Exploring the Deoxy-D-xylulose-5-phosphate Synthase Gene Family in Tomato (Solanum lycopersicum)
Abstract
1. Introduction
2. Results
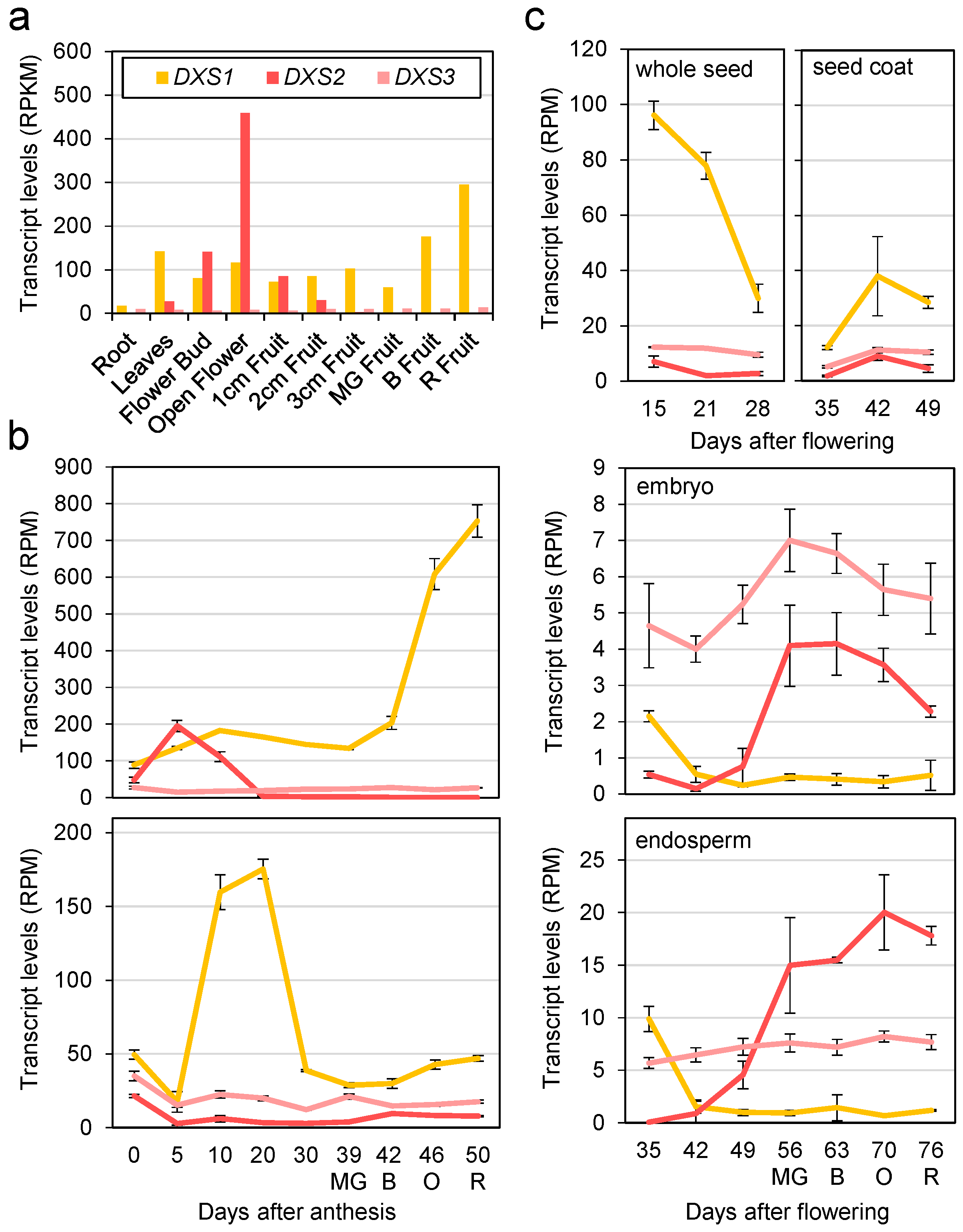
2.1. DXS-Encoding Tomato Transcripts Are Differentially Expressed but Overlap in Some Tissues
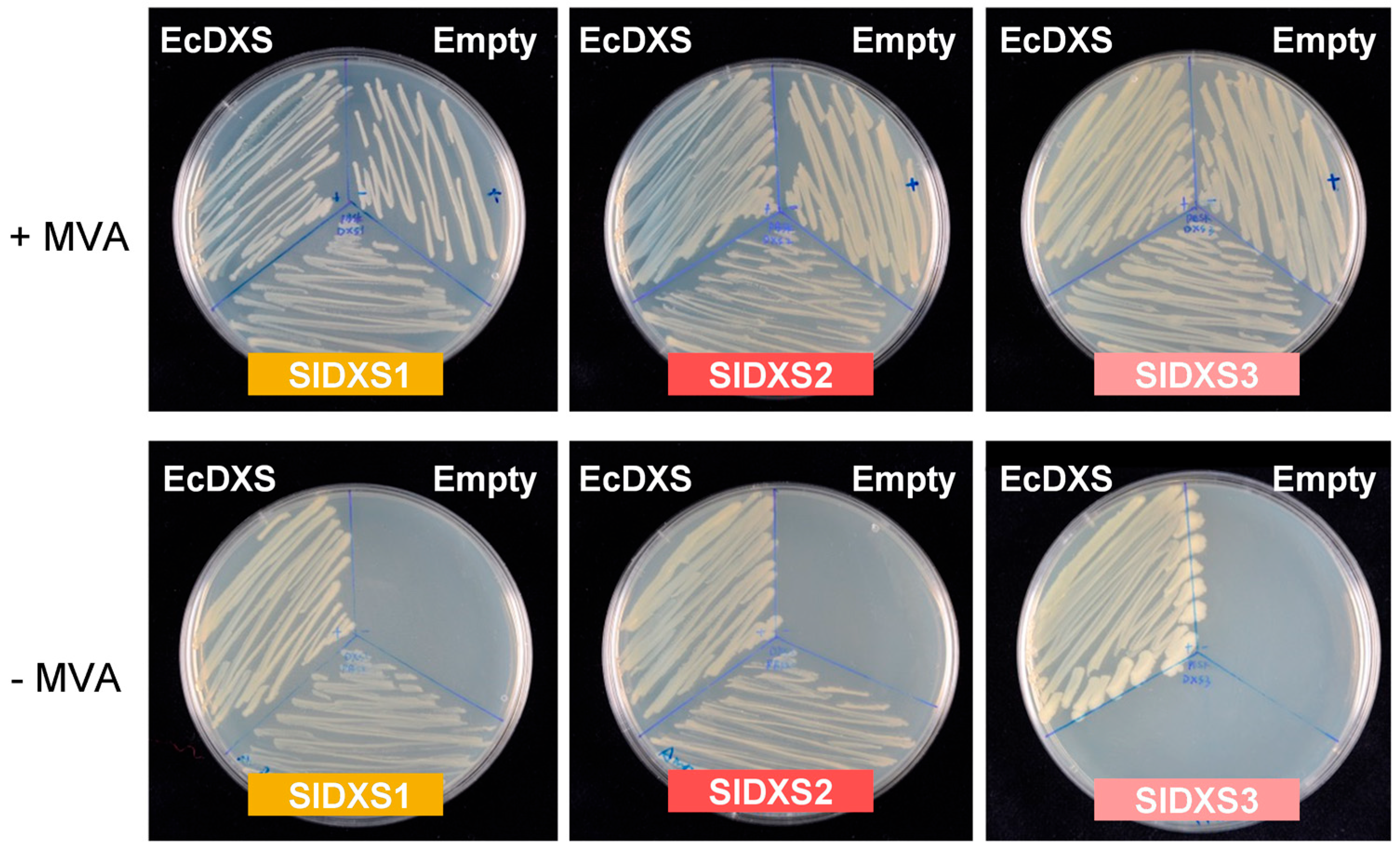
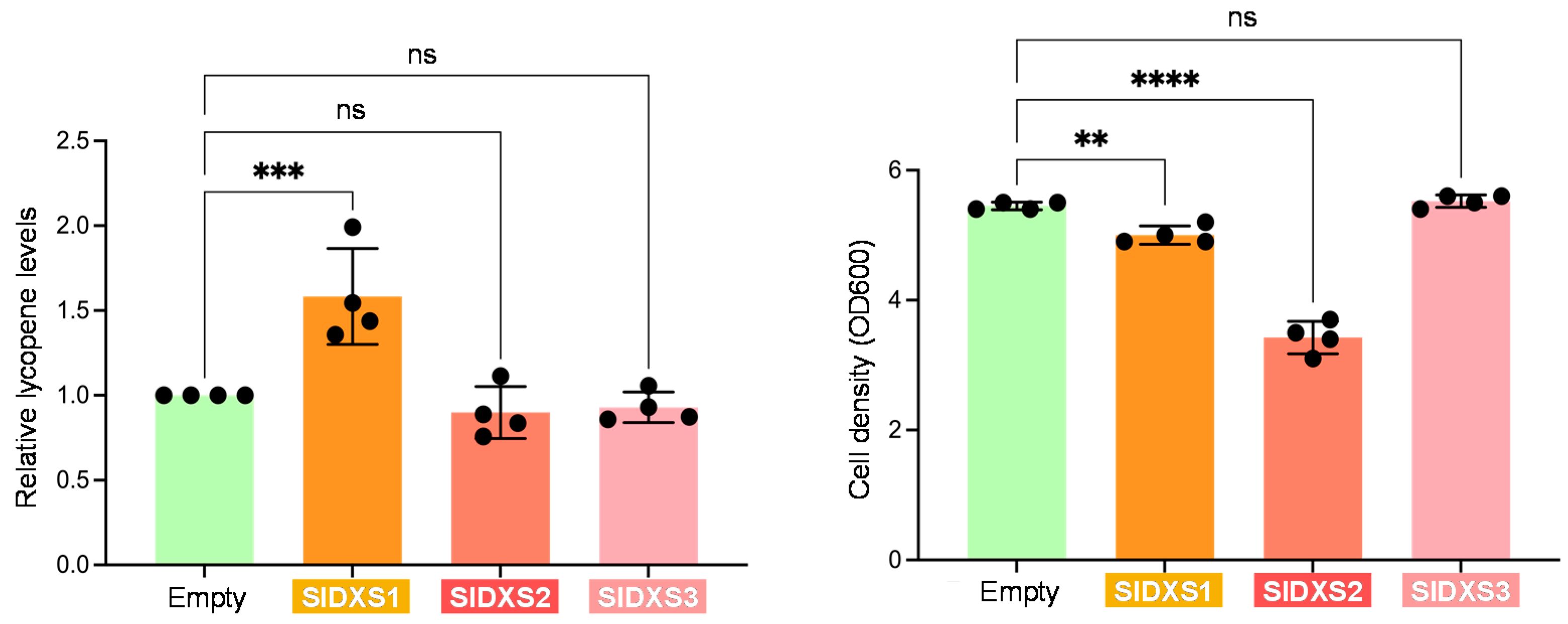
2.2. SlDXS1 and SlDXS2, but Not SlDXS3, Are True DXS Enzymes
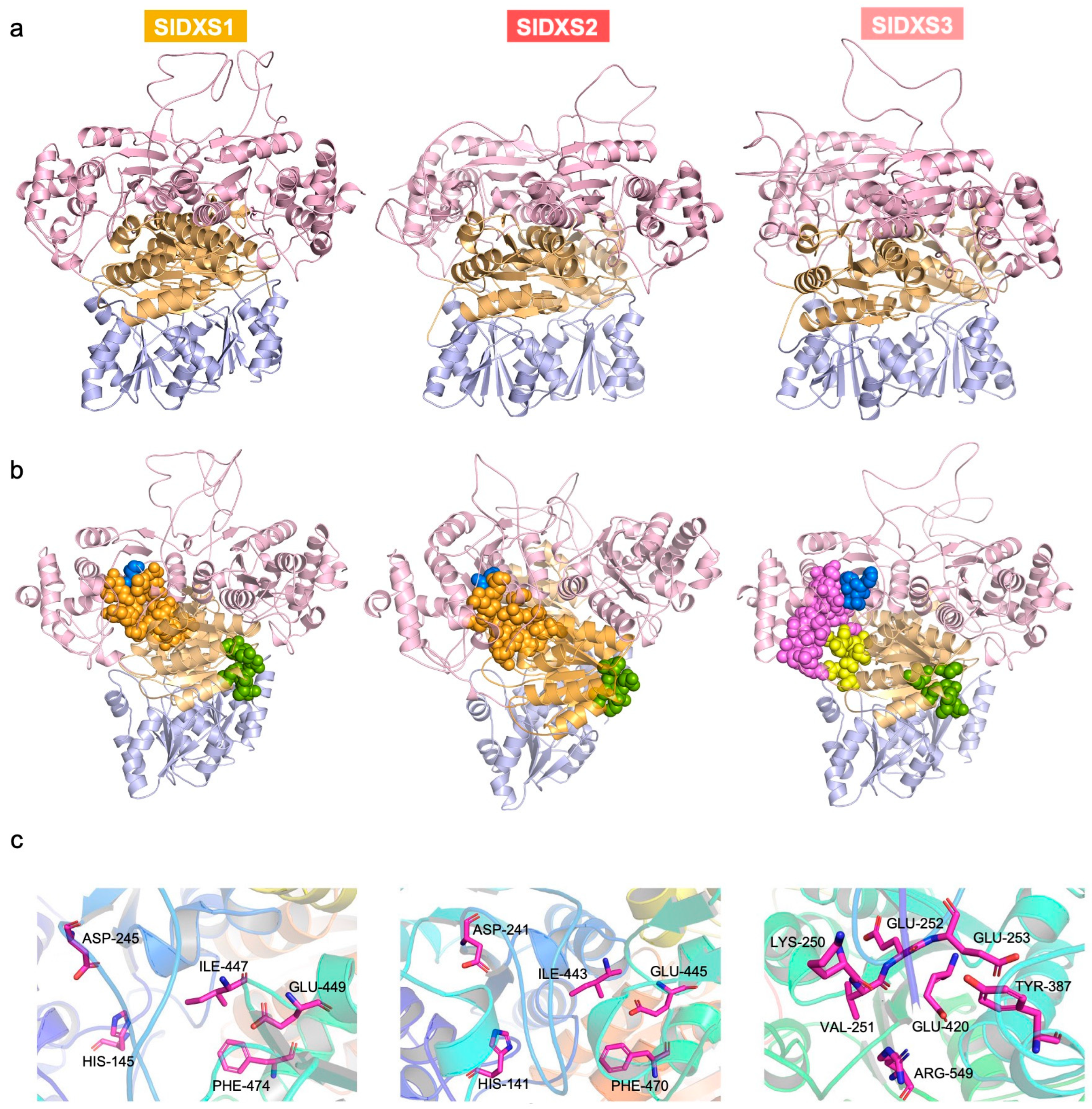
2.3. SlDXS3 Protein Lacks Structural Features Required for DXS Activity
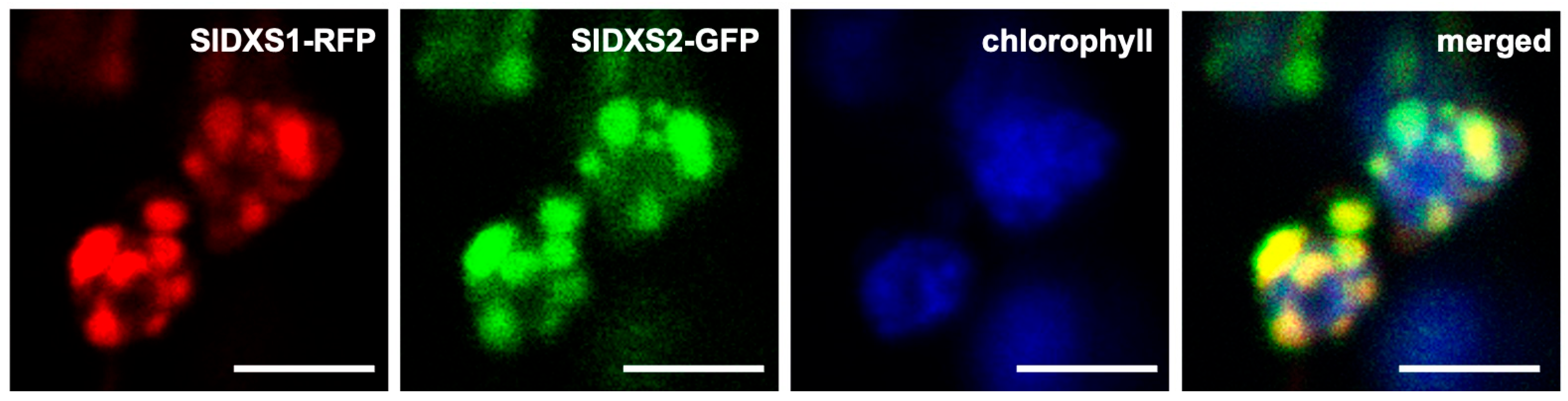
2.4. SlDXS1 and SlDXS2 Co-Localize in Chloroplasts
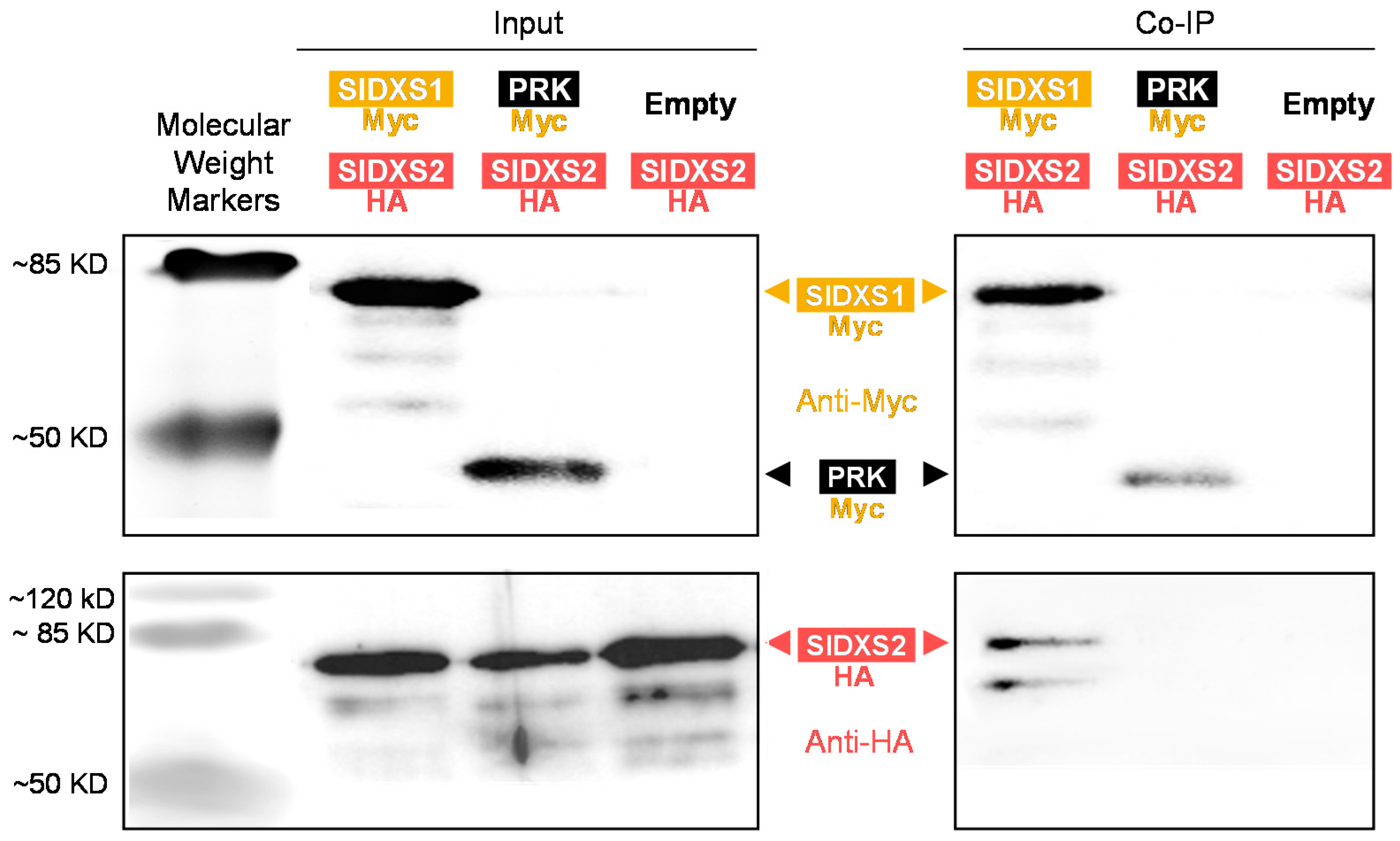
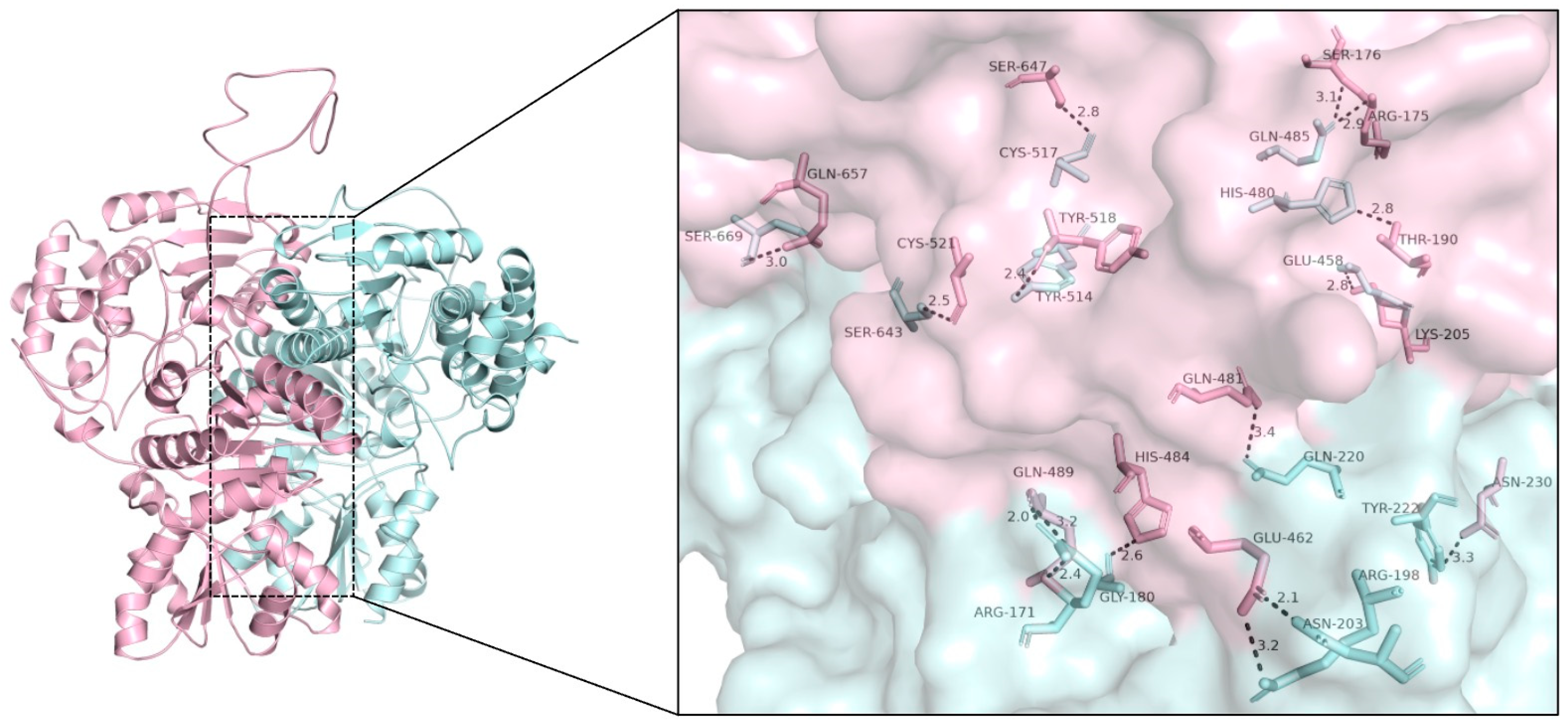
2.5. Heterodimers of SlDXS1 and SlDXS2 Can Be Potentially Active
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plant Material and Growth Conditions
4.2. Gene Constructs
4.3. Complementation and Carotenoid Production Assays
4.4. Computational Modeling
4.5. Transient Expression Assays
4.6. Subcellular Localization Assays
4.7. Co-Immunoprecipitation Assays
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nogueira, M.; Enfissi, E.M.; Almeida, J.; Fraser, P.D. Creating plant molecular factories for industrial and nutritional isoprenoid production. Curr. Opin. Biotechnol. 2017, 49, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Concepción, M.; Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002, 130, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Concepción, M.; Boronat, A. Breaking new ground in the regulation of the early steps of plant isoprenoid biosynthesis. Curr. Opin. Plant Biol. 2015, 25, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Volke, D.C.; Rohwer, J.; Fischer, R.; Jennewein, S. Investigation of the methylerythritol 4-phosphate pathway for microbial terpenoid production through metabolic control analysis. Microb. Cell Factories 2019, 18, 192. