Chemophenetic Approach to Selected Senecioneae Species, Combining Morphometric and UHPLC-HRMS Analyses
Abstract
1. Introduction
2. Results and Discussion
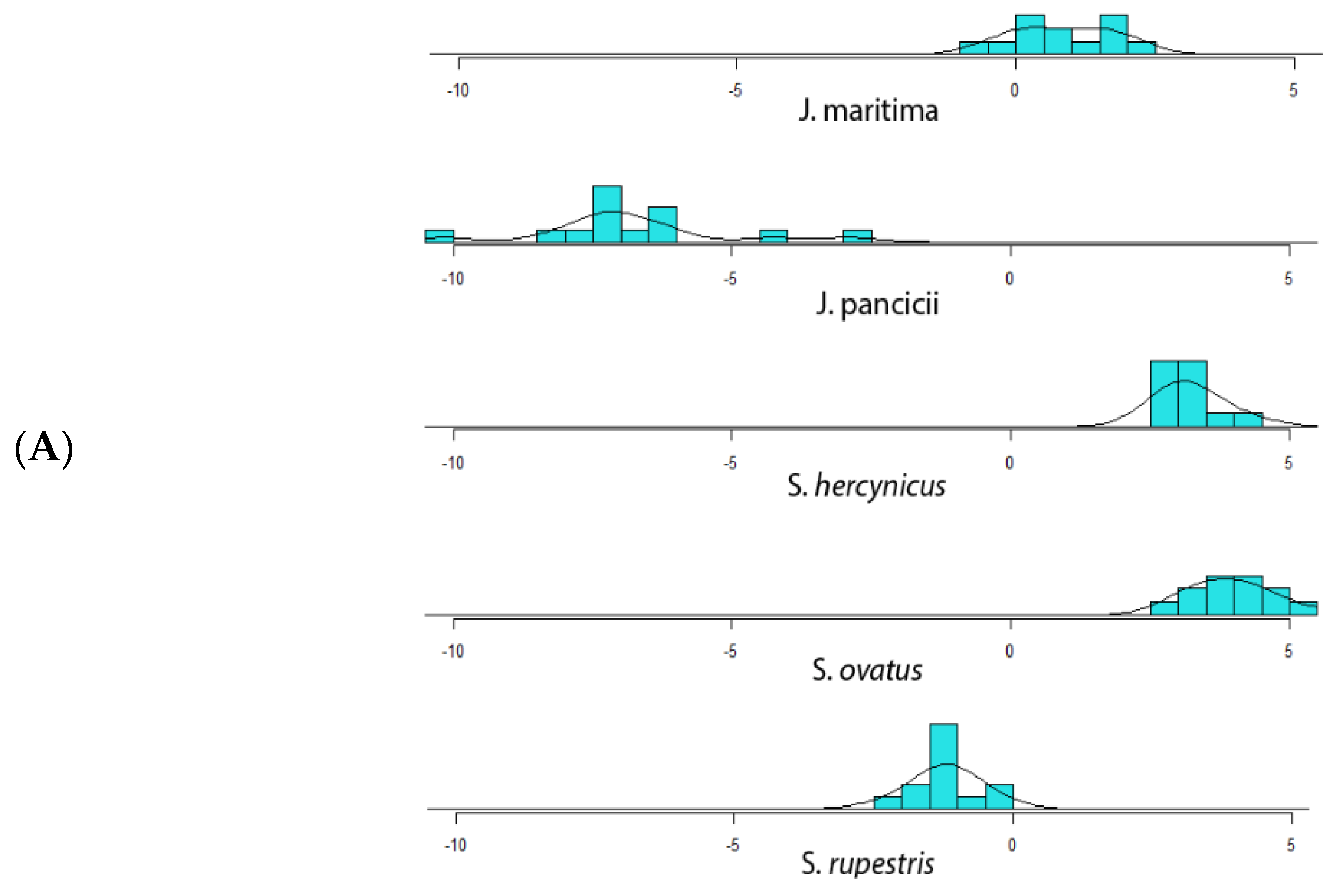
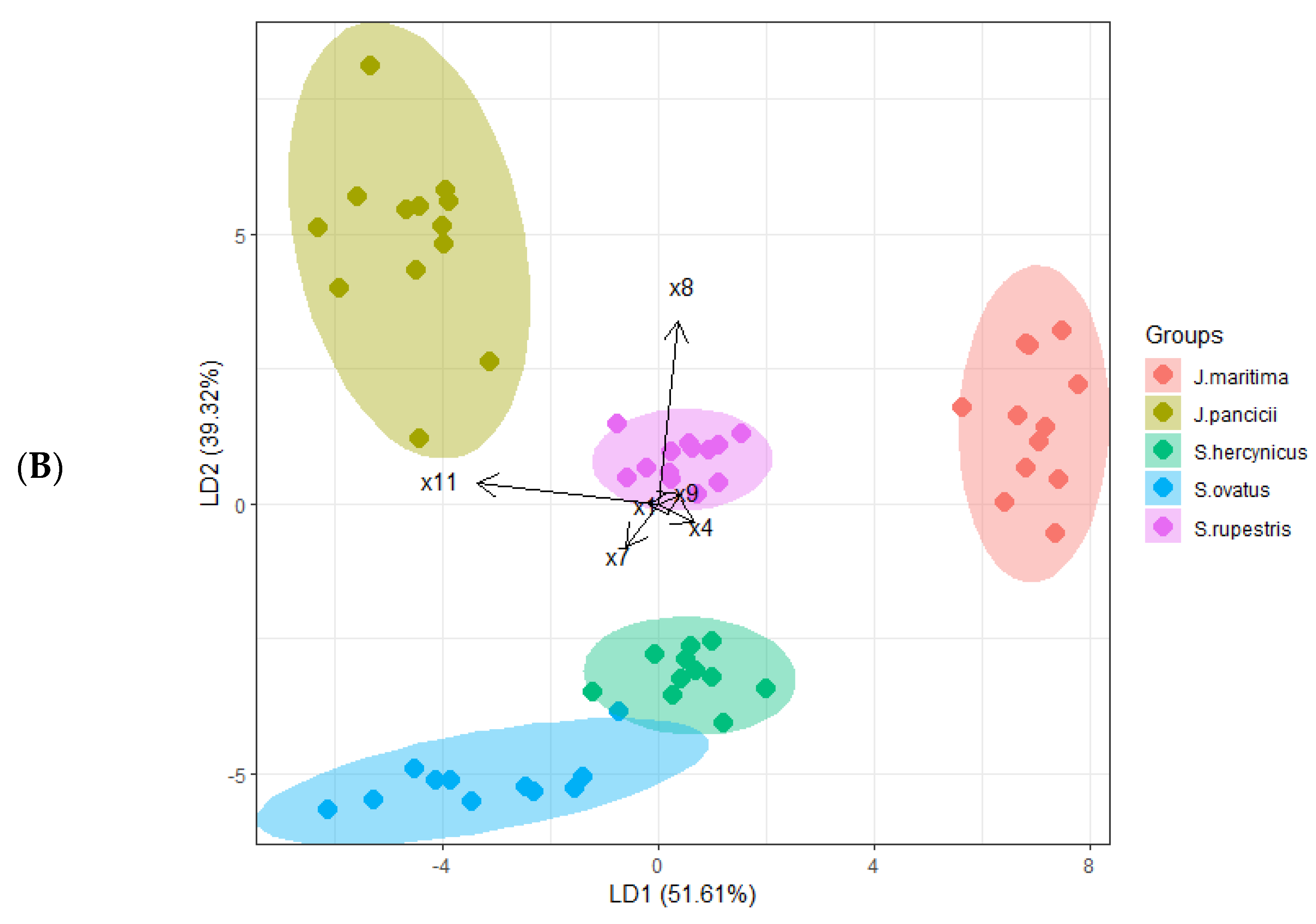
2.1. Morphometric Analysis
Correlation and Linear Discriminant Analysis (LDA)
2.2. UHPLC-HRMS Identification and Tentative Annotation of Specialized Natural Products
2.2.1. Hydroxybenzoic, Hydroxycinnamic Acids, and Their Glycosides
2.2.2. Acylquinic Acids
2.2.3. Flavonoids
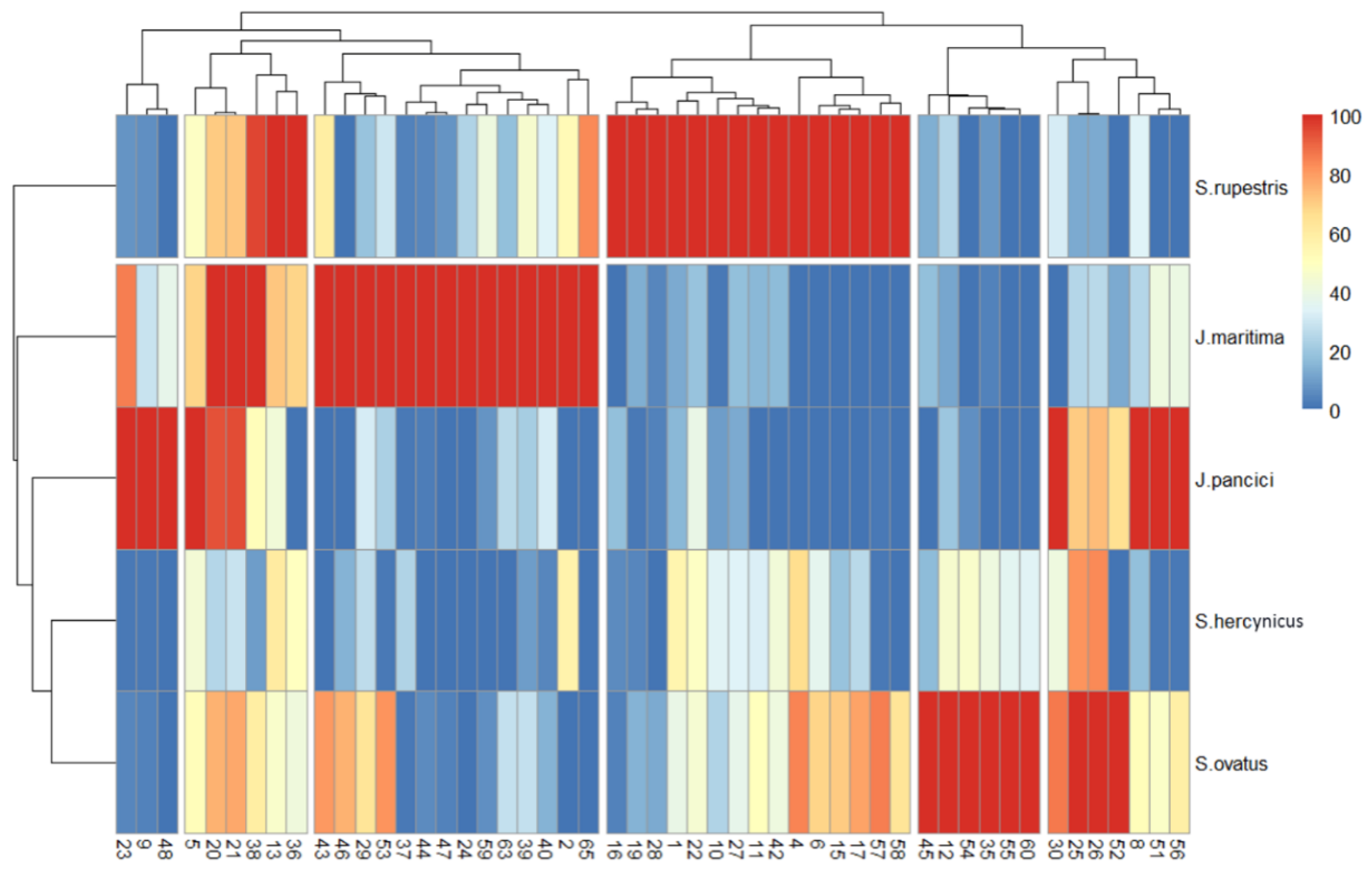
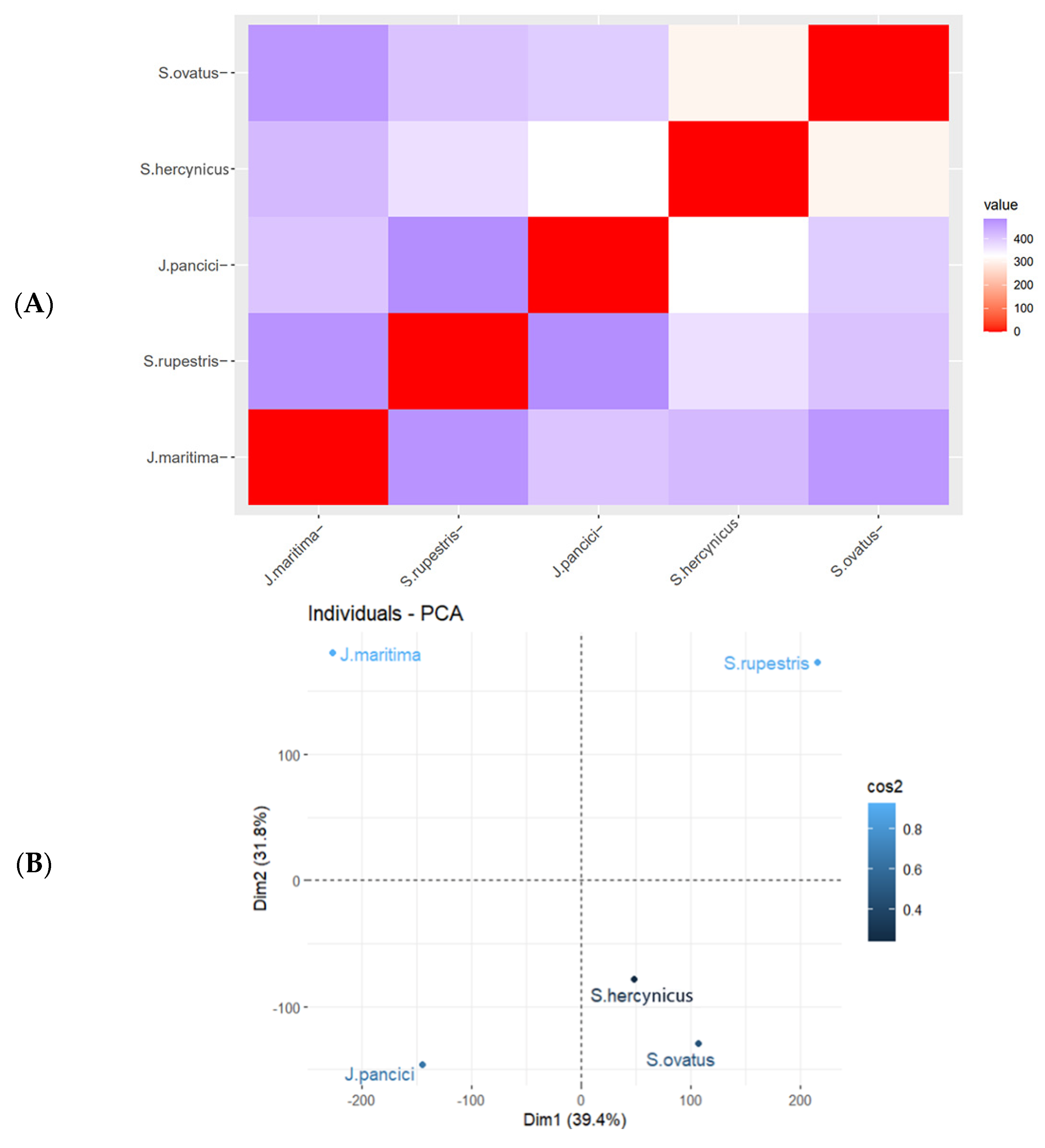
2.3. Chemophenetic Significance
3. Materials and Methods
3.1. Plant Material
3.2. Morphometric Measurements
3.3. Chemicals and Reagents
3.4. Sample Extraction and Sample Preparation
3.5. Ultra-High-Performance Liquid Chromatography—High Resolution Mass Spectrometry (UHPLC-HRMS)
3.6. File Conversions and Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pelser, P.B.; Nordenstam, B.; Kadereit, J.W.; Watson, L.E. An ITS phylogeny of tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon 2007, 56, 1077–1104. [Google Scholar] [CrossRef]
- Rola, K. Morphometry and distribution of Senecio nemorensis agg. species (Asteraceae) in Poland. Pol. Bot. J. 2014, 59, 37–54. [Google Scholar] [CrossRef]
- Galasso, G.; Bartolucci, F. Four new combinations in Jacobaea Mill.(Asteraceae, Senecioneae) for the European flora. Nat. Hist. Sci. 2015, 2, 95–96. [Google Scholar] [CrossRef]
- Bog, M.; Elmer, M.; Doppel, M.; Ehrnsberger, H.F.; Beuerle, T.; Heilmann, J.; Oberprieler, C. Phytochemical investigations and food-choice experiments with two mollusc species in three central European Senecio L.(Asteraceae, Senecioneae) species and their hybrids. Chemoecology 2017, 27, 155–169. [Google Scholar] [CrossRef]
- Hodalova, I.; Grulich, V.; Marhold, K. A multivariate morphometric study of Senecio paludosus L.(Asteraceae) in Central and Western Europe. Bot. Helv. 2002, 112, 137–152. [Google Scholar]
- Podsiedlik, M.; Nowinska, R.; Bednorz, L. A morphometric study on Senecio erucifolius (Asteraceae) from Poland and its taxonomic implications. Acta Soc. Bot. Pol. 2016, 85, 2305. [Google Scholar] [CrossRef]
- Abbott, R.; James, J.; Forbes, D.; Comes, H.-P. Hybrid origin of the Oxford Ragwort, Senecio squalidus L: Morphological and allozyme differences between S. squalidus and S. rupestris Waldst. and Kit. Watsonia 2002, 24, 17–30. [Google Scholar]
- Fei, D.-Q.; Zhang, Z.-X.; Chen, J.-J.; Gao, K. Eremophilane-type sesquiterpenes from Senecio nemorensis. Planta Med. 2007, 73, 1292–1297. [Google Scholar] [CrossRef]
