholar] [CrossRef]
- Wan, Z.; Liu, Y.; Guo, D.; Fan, R.; Liu, Y.; Xu, K.; Zhu, J.; Quan, L.; Lu, W.; Bai, X.; et al. CRISPR/Cas9-mediated targeted mutation of the E1 decreases photoperiod sensitivity, alters stem growth habits, and decreases branch number in soybean. Front. Plant Sci. 2022, 13, 1066820. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Wan, Z.; Jiao, S.; Zhou, J.; Xu, K.; Nan, H.; Liu, Y.; Xiong, S.; Fan, R.; Zhu, J.; et al. GmMDE genes bridge the maturity gene E1 and florigens in photoperiodic regulation of flowering in soybean. Plant Physiol. 2022, 189, 1021–1036. [Google Scholar] [CrossRef]
- Cai, Y.; Chen, L.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 2018, 16, 176–185. [Google Scholar] [CrossRef]
- Wang, Y.; Yuan, L.; Su, T.; Wang, Q.; Gao, Y.; Zhang, S.; Jia, Q.; Yu, G.; Fu, Y.; Cheng, Q.; et al. Light- and temperature-entrainable circadian clock in soybean development. Plant Cell Environ. 2020, 43, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.-H.; Li, Y.; Lu, H.; Hong, H.; Tian, Y.; Li, H.; Zhao, T.; Zhou, X.; Liu, J.; et al. A Domestication-Associated Gene GmPRR3b Regulates the Circadian Clock and Flowering Time in Soybean. Mol. Plant 2020, 13, 745–759. [Google Scholar] [CrossRef]
- Chen, L.; Nan, H.; Kong, L.; Yue, L.; Yang, H.; Zhao, Q.; Fang, C.; Li, H.; Cheng, Q.; Lu, S.; et al. Soybean AP1 homologs control flowering time and plant height. J. Integr. Plant Biol. 2020, 62, 1868–1879. [Google Scholar] [CrossRef]
- Zhao, F.; Lyu, X.; Ji, R.; Liu, J.; Zhao, T.; Li, H.; Liu, B.; Pei, Y. CRISPR/Cas9-engineered mutation to identify the roles of phytochromes in regulating photomorphogenesis and flowering time in soybean. Crop J. 2022, 10, 1654–1664. [Google Scholar] [CrossRef]
- Schmidt, F.J.; Zimmermann, M.M.; Wiedmann, D.R.; Lichtenauer, S.; Grundmann, L.; Muth, J.; Twyman, R.M.; Prüfer, D.; Noll, G.A. The Major Floral Promoter NtFT5 in Tobacco (Nicotiana tabacum) Is a Promising Target for Crop Improvement. Front. Plant Sci. 2020, 10, 1666. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Wang, K.; Huang, B.; Mila, I.; Frasse, P.; Maza, E.; Djari, A.; Hernould, M.; Zouine, M.; Li, Z.; et al. The auxin-responsive transcription factor SlDOF9 regulates inflorescence and flower development in tomato. Nat. Plants 2022, 8, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Moreira, J.d.R.; Quiñones, A.; Lira, B.S.; Robledo, J.M.; Curtin, S.J.; Vicente, M.H.; Ribeiro, D.M.; Ryngajllo, M.; Jiménez-Gómez, J.M.; Peres, L.E.P.; et al. SELF PRUNING 3C is a flowering repressor that modulates seed germination, root architecture, and drought responses. J. Exp. Bot. 2022, 73, 6226–6240. [Google Scholar] [CrossRef]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Minerva Anestesiol. 2016, 30, 2048–2061. [Google Scholar] [CrossRef]
- Lin, W.; Gupta, S.K.; Arazi, T.; Spitzer-Rimon, B. MIR172d Is Required for Floral Organ Identity and Number in Tomato. Int. J. Mol. Sci. 2021, 22, 4659. [Google Scholar] [CrossRef]
- Kwon, C.-T.; Heo, J.; Lemmon, Z.H.; Capua, Y.; Hutton, S.F.; Van Eck, J.; Park, S.J.; Lippman, Z.B. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 2020, 38, 182–188. [Google Scholar] [CrossRef]