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.P.; Rohwer, J.M.; Ghirardo, A.; Hammerbacher, A.; Ortiz-Alcaide, M.; Raguschke, B.; Schnitzler, J.-P.; Gershenzon, J.; Phillips, M.A. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in arabidopsis. Plant Physiol. 2014, 165, 1488–1504. [Google Scholar] [CrossRef] [PubMed]
- EEnfissi, E.M.A.; Fraser, P.D.; Lois, L.-M.; Boronat, A.; Schuch, W.; Bramley, P.M. Metabolic engineering of the mevalonate and non-mevalonate isopentenyl diphosphate-forming pathways for the production of health-promoting isoprenoids in tomato. Plant Biotechnol. J. 2005, 3, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Estévez, J.M.; Cantero, A.; Reindl, A.; Reichler, S.; León, P. 1-Deoxy-D-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J. Biol. Chem. 2001, 276, 22901–22909. [Google Scholar] [CrossRef]
- Morris, W.L.; Ducreux, L.J.M.; Hedden, P.; Millam, S.; Taylor, M.A. Overexpression of a bacterial 1-deoxy-D-xylulose 5-phosphate synthase gene in potato tubers perturbs the isoprenoid metabolic network: Implications for the control of the tuber life cycle. J. Exp. Bot. 2006, 57, 3007–3018. [Google Scholar] [CrossRef]
- Estévez, J.M.; Cantero, A.; Romero, C.; Kawaide, H.; Jiménez, L.F.; Kuzuyama, T.; Seto, H.; Kamiya, Y.; León, P. Analysis of the expression of CLA1, a gene that encodes the 1-deoxyxylulose 5-phosphate synthase of the 2-C-methyl-D-erythritol-4-phosphate pathway in Arabidopsis. Plant Physiol. 2000, 124, 95–103. [Google Scholar] [CrossRef]
- Bouvier, F.; D’Harlingue, A.; Suire, C.; Backhaus, R.A.; Camara, B. Dedicated roles of plastid transketolases during the early onset of isoprenoid biogenesis in pepper fruits. Plant Physiol. 1998, 117, 1423–1431. [Google Scholar] [CrossRef][Green Version]