- Yang, Y.; Zhao, L.; Wang, Y.F.; Chang, M.L.; Huo, C.H.; Gu, Y.C.; Shi, Q.W.; Kiyota, H. Chemical and pharmacological research on plants from the genus Senecio. Chem. Biodivers. 2011, 8, 13–72. [Google Scholar] [CrossRef]
- Mandić, B.M.; Gođevac, D.M.; Vujisić, L.V.; Trifunović, S.S.; Tesević, V.V.; Vajs, V.V.; Milosavljević, S.M. Semiquinol and phenol compounds from seven Senecio species. Chem. Pap. 2011, 65, 90–92. [Google Scholar] [CrossRef]
- Albayrak, S.; Aksoy, A.; Yurtseven, L.; Yaşar, A. A comparative study on phenolic components and biological activity of some Senecio species in Turkey. J. Pharm. Pharmacol. 2014, 66, 1631–1640. [Google Scholar] [CrossRef]
- Mohamed, S. Phytochemical and biological study of (Senecio glaucus subsp. coronopifolius)(Maire) c. alexander growing in Egypt. Al-Azhar J. Pharm. Sci. 2015, 52, 283–298. [Google Scholar] [CrossRef]
- Bousetla, A.; Keskinkaya, H.B.; Bensouici, C.; Lefahal, M.; Atalar, M.N.; Akkal, S. LC-ESI/MS-phytochemical profiling with antioxidant and antiacetylcholinesterase activities of Algerian Senecio angulatus Lf extracts. Nat. Prod. Res. 2021, 37, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Balabanova, V.; Voynikov, Y.; Zengin, G.; Gevrenova, R.; Zheleva-Dimitrova, D. A view on antioxidant and enzyme inhibitory activity of senecio hercynicus herbal drugs. Comptes Rendus L Acad. Bulg. Des. Sci. 2020, 73, 1673–1680. [Google Scholar]
- Arab, Y.; SAHIN, B.; CEYLAN, O.; Zellagui, A.; OLMEZ, O.T.; Kucukaydin, S.; Tamfu, A.N.; Ozturk, M.; Gherraf, N. Assessment of in vitro activities and chemical profiling of Senecio hoggariensis growing in Algerian Sahara. Biodiversitas J. Biol. Divers. 2022, 23, 3498–3506. [Google Scholar] [CrossRef]
- Shi, B.-J.; Xiong, A.-Z.; Zheng, S.-S.; Chou, G.-X.; Wang, Z.-T. Two new pyrrolizidine alkaloids from Senecio nemorensis. Nat. Prod. Res. 2010, 24, 1897–1901. [Google Scholar] [CrossRef]
- Dekić, M.; Radulović, N.; Stojanović, N.; Mladenović, M. Analgesic activity of dehydrofukinone, sesquiterpene ketone from Senecio nemorensis L.(Asteraceae). Facta Univ. Ser. Phys. Chem. Technol. 2018, 16, 119. [Google Scholar]
- Tundis, R.; Loizzo, M.; Menichini, F.; Bonesi, M.; Conforti, F.; Statti, G.; Passalacqua, N.; Curini, M. In vitro hypoglycaemic activity of Senecio nemorensis subsp. stabianus Lacaita (Asteraceae). Planta Med. 2009, 75, PH20. [Google Scholar] [CrossRef]
- Christov, V.; Simeonov, M.; Velcheva, N.; Karadjova, O.; Atanassov, N.; Ivanova, I.; Evstatieva, L. Pyrrolizidine Alkaloids from Bulgarian Species—Genus Senecio and their Insecticidal Properties. Biotechnol. Biotechnol. Equip. 1997, 11, 53–59. [Google Scholar] [CrossRef]