- Gupta, A.; Hua, L.; Zhang, Z.; Yang, B.; Li, W. CRISPR-induced miRNA156-recognition element mutations in TaSPL13 improve multiple agronomic traits in wheat. Plant Biotechnol. J. 2023, 21, 536–548. [Google Scholar] [CrossRef]
- Sun, J.; Bie, X.M.; Chu, X.L.; Wang, N.; Zhang, X.S.; Gao, X.-Q. Genome-edited TaTFL1-5 mutation decreases tiller and spikelet numbers in common wheat. Front. Plant Sci. 2023, 14, 1142779. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ke, W.; He, F.; Chai, L.; Cheng, X.; Xu, H.; Wang, X.; Du, D.; Zhao, Y.; Chen, X.; et al. A single nucleotide deletion in the third exon of FT-D1 increases the spikelet number and delays heading date in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2022, 20, 920–933. [Google Scholar] [CrossRef] [PubMed]
- Errum, A.; Rehman, N.; Uzair, M.; Inam, S.; Ali, G.M.; Khan, M.R. CRISPR/Cas9 editing of wheat Ppd-1 gene homoeologs alters spike architecture and grain morphometric traits. Funct. Integr. Genom. 2023, 23, 66. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Tang, H.; Gong, Q.; Du, L.; Pei, X.; Ye, X. CRISPR/Cas9 editing of wheat TaQ genes alters spike morphogenesis and grain threshability. J. Genet. Genom. 2020, 47, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Anand, A.; Bass, S.H.; Wu, E.; Wang, N.; McBride, K.E.; Annaluru, N.; Miller, M.; Hua, M.; Jones, T.J. An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol. Biol. 2018, 97, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, K.; Finley, T.; Chappell, H.; Veena, V.; Wang, K. An Improved Agrobacterium-Mediated Transformation and Genome-Editing Method for Maize Inbred B104 Using a Ternary Vector System and Immature Embryos. Front. Plant Sci. 2022, 13, 860971. [Google Scholar] [CrossRef]
- Uranga, M.; Daròs, J. Tools and targets: The dual role of plant viruses in CRISPR–Cas genome editing. Plant Genome 2023, 16, e20220. [Google Scholar] [CrossRef]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef]
- Lee, K.; Wang, K. Strategies for genotype-flexible plant transformation. Curr. Opin. Biotechnol. 2023, 79, 102848. [Google Scholar] [CrossRef]
- Impens, L.; Lorenzo, C.D.; Vandeputte, W.; Wytynck, P.; Debray, K.; Haeghebaert, J.; Herwegh, D.; Jacobs, T.B.; Ruttink, T.; Nelissen, H.; et al. Combining multiplex gene editing and doubled haploid technology in maize. New Phytol. 2023, 239, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Gaillochet, C.; Develtere, W.; Jacobs, T.B. CRISPR screens in plants: Approaches, guidelines, and future prospects. Plant Cell 2021, 33, 794–813. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hodaei, A.; Werbrouck, S.P.O. Unlocking Nature’s Clock: CRISPR Technology in Flowering Time Engineering. Plants 2023, 12, 4020. https://doi.org/10.3390/plants12234020
Hodaei A, Werbrouck SPO. Unlocking Nature’s Clock: CRISPR Technology in Flowering Time Engineering. Plants. 2023; 12(23):4020. https://doi.org/10.3390/plants12234020
Chicago/Turabian StyleHodaei, Ashkan, and Stefaan P. O. Werbrouck. 2023. "Unlocking Nature’s Clock: CRISPR Technology in Flowering Time Engineering" Plants 12, no. 23: 4020. https://doi.org/10.3390/plants12234020
APA StyleHodaei, A., & Werbrouck, S. P. O. (2023). Unlocking Nature’s Clock: CRISPR Technology in Flowering Time Engineering. Plants, 12(23), 4020. https://doi.org/10.3390/plants12234020