- Xiang, S.; Usunow, G.; Lange, G.; Busch, M.; Tong, L. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 2007, 282, 2676–2682. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Leung, S.K.P.; Zhang, W.; Lai, L.T.F.; Chan, Y.K.; Wong, M.C.; Benlekbir, S.; Cui, Y.; Jiang, L.; Lau, W.C.Y. Structural basis of substrate recognition and thermal protection by a small heat shock protein. Nat. Commun. 2021, 12, 3007. [Google Scholar] [CrossRef] [PubMed]
- Walter, M.H.; Hans, J.; Strack, D. Two distantly related genes encoding 1-deoxy-d-xylulose 5-phosphate synthases: Differential regulation in shoots and apocarotenoid-accumulating mycorrhizal roots. Plant J. 2002, 31, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Cordoba, E.; Porta, H.; Arroyo, A.; Román, C.S.; Medina, L.; Rodríguez-Concepción, M.; León, P. Functional characterization of the three genes encoding 1-deoxy-D-xylulose 5-phosphate synthase in maize. J. Exp. Bot. 2011, 62, 2023–2038. [Google Scholar] [CrossRef]
- Saladié, M.; Wright, L.P.; Garcia-Mas, J.; Rodriguez-Concepcion, M.; Phillips, M.A. The 2-C-methylerythritol 4-phosphate pathway in melon is regulated by specialized isoforms for the first and last steps. J. Exp. Bot. 2014, 65, 5077–5092. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; León, P.; Boronat, A.; Rodriguezconcepcion, M. The plastidial MEP pathway: Unified nomenclature and resources. Trends Plant Sci. 2008, 13, 619–623. [Google Scholar] [CrossRef] [PubMed]
- de Luna-Valdez, L.; Chenge-Espinosa, M.; Hernández-Muñoz, A.; Cordoba, E.; López-Leal, G.; Castillo-Ramírez, S.; León, P. Reassessing the evolution of the 1-deoxy-D-xylulose 5-phosphate synthase family suggests a possible novel function for the DXS class 3 proteins. Plant Sci. 2021, 310, 110960. [Google Scholar] [CrossRef]