- Vladimirov, V. Asteraceae. Senecio nemorensis L. group, Senecio hercynicus Herborg. In Flora Republicae Bulgaricae; Peev, D., Ed.; Marin Drinov: Sofia, Bulgaria, 2012; Volume XI, pp. 432–434. [Google Scholar]
- Christov, V.; Evstatieva, L. Alkaloid profile of Bulgarian species from genus Senecio L. Z. Für Nat. C 2003, 58, 300–302. [Google Scholar] [CrossRef]
- Christov, V.; Kostova, N.; Evstatieva, L. 6α-Angeloylplatynecine: A new alkaloid from Senecio nemorensis subsp. fuchsii (CC Gmelin) celak. Nat. Prod. Res. 2005, 19, 389–392. [Google Scholar] [CrossRef]
- Güner, A.; Akyıldırım, B.; Alkayış, M.; Çıngay, B.; Kanoğlu, S.; Özkan, A.; Öztekin, M.; Tuğ, G.; Güner, A.; Aslanl, S.; et al. Türkiye Bitkileri Listesi (Damarlı Bitkiler); Nezahat Gökyiğit Botanik Bahçesi ve Flora Araştırmaları Derneği Yayını: Istanbul, Turkey, 2012. [Google Scholar]
- Zidorn, C. Plant chemophenetics−A new term for plant chemosystematics/plant chemotaxonomy in the macro-molecular era. Phytochemistry 2019, 163, 147–148. [Google Scholar] [CrossRef] [PubMed]
- Dewick, P.M. Medicinal Natural Products: A Biosynthetic Approach; John Wiley & Sons: Hoboken, NJ, USA, 2002. [Google Scholar]
- Hodálová, I. Taxonomy of the Senecio nemorensis group (Compositae) in the Carpathians. Biologia 1999, 54, 395–404. [Google Scholar]
- Oberprieler, C.; Barth, A.; Schwarz, S.; Heilmann, J. Morphological and phytochemical variation, genetic structure and phenology in an introgressive hybrid swarm of Senecio hercynicus and S. ovatus (Compositae, Senecioneae). Plant Syst. Evol. 2010, 286, 153–166. [Google Scholar] [CrossRef]
- Oberprieler, C.; Hartl, S.; Schauer, K.; Meister, J.; Heilmann, J. Morphological, phytochemical and genetic variation in mixed stands and a hybrid swarm of Senecio germanicus and S. ovatus (Compositae, Senecioneae). Plant Syst. Evol. 2011, 293, 177–191. [Google Scholar] [CrossRef]
- Gareth, J.; Daniela, W.; Trevor, H.; Robert, T. An Introduction to Statistical Learning: With Applications in R; Spinger: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Denis, D.J. Univariate, Bivariate, and Multivariate Statistics Using R: Quantitative Tools for Data Analysis and Data Science; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Barkley, T.M. Senecio. In North American Flora, Series II; AbeBooks: Victoria, BC, Canada, 1978; Volume 10, pp. 50–139. [Google Scholar]