- Paetzold, H.; Garms, S.; Bartram, S.; Wieczorek, J.; Urós-Gracia, E.-M.; Rodríguez-Concepción, M.; Boland, W.; Strack, D.; Hause, B.; Walter, M.H. The Isogene 1-Deoxy-D-Xylulose 5-Phosphate Synthase 2 Controls Isoprenoid Profiles, Precursor Pathway Allocation, and Density of Tomato Trichomes. Mol. Plant 2010, 3, 904–916. [Google Scholar] [CrossRef]
- García-Alcázar, M.; Giménez, E.; Pineda, B.; Capel, C.; García-Sogo, B.; Sánchez, S.; Yuste-Lisbona, F.J.; Angosto, T.; Capel, J.; Moreno, V.; et al. Albino T-DNA tomato mutant reveals a key function of 1-deoxy-D-xylulose-5-phosphate synthase (DXS1) in plant development and survival. Sci. Rep. 2017, 7, 45333. [Google Scholar] [CrossRef]
- Lois, L.M.; Rodríguez-Concepción, M.; Gallego, F.; Campos, N.; Boronat, A. Carotenoid biosynthesis during tomato fruit development: Regulatory role of 1-deoxy-D-xylulose 5-phosphate synthase. Plant J. 2000, 22, 503–513. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Salvatore, M.; Emanuelsson, O.; Winther, O.; von Heijne, G.; Elofsson, A.; Nielsen, H. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2019, 2, e201900429. [Google Scholar] [CrossRef] [PubMed]
- Sauret-Güeto, S.; Urós, E.M.; Ibáñez, E.; Boronat, A.; Rodríguez-Concepción, M. A mutant pyruvate dehydrogenase E1 subunit allows survival of Escherichia coli strains defective in 1-deoxy-D-xylulose 5-phosphate synthase. FEBS Lett. 2006, 580, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Arranz, S.; Perez-Gil, J.; Marshall-Sabey, D.; Rodriguez-Concepcion, M. Engineering Pseudomonas putida for isoprenoid production by manipulating endogenous and shunt pathways supplying precursors. Microb. Cell Factories 2019, 18, 152. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.; DeColli, A.; Meyers, C.F.; Nemeria, N.S.; Jordan, F. Conformational dynamics of 1-deoxy-D-xylulose 5-phosphate synthase on ligand binding revealed by H/D exchange MS. Proc. Natl. Acad. Sci. USA 2017, 114, 9355–9360. [Google Scholar] [CrossRef] [PubMed]
- DeColli, A.A.; Zhang, X.; Heflin, K.L.; Jordan, F.; Meyers, C.L.F. Active Site Histidines Link Conformational Dynamics with Catalysis on Anti-Infective Target 1-Deoxy-d-xylulose 5-Phosphate Synthase. Biochemistry 2019, 58, 4970–4982. [Google Scholar] [CrossRef] [PubMed]
- Altincicek, B.; Hintz, M.; Sanderbrand, S.; Wiesner, J.; Beck, E.; Jomaa, H. Tools for discovery of inhibitors of the 1-deoxy-D-xylulose 5-phosphate (DXP) synthase and DXP reductoisomerase: An approach with enzymes from the pathogenic bacterium Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2000, 190, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Gierse, R.M.; Reddem, E.R.; Alhayek, A.; Baitinger, D.; Hamid, Z.; Jakobi, H.; Laber, B.; Lange, G.; Hirsch, A.K.; Groves, M.R. Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties. Biochem. Biophys. Res. Commun. 2021, 539, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Pieper, U. MODBASE: A database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006, 34, D291–D295. [Google Scholar] [CrossRef]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.Y.; Sali, A. Protein structure modeling with MODELLER. Struct. Proteom. High-Throughput Methods 2008, 2008, 145–159. [Google Scholar]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Sprenger, G.A.; Schörken, U.; Wiegert, T.; Grolle, S.; de Graaf, A.A.; Taylor, S.V.; Begley, T.P.; Bringer-Meyer, S.; Sahm, H. Identification of a thiamin-dependent synthase in Escherichia coli required for the formation of the 1-deoxy-D-xylulose 5-phosphate precursor to isoprenoids, thiamin, and pyridoxol. Proc. Natl. Acad. Sci. USA 1997, 94, 12857–12862. [Google Scholar] [CrossRef] [PubMed]