- Gevrenova, R.; Zheleva-Dimitrova, D.; Balabanova, V.; Voynikov, Y.; Sinan, K.I.; Mahomoodally, M.F.; Zengin, G. Integrated phytochemistry, bio-functional potential and multivariate analysis of Tanacetum macrophyllum (Waldst. & Kit.) Sch. Bip. and Telekia speciosa (Schreb.) Baumg.(Asteraceae). Ind. Crop. Prod. 2020, 155, 112817. [Google Scholar]
- Gevrenova, R.; Zengin, G.; Sinan, K.I.; Yıldıztugay, E.; Zheleva-Dimitrova, D.; Picot-Allain, C.; Mahomoodally, M.F.; Imran, M.; Dall’Acqua, S. UHPLC-MS Characterization and biological insights of different solvent extracts of two Achillea species (A. aleppica and A. santolinoides) from Turkey. Antioxidants 2021, 10, 1180. [Google Scholar] [CrossRef]
- Clifford, M.N.; Knight, S.; Kuhnert, N. Discriminating between the six isomers of dicaffeoylquinic acid by LC-MS n. J. Agric. Food Chem. 2005, 53, 3821–3832. [Google Scholar] [CrossRef]
- Zheleva-Dimitrova, D.; Zengin, G.; Balabanova, V.; Voynikov, Y.; Lozanov, V.; Lazarova, I.; Gevrenova, R. Chemical characterization with in vitro biological activities of Gypsophila species. J. Pharm. Biomed. Anal. 2018, 155, 56–69. [Google Scholar] [CrossRef]
- Venables, W.; Ripley, B.D. Statistics Complements to Modern Applied Statistics with S, 4th ed.; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Weihs, C.; Ligges, U.; Luebke, K.; Raabe, N. klaR analyzing German business cycles. In Data Analysis and Decision Support; Springer: Berlin/Heidelberg, Germany, 2005; pp. 335–343. [Google Scholar]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Lumley, T. Package ‘Leaps’. Regression Subset Selection Based on Fortran Code by Alan Miller. 2013. Available online: https://cran.microsoft.com/snapshot/2016-08-29/web/packages/leaps/leaps.pdf (accessed on 11 November 2022).
- Kassambara, A.; Mundt, F. Package ‘Factoextra’. Extract and Visualize the Results of Multivariate Data Analyses. 2017, 76. Available online: https://cran.microsoft.com/snapshot/2016-11-30/web/packages/factoextra/factoextra.pdf (accessed on 11 November 2022).
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K. Cluster: Cluster analysis basics and extensions. R Package Version 2012, 1, 56. [Google Scholar]
- Berkelaar, M. Package ‘lpSolve’. 2015. Available online: https://cran.uib.no/web/packages/lpSolve/lpSolve.pdf (accessed on 11 November 2022).
- Signorell, A.; Aho, K.; Alfons, A.; Anderegg, N.; Aragon, T.; Arppe, A.; Baddeley, A.; Barton, K.; Bolker, B.; Borchers, H. DescTools: Tools for descriptive statistics. R Package Version 0.99 2019, 28, 17. [Google Scholar]
- Kolde, R.; Kolde, M.R. Package ‘Pheatmap’. R Package. 2018. Available online: https://cran.microsoft.com/snapshot/2018-06-22/web/packages/pheatmap/pheatmap.pdf (accessed on 11 November 2022).