- Lois, L.M.; Campos, N.; Putra, S.R.; Danielsen, K.; Rohmer, M.; Boronat, A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc. Natl. Acad. Sci. USA 1998, 95, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M.; Wildung, M.R.; McCaskill, D.; Croteau, R. A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc. Natl. Acad. Sci. USA 1998, 95, 2100–2104. [Google Scholar] [CrossRef] [PubMed]
- McGreig, J.E.; Uri, H.; Antczak, M.; Sternberg, M.J.E.; Michaelis, M.; Wass, M.N. 3DLigandSite: Structure-based prediction of protein—Ligand binding sites. Nucleic Acids Res. 2022, 50, W13–W20. [Google Scholar] [CrossRef] [PubMed]
- Wass, M.N.; Sternberg, M.J.E. Prediction of ligand binding sites using homologous structures and conservation at CASP8. Proteins Struct. Funct. Bioinform. 2009, 77, 147–151. [Google Scholar] [CrossRef] [PubMed]
- Pulido, P.; Toledo-Ortiz, G.; Phillips, M.A.; Wright, L.P.; Rodríguez-Concepción, M. Arabidopsis J-Protein J20 delivers the first enzyme of the plastidial isoprenoid pathway to protein quality control. Plant Cell 2013, 25, 4183–4194. [Google Scholar] [CrossRef] [PubMed]
- Pulido, P.; Llamas, E.; Llorente, B.; Ventura, S.; Wright, L.P.; Rodríguez-Concepción, M. Specific Hsp100 Chaperones Determine the Fate of the First Enzyme of the Plastidial Isoprenoid Pathway for Either Refolding or Degradation by the Stromal Clp Protease in Arabidopsis. PLoS Genet. 2016, 12, e1005824. [Google Scholar] [CrossRef]
- Perello, C.; Llamas, E.; Burlat, V.; Ortiz-Alcaide, M.; Phillips, M.A.; Pulido, P.; Rodriguez-Concepcion, M. Differential subplastidial localization and turnover of enzymes involved in isoprenoid biosynthesis in chloroplasts. PLoS ONE 2016, 11, e0150539. [Google Scholar] [CrossRef]
- Guirimand, G.; Guihur, A.; Perello, C.; Phillips, M.; Mahroug, S.; Oudin, A.; de Bernonville, T.D.; Besseau, S.; Lanoue, A.; Giglioli-Guivarc’h, N.; et al. Cellular and subcellular compartmentation of the 2C-methyl-D-erythritol 4-phosphate pathway in the madagascar periwinkle. Plants 2020, 9, 462. [Google Scholar] [CrossRef]