- Heinzen, E.; Sinnwell, J.; Atkinson, E.; Gunderson, T.; Votruba, P.; Dougherty, G.; Lennon, R.; Hanson, A.; Goergen, K.; Lundt, E. An Arsenal of “R” Functions for Large-Scale Statistical Summaries: R Package Arsenal Version 3.3.0; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]

| № | Annotated Compounds | Molecular Formula | Exact Mass [M−H]− | MS2 | tR (Min) | Distribution |
|---|---|---|---|---|---|---|
| Hydroxybenzoic, Hydroxycinnamic and Acylquinic Acids, Their Derivatives and Coumarin | ||||||
| 1 | protocatechuic acid-O-hexoside | C13H16O9 | 315.0722 | 315.0726 (100), 153.0182 (29.5), 152.0104 (60.9), 109.0284 (10.1) | 1.74 | A, B, C, D, E |
| 2 | vanillic acid 4-O-hexoside | C14H18O9 | 329.0878 | 329.0885 (1.8), 167.034 (100), 152.0103 (23), 123.0438 (14.3), 108.0202 (37.8) | 1.81 | A, B, C, E |
| 3 | syringic acid * | C9H10O5 | 197.0456 | 197.0449 (16.5), 182.0211 (3.2), 153.0549 (8.9), 138.0314 (3.3), 123.0437 (58.3) | 1.76 | C |
| 4 | vanillic acid * | C8H8O4 | 167.0350 | 167.0339 (31.1), 152.0103 (100), 123.0438 (32.1), 108.0202 (52.9), 95.0486 (8) | 1.82 | A, B, C, D, E |
| 5 | protocatechuic acid * | C7H6O4 | 153.0193 | 153.0182 (15.2), 109.0281 (100), 91.0173 (1.2), 81.033 (1.4) | 2.04 | A, B, C, D, E |
| 6 | p-hydroxyphenylacetic acid-O-hexoside | C14H18O8 | 313.0929 | 313.0923 (13), 151.0389 (100), 133.0284 (0.2), 123.0075 (0.8), 109.0281 (4.2) | 3.00 | A, B, C |
| 7 | gluconic acid-O-hexoside | C15H18O10 | 357.0827 | 357.083 (100), 195.0293 (10.8), 177.0183 (8), 151.039 (71.8) | 2.25 | C |
| 8 | syringic acid 4-O-hexoside | C15H20O10 | 359.0984 | 359.0985 (8), 197.0448 (100), 182.0212 (21.7), 153.0546 (16.1), 138.031 (29.3), 123.0074 (33.1) | 2.27 | A, B, C, D, E |
| 9 | neochlorogenic acid * | C16H18O9 | 353.0878 | 353.0882 (46.1), 191.0553 (100), 179.0341 (68.1), 173.0444 (4.1), 161.0235 (5.9), 135.0439 (54.4), 127.0385 (0.9), 111.0438 (0.7), 93.0329 (3.7), 85.0279 (8.5) | 2.37 | A, B, C, D, E |
| 10 | caffeic acid-O-hexoside | C15H18O9 | 341.0878 | 341.0884 (2.2), 179.034 (2.9), 135.0438 (100), 107.0488 (0.7) | 2.54 | A, B, C, D, E |
| 11 | 4-hydroxybenzoic acid-O-hexoside | C13H16O8 | 299.0773 | 299.0775 (13.7), 137.0231 (100), 93.033 (0.2) | 2.46 | A, B, C, E |
| 12 | esculetin-O-hexoside | C15H16O9 | 339.0722 | 339.0721 (10.6), 177.0184 (100), 149.0233 (0.9), 133.0282 (8), 105.0331 (3.3), 89.0381 (2.4) | 2.72 | A, B, C, D, E |
| 13 | 4-hydroxybenzoic acid | C7H6O3 | 137.0244 | 137.0232 (100), 108.0203 (11.2), 93.0333 (3.3) | 2.84 | A, B, C, D, E |
| 14 | ferulic acid * | C10H10O4 | 193.0506 | 193.05 (100), 178.0264 (74.8), 163.0391 (34.4), 149.0598 (38), 134.036 (82.5) | 2.96 | A, B, C |
| 15 | ferulic acid-O-hexoside | C16H20O9 | 355.1035 | 355.1048 (1), 193.0499 (100), 178.0263 (10.9), 149.0596 (21.4), 134.036 (62.1) | 2.96 | A, B, C |
| 16 | gentisic acid * | C7H6O4 | 153.0193 | 153.0182 (46.7), 123.0074 (20.8), 109.0283 (40.6), 81.0331 (5.4) | 2.98 | A, B, C, D |
| 17 | 4-hydroxybenzoic acid-O-hexoside isomer | C13H16O8 | 299.0773 | 299.0783 (1.3), 137.0231 (100), 93.033 (50.8) | 3.00 | A, B, C |
| 18 | 3-p-coumaroylquinic acid | C16H18O8 | 337.1500 | 191.0554 (19.6), 163.039 (100), 161.0443 (4.2), 119.0488 (23.7) | 3.04 | C |
| 19 | caffeic acid-O-hexoside | C15H18O9 | 341.0878 | 341.088 (26.8), 179.034 (100), 135.0438 (77), 107.0486 (0.8) | 3.07 | A, B, C, D, E |
| 20 | quinic acid | C7H12O6 | 191.0561 | 191.0553 (100), 173.0446 (2), 155.0338 (0.2), 127.0388 (4.3), 111.0437 (1.9), 93.0331 (6.4), 85.0279 (18.1) | 3.19 | A, B, C, D, E |
| 21 | chlorogenic acid | C16H18O9 | 353.0878 | 353.0881 (3.9), 191.0553 (100), 179.0343 (1.1), 173.0449 (0.4), 161.0232 (1.6), 135.0439 (0.5), 127.0386 (1.3), 111.0433 (0.3), 93.033 (2.2), 85.0279 (7.2) | 3.19 | A, B, C, D, E |
| 22 | caffeic acid-O-hexoside isomer I | C15H18O9 | 341.0878 | 341.0881 (9.5), 179.034 (100), 135.0438 (60.8), 107.049 (0.6) | 3.27 | A, B, C, D, E |
| 23 | 4-caffeoylquinic acid | C16H18O9 | 353.0878 | 353.0882 (32.1), 191.0554 (97.5), 179.0341 (72.6), 173.0446 (100), 135.0439 (56.3), 127.0387 (1.8), 111.0435 (3.3), 93.0331 (22), 85.028 (11.3) | 3.37 | A, B, C, D, E |
| 24 | p-hydroxyphenylacetic acid | C8H8O3 | 151.0401 | 151.0389 (100), 136.0154 (2), 123.0074 (4.2), 109.028 (15) | 3.47 | C, E |
| 25 | coumaric acid-O-hexoside | C15H18O8 | 325.0929 | 325.0923 (1.7), 163.039 (100), 145.0284 (3.5), 119.0488 (92.1), 93.0333 (0.8) | 3.33 | A, B, C, D, E |