- Di, X.; Ortega-Alarcon, D.; Kakumanu, R.; Iglesias-Fernandez, J.; Diaz, L.; Baidoo, E.E.; Velazquez-Campoy, A.; Rodríguez-Concepción, M.; Perez-Gil, J. MEP pathway products allosterically promote monomerization of deoxy-D-xylulose-5-phosphate synthase to feedback-regulate their supply. Plant Commun. 2023, 4, 100512. [Google Scholar] [CrossRef]
- Barja, M.V.; Ezquerro, M.; Beretta, S.; Diretto, G.; Florez-Sarasa, I.; Feixes, E.; Fiore, A.; Karlova, R.; Fernie, A.R.; Beekwilder, J.; et al. Several geranylgeranyl diphosphate synthase isoforms supply metabolic substrates for carotenoid biosynthesis in tomato. New Phytol. 2021, 231, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Pierce, B.G.; Wiehe, K.; Hwang, H.; Kim, B.-H.; Vreven, T.; Weng, Z. ZDOCK server: Interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 2014, 30, 1771–1773. [Google Scholar] [CrossRef] [PubMed]
- de Castro, R.D.; Hilhorst, H.W. Hormonal control of seed development in GA- and ABA-deficient tomato (Lycopersicon esculentum Mill. cv. Moneymaker) mutants. Plant Sci. 2006, 170, 462–470. [Google Scholar] [CrossRef]
- Ezquerro, M.; Burbano-Erazo, E.; Rodriguez-Concepcion, M. Overlapping and specialized roles of tomato phytoene synthases in carotenoid and abscisic acid production. Plant Physiol. 2023, 193, 2021–2036. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Villalón, A.; Pérez-Gil, J.; Rodríguez-Concepción, M. Carotenoid accumulation in bacteria with enhanced supply of isoprenoid precursors by upregulation of exogenous or endogenous pathways. J. Biotechnol. 2008, 135, 78–84. [Google Scholar] [CrossRef]
| Gene | Template | Identity | Coverage | Range | Ramachandran Favoured | QMEAN | QMEANDisCo Global |
|---|---|---|---|---|---|---|---|
| SIDXS1 | 7bzx.1. A | 88.75% | 0.92 | 72–706 | 88.13% | 0.72 | 0.73 ± 0.05 |