| 26 | p-coumaric acid * | C9H8O3 | 163.0401 | 163.0389 (6.7), 135.0438 (0.7), 119.0488 (100) | 3.33 | A, B, C, D, E |
| 27 | caffeic acid * | C9H8O4 | 179.0350 | 179.0341 (20.5), 135.0438 (100), 117.0332 (0.7), 107.0489 (1.4) | 3.53 | A, B, C, D, E |
| 28 | caffeic acid-O-hexoside isomer II | C15H18O9 | 341.0878 | 341.088 (24.5), 179.0341 (100), 135.0439 (85.6), 107.0489 (0.5) | 3.79 | B, C, D, E |
| 29 | 5-p-coumaroylquinic acid | C16H18O8 | 337.0929 | 337.0933 (8.7), 191.0554 (100), 173.0449 (6), 163.0389 (5.7), 127.0391 (1), 119.0489 (4.8), 111.0437 (1.9), 93.033 (17.2), 85.028 (4.9) | 3.95 | A, B, C, D, E |
| 30 | 3-hydroxy-dihydrocaffeoyl-5-caffeoylquinic acid | C25H26O13 | 533.1301 | 533.1306 (100), 191.0554 (83.4), 173.0447 (10.1), 161.0596 (3.2), 127.0387 (3.3), 93.033 (17.8), 85.028 (11.8) | 3.09 | A, B, C, D, E |
| 31 | isoferulic acid | C10H10O4 | 193.0506 | 193.0499 (100), 178.0265 (0.8), 163.0391 (41.6), 149.0597 (18.8), 135.0439 (38.8) | 4.10 | C |
| 32 | syiringaldehide | C9H10O4 | 181.0506 | 181.0497 (15.2), 166.0261 (100), 151.0025 (58.4), 123.0074 (15.7) | 4.22 | C |
| 33 | acetoxy-hydroxyacetophenone-O-hexoside | C16H20O9 | 355.1035 | 355.1039 (13.7), 193.0494 (1.2), 151.0389 (100), 123.0076 (0.9), 109.0281 (4.4) | 4.27 | C |
| 34 | taraxafolin B-(caffeoyl)-hexoside | C26H28O16 | 595.1305 | 595.1308 (100), 341.0883 (25.4), 253.0353 (23.5), 235.0245 (3.5), 209.0446 (1.4), 191.0341 (31.1), 179.0341 (93.7), 165.0545 (16.4), 135.0438 (56) | 4.41 | C |
| 35 | 5-feruoylquinic acid | C17H20O9 | 367.1035 | 367.1038 (15.4), 191.0554 (100), 173.0445 (10.5), 134.0359 (13.1), 111.0437 (3.8), 93.0331 (25.5), 85.028 (5.1) | 4.42 | A, B, C, E |
| 36 | m-coumaric acid * | C9H8O3 | 163.0401 | 163.039 (7.6), 135.0439 (0.5), 119.0489 (100) | 4.56 | A, B, C, E |
| 37 | 3,4-dicaffeoylquinic acid * | C25H24O12 | 515.1195 | 515.1198 (100), 353.0881 (15), 335.0773 (5.3), 203.0344 (0.5), 191.0555 (29.1), 179.0341 (53), 173.0446 (58.7), 161.0233 (17), 135.0439 (53.1), 127.0386 (2.2), 111.0437 (3.6), 93.0331 (16.8), 85.028 (3.7) | 5.69 | A, B, C, D, E |
| 38 | 3,5-dicaffeoylquinic acid * | C25H24O12 | 515.1195 | 515.1202 (13.5), 353.0881 (100), 191.0554 (91.3), 179.0341 (49.4), 173.0443 (3.7), 161.0234 (4.2), 135.0439 (55.8), 111.0437 (1.3), 93.0332 (4.2), 85.028 (9.8) | 5.86 | A, B, C, D, E |
| 39 | 1,5-dicaffeoylquinic acid * | C25H24O12 | 515.1195 | 515.1199 (25.5), 353.088 (92.4), 335.0777 (1.9), 191.0554 (100), 179.0341 (53.3), 173.0446 (8.7), 135.0439 (65), 127.0387 (4.2), 111.0437 (2.1), 93.0332 (6.5), 85.028 (10.2) | 6.02 | A, B, C, D, E |
| 40 | 4,5-dicaffeoylquinic acid * | C25H24O12 | 515.1195 | 515.1197 (100), 353.0883 (72.3), 203.0341 (1.5), 191.0553 (38.9), 179.0341 (66.6), 173.0446 (98.1), 135.0439 (69.5), 111.0435 (5.2), 93.033 (30.8), 85.0279 (8.3) | 6.22 | A, B, C, D, E |
| 41 | shikimic acid | C7H10O5 | 173.0456 | 173.0444 (100), 111.0437 (10), 93.033 (68.4) | 6.22 | E |
| 42 | salicilic acid * | C7H6O3 | 137.0244 | 137.023 (8.7), 93.0331 (100) | 6.29 | A, C |
| 43 | 3-p-coumaroyl-5-caffeoylquinic acid | C25H24O11 | 499.1246 | 499.1238 (16.4), 353.0901 (1.5), 337.0933 (73.9), 335.0797 (1.7), 191.0553 (12.4), 173.0449 (7.9), 163.039 (100), 135.0441 (4.2), 119.0489 (34.4), 93.0334 (4.4) | 6.51 | B, C, E |
| 44 | 3-caffeoyl-5-p-coumaroylquinic acid | C25H24O11 | 499.1246 | 499.125 (26.1), 353.0882 (64.8), 337.0938 (17.5), 191.0554 (100), 179.0341 (34.5), 173.0446 (6.9), 163.0389 (2.9), 161.0231 (5.5), 135.0439 (36.8), 119.0488 (2.8), 111.0436 (1.4), 93.0331 (10.5), 85.0279 (7.1) | 6.57 | B, E |
| 45 | 3-feruoyl-5-caffeoylquinic acid | C26H26O12 | 529.1352 | 529.1296 (2.3), 367.1036 (99.2), 335.078 (1.1), 193.0499 (100), 191.0557 (3), 173.0443 (6.9), 161.0235 (2), 134.036 (68.5), 93.0331 (3.2) | 6.82 | A, B, C, E |
| 46 | 3-caffeoyl-5-feruoylquinic acid | C26H26O12 | 529.1352 | 529.1353 (41.1), 367.1037 (0.7), 353.0882 (43.5), 335.0794 (0.8), 191.0555 (100), 179.0342 (40.7), 173.0446 (11.5), 161.0238 (5.4), 135.0439 (37.7), 134.0361 (12.9), 127.0383 (1.3), 111.0437 (1.3), 93.0331 (15.5), 85.028 (7.5) | 6.89 | A, B, C, E |
| 47 | 3,4,5-tricaffeoylquinic acid | C34H30O15 | 677.1512 | 677.1517 (100), 515.1202 (46.2), 353.0883 (47.1), 335.0783 (13.9), 191.0554 (45.2), 179.0342 (65.2), 173.0446 (90.2), 161.0234 (24.1), 135.0439 (72.1), 111.0436 (5.6), 93.0331 (21.7) | 7.77 | B, C, E |
| Flavonoids | ||||||
| 48 | 6,8-di-C-hexosyl-naringenin | C27H32O15 | 595.1669 | 595.1677 (100), 475.1247 (3.8), 457.1138 (1.3), 427.1055 (1.2), 415.1037 (11.6), 385.0933 (36.1), 355.0822 (38.9), 343.0825 (3.9), 313.0722 (6.1), 119.0489 (15.5), 107.0123 (3.5), | 3.63 | D, E |
| 49 | kaempferol-O-dihexoside | C27H30O16 | 609.1461 | 609.1462 (100), 447.0931 (24.7), 285.0405 (50.3), 284.0325 (7.6), 255.0300 (33.4), 227.0347 (5.7), 211.0391 (2.2) | 3.81 | C |
| 50 | isorhamnetin-O-dihexoside | C28H32O17 | 639.1567 | 639.1575 (100), 477.1039 (34.6), 315.0514 (56.7), 300.0275 (11.8), 314.0429 (12.6), 285.0408 (6.4), 270.0172 (20.8), 242.0218 (14.0), 227.0344 (0.7), 151.0027 (5.5), 107.0124 (1.3) | 4.04 | C |
| 51 | rutin * | C27H30O16 | 609.1461 | 609.1469 (100), 301.0352 (39.6), 300.0279 (64.0), 271.0249 (29.6), 255.0298 (14.6), 243.0296 (6.4), 227.0345 (2.2), 178.9975 (3.4), 163.0015 (0.8), 151.0025 (5.6), 121.0286 (0.9), 107.0125 (1.0) | 5.07 | A, B, C, D, E |
| 52 | isorhamnetin-O-pentosylhexoside | C27H30O16 | 609.1461 | 609.1464 (100), 315.0504 (19.9), 314.0436 (85.1), 299.0196 (18.4), 271.0252 (22.0), 243.0297 (20.1), 227.0350 (5.1), 178.9978 (1.3), 151.0023 (34.0) | 5.16 | A, B, C, D, E |
| 53 | isoquercitrin * | C21H20O12 | 463.0882 | 463.0887 (100), 301.0352 (44.4), 300.0278 (69.8), 271.0250 (46.2), 255.0300 (20.0), 243.0298 (11.0), 227.0346 (4.6), 211.0389 (1.4), 178.9976 (4.4), 151.0026 (7.8), 121.0280 (1.5), 107.0123 (2.8) | 5.27 | A, B, C, D, E |
| 54 | quercetin 7-O-hexuronide | C21H18O13 | 477.0675 | 477.0671 (47.4), 301.0356 (100), 283.0245 (2.1), 255.0302 (3.1), 227.0343 (2.0), 211.0396 (1.8), 178.9976 (9.9), 163.0028 (2.4), 151.0025 (20.2), 121.0281 (6.3), 107.0124 (8.9) | 5.22 | A, B, C, D |
| 55 | luteolin-O-hexuronide | C21H18O12 | 461.0726 | 461.0730 (39.5), 285.0406 (100), 257.0457 (4.6), 229.0505 (6.0), 213.0544 (2.0), 175.0242 (5.8), 151.0023 (0.9), 107.0125 (2.5) | 5.84 | A, B |
| 56 | kaempferol 7-O-rutinoside * | C27H30O15 | 593.1512 | 593.1516 (100), 285.0405 (90.5), 255.0298 (42.9), 227.0346 (31.3), 163.0025 (1.1) | 5.62 | A, B, D, E |
| 57 | quercetin 3-O-acetylhexoside | C23H22O13 | 505.0988 | 505.0996 (100), 463.0891 (0.8), 301.0351 (34.1), 300.0278 (88.9), 271.0251 (42.8), 255.0299 (21.5), 243.0297 (11.7), 227.0343 (3.1), 178.9976 (2.5), 163.0027 (2.8), 151.0024 (7.8), 121.0283 (1.1), 107.0124 (2.4) | 5.61 | A, B, C, D |
| 58 | kaempferol-3-O-glucoside * | C21H20O11 | 447.0933 | 447.0938 (100), 285.0401 (21.4), 284.0329 (51.3), 255.0300(38.4), 227.0347 (40.6), 151.0024 (2.7), | 5.87 | B, C |
| 59 | isorhamnetin 3-O-glucoside * | C22H22O12 | 477.1039 | 477.1041 (100), 315.0495 (9.7), 314.0437 (51.2), 299.0213 (3.2), 271.0251 (18.8), 257.0460 (3.9), 243.0299 (22.3), 227.0341 (2.9), 215.0340 (3.7), 178.9972 (0.6), 151.0021 (1.7) | 6.02 | A, B, C, D, E |
| 60 | isorhamnetin-O-hexuronide | C22H20O13 | 491.0831 | 491.0836 (48.3), 315.0515 (100), 300.0278 (29.1), 271.0251 24.7), 255.0299 (10.7), 227.0347 (1.8), 175.0238 (5.7), 151.0029 (2.6), 107.0122 (0.8) | 6.09 | A, B |
| 61 | quercetin-3-O-rhamnoside (quercitrin) * | C21H20O11 | 447.0933 | 447.0936 (100), 301.0355 (81.9), 300.0278 (22.24), 271.0248 (1.7), 255.0298 (2.0), 227.0352 (1.6), 178.9974 (2.6), 151.0025 (40.6), 121.0281 (8.9), 107.0124 (15.2) | 6.76 | B |
| 62 | luteolin * | C15H10O6 | 285.0405 | 285.0406 (100), 175.0392 (3.0), 151.0024 (4.7), 133.0282 (22.8), 107.0124 (3.7) | 7.56 | C |
| 63 | Quercetin * | C15H10O7 | 301.0354 | 301.0356 (100), 273.0411 (3.3), 257.0469 (1.8), 245.0444 (0.8), 229.0500 (0.6), 215.1699 (0.3), 178.9977 (21.3), 151.0024 (49.4), 121.0281 (14.2), 107.0123 (12.9) | 7.61 | B, C, D, E |
| 64 | apigenin * | C15H10O5 | 269.0456 | 269.0458 (100), 225.0549(1.9), 201.0550 (0.9), 151.0025 (5.7), 121.0282 (1.3), 117.0332 (18.4), 107.0124 (5.3) | 8.62 | C |
| 65 | kaempferol * | C15H10O6 | 285.0405 | 285.0406 (100), 257.0465 (0.8), 243.0298 (0.2), 227.0353 (0.9), 211.0397 (1.3), 151.0025 (1.3), 107.0123 (1.2) | 8.85 | C, E |
| 66 | chrysoeriol * | C16H12O6 | 299.0561 | 299.0564 (63.8), 284.0329 (100), 255.0300 (46.5), 227.0344 (38.2), 211.0394 (1.5), 151.0024 (0.3) | 9.32 | C |
| 67 | cirsiliol | C17H14O7 | 329.0667 | 329.0672 (100), 314.0441 (43.6), 299.0199(89.6), 271.0248 (55.9), 243.0296 (6.5), 227.0345 (3.4), 211.1333 (6.8) | 9.60 | C |
| 68 | cirsimaritin | C17H14O6 | 313.0718 | 313.0721 (100), 298.0483 (65.2), 283.0251 (52.5), 255.0298 (64.1), 227.0341 (4.4), 211.0396 (5.7) | 12.27 | C |
| № | Compounds | Species | ||||
|---|---|---|---|---|---|---|
| J.maritima | J.pancici | S.hercynicus | S.ovatus | S.rupestris | ||