| SIDXS2 | 7bzx.1. A | 73.93% | 0.91 | 68–702 | 87.28% | 0.70 | 0.72 ± 0.05 |
| SIDXS3 | 7bzx.1. A | 58.24% | 0.92 | 69–696 | 86.18% | 0.65 | 0.68 ± 0.05 |
| Gene ID | Primer Name | Sequence (5′ to 3′) |
|---|---|---|
| Solyc01g067890 | SlDXS1-F | ATCATGGCTTTGTGTGCTTATGCATTTCCTG |
| SlDXS1-R | CTTACTCGAGTGTCATGACCTCTAGAGCCTCTC | |
| Solyc11g010850 | SlDXS2-F | CAGATGGCAGTTTCTTCAGGCACTGTATTTAG |
| SlDXS2I-R | CTTAGTCGACGTTTACTACCATAGCTTCTTTTG | |
| Solyc08g066950 | SlDXS3-F | TGATGGTTGCTGTTACTGCTCAGTACCCATTTG |
| SlDXS3L-R | GTTAGTCGACGCACATCAAAAGAAGAGCTTCTC | |
| Solyc01g067890 | GW-SIDXS1-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGCTTTGTGTGCTTATGCAT |
| GW-SIDXS1-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCTGTCATGACCTCTAGAGC | |
| Solyc11g010850 | GW-SIDXS2-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGATGGCAGTTTCTTCAGGCACT |
| GW-SIDXS2-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCGTTTACTACCATAGCTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di, X.; Rodriguez-Concepcion, M. Exploring the Deoxy-D-xylulose-5-phosphate Synthase Gene Family in Tomato (Solanum lycopersicum). Plants 2023, 12, 3886. https://doi.org/10.3390/plants12223886
Di X, Rodriguez-Concepcion M. Exploring the Deoxy-D-xylulose-5-phosphate Synthase Gene Family in Tomato (Solanum lycopersicum). Plants. 2023; 12(22):3886. https://doi.org/10.3390/plants12223886
Chicago/Turabian StyleDi, Xueni, and Manuel Rodriguez-Concepcion. 2023. "Exploring the Deoxy-D-xylulose-5-phosphate Synthase Gene Family in Tomato (Solanum lycopersicum)" Plants 12, no. 22: 3886. https://doi.org/10.3390/plants12223886
APA StyleDi, X., & Rodriguez-Concepcion, M. (2023). Exploring the Deoxy-D-xylulose-5-phosphate Synthase Gene Family in Tomato (Solanum lycopersicum). Plants, 12(22), 3886. https://doi.org/10.3390/plants12223886