| 1 | protocatechuic acid-O-hexoside | 12.64 | 15.27 | 55.32 | 38.45 | 100 |
| 2 | vanillic acid 4-O-hexoside | 100 | 0 | 56.87 | 0 | 52.35 |
| 4 | vanillic acid | 0 | 0 | 66.47 | 84.99 | 100 |
| 5 | protocatechuic acid | 68.03 | 100 | 43.93 | 51.5 | 48.22 |
| 6 | p-hydroxyphenylacetic acid-O-hexoside | 0 | 0 | 36.05 | 68.05 | 100 |
| 8 | syringic acid 4-O-hexoside | 24.14 | 100 | 17.32 | 51.69 | 33.42 |
| 9 | neochlorogenic acid | 28.06 | 100 | 1.42 | 3.21 | 6.19 |
| 10 | caffeic acid-O-hexoside | 1.86 | 9.82 | 34.24 | 23.69 | 100 |
| 11 | 4-hydroxybenzoic acid-O-hexoside | 15.24 | 0 | 32.1 | 50.1 | 100 |
| 12 | esculetin-O-hexoside | 12 | 19.44 | 45.48 | 100 | 24.71 |
| 13 | 4-hydroxybenzoic acid | 71.02 | 42.4 | 60.25 | 46.55 | 100 |
| 15 | ferulic acid-O-hexoside | 0 | 0 | 18.03 | 70.66 | 100 |
| 16 | gentisic acid | 0 | 17.77 | 5.99 | 3.02 | 100 |
| 17 | 4-hydroxybenzoic acid-O-hexoside isomer | 0 | 0 | 25.97 | 78.36 | 100 |
| 19 | caffeic acid-O-hexoside | 13.08 | 1.24 | 4.61 | 14.54 | 100 |
| 20 | quinic acid | 100 | 93.53 | 24.24 | 75.9 | 70.84 |
| 21 | chlorogenic acida | 100 | 94.47 | 28.47 | 77.32 | 71.97 |
| 22 | caffeic acid-O-hexoside isomer I | 18.62 | 38.53 | 48.92 | 44.77 | 100 |
| 23 | 4-caffeoylquinic acid | 85.4 | 100 | 1.78 | 4.3 | 7.54 |
| 24 | p-hydroxyphenylacetic acid | 100 | 0 | 0 | 0 | 23.6 |
| 25 | coumaric acid-O-hexoside | 24.08 | 70.79 | 81.31 | 100 | 12.01 |
| 26 | p-coumaric acid | 25.08 | 73.49 | 83.38 | 100 | 12.42 |
| 27 | caffeic acid | 17.16 | 12.38 | 33.55 | 37.96 | 100 |
| 28 | caffeic acid-O-hexoside isomer II | 4.28 | 3.58 | 0 | 12.49 | 100 |
| 29 | 5-p-coumaroylquinic acid | 100 | 31.1 | 26.13 | 63.12 | 18.11 |
| 30 | 3-hydroxy-dihydroxy-5-caffeoylquinic acid | 0 | 100 | 40.05 | 86.58 | 31.91 |
| 35 | 5-feruoylquinic acid | 3.74 | 0 | 40.78 | 100 | 8.11 |
| 36 | m-coumaric acid | 68.8 | 0 | 49.92 | 41.24 | 100 |
| 37 | 3,4-dicaffeoylquinic acid | 100 | 1.7 | 23.32 | 1.57 | 3.38 |
| 38 | 3,5-dicaffeoylquinic acid | 100 | 51.54 | 9.21 | 59.82 | 95.36 |
| 39 | 1,5-dicaffeoylquinic acid | 100 | 21.84 | 9.53 | 27.34 | 45.34 |
| 40 | 4,5-dicaffeoylquinic acid | 100 | 31.87 | 4.14 | 14.58 | 33.42 |
| 42 | salicilic acid | 16.54 | 0 | 42.11 | 40.7 | 100 |
| 43 | 3-p-coumaroyl-5-caffeoylquinic acid | 100 | 0 | 0 | 80.53 | 59.01 |
| 44 | 3-caffeoyl-5-p-coumaroylquinic acid | 100 | 2.33 | 0 | 5.89 | 4.4 |
| 45 | 3-feruoyl-5-caffeoylquinic acid | 17.99 | 0 | 16.99 | 100 | 13.26 |
| 46 | 3-caffeoyl-5-feruoylquinic acid | 100 | 0 | 14.92 | 75.24 | 0 |
| 47 | 3,4,5-tricaffeoylquinic acid | 100 | 0 | 0 | 3.95 | 7.03 |
| 48 | 6, 8-di-C-hexosyl-naringenin | 37.7 | 100 | 0 | 0 | 0 |
| 51 | rutin | 40.39 | 100 | 1.62 | 47.2 | 0.13 |
| 52 | isorhamnetin-O-pentosylhexoside | 12.03 | 65.54 | 0 | 100 | 0.64 |
| 53 | isoquercitrin | 100 | 22.06 | 1.19 | 81.87 | 29.95 |
| 54 | quercetin 7-O-hexuronide | 0 | 6.91 | 46.7 | 100 | 0.98 |
| 55 | luteolin-O-hexuronide | 0 | 0 | 35.07 | 100 | 0 |
| 56 | kaempferol 7-O-rutinoside | 39.27 | 100 | 1.89 | 58.25 | 0 |
| 57 | quercetin 3-O-acetylhexoside | 0 | 1.07 | 1.66 | 85.28 | 100 |
| 58 | kaempferol-3-O-glucoside | 0 | 0 | 0 | 63.53 | 100 |
| 59 | isorhamnetin 3-O-glucoside | 100 | 7.5 | 0.73 | 6.28 | 40.77 |
| 60 | isorhamnetin-O-hexuronide | 0 | 0 | 34.34 | 100 | 0 |
| 63 | quercetin | 100 | 25.2 | 0 | 27.19 | 17.63 |
| 65 | kaempferol | 100 | 0 | 0 | 0 | 83.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voynikov, Y.; Balabanova, V.; Gevrenova, R.; Zheleva-Dimitrova, D. Chemophenetic Approach to Selected Senecioneae Species, Combining Morphometric and UHPLC-HRMS Analyses. Plants 2023, 12, 390. https://doi.org/10.3390/plants12020390
Voynikov Y, Balabanova V, Gevrenova R, Zheleva-Dimitrova D. Chemophenetic Approach to Selected Senecioneae Species, Combining Morphometric and UHPLC-HRMS Analyses. Plants. 2023; 12(2):390. https://doi.org/10.3390/plants12020390
Chicago/Turabian StyleVoynikov, Yulian, Vessela Balabanova, Reneta Gevrenova, and Dimitrina Zheleva-Dimitrova. 2023. "Chemophenetic Approach to Selected Senecioneae Species, Combining Morphometric and UHPLC-HRMS Analyses" Plants 12, no. 2: 390. https://doi.org/10.3390/plants12020390
APA StyleVoynikov, Y., Balabanova, V., Gevrenova, R., & Zheleva-Dimitrova, D. (2023). Chemophenetic Approach to Selected Senecioneae Species, Combining Morphometric and UHPLC-HRMS Analyses. Plants, 12(2), 390. https://doi.org/10.3390/plants12020390










