Silencing of a Pectin Acetylesterase (PAE) Gene Highly Expressed in Tobacco Pistils Negatively Affects Pollen Tube Growth
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA Extractions and RT-qPCR
2.3. In Situ Hybridization
2.4. Cloning Procedures
2.5. Plant Transformation
2.6. Scanning Electron Microscopy (SEM)
2.7. Pollen Tube Growth Analyses
2.8. Bioinformatics Analyses
2.8.1. Sequence Retrieval
2.8.2. Phylogenetic Analyses
2.8.3. Mining of Expression Data
2.8.4. Structure Prediction and Structural Alignment
3. Results
3.1. NtPAE1 Encodes a Flower-Specific Pectin Acetylesterase Preferentially Expressed in Pistils
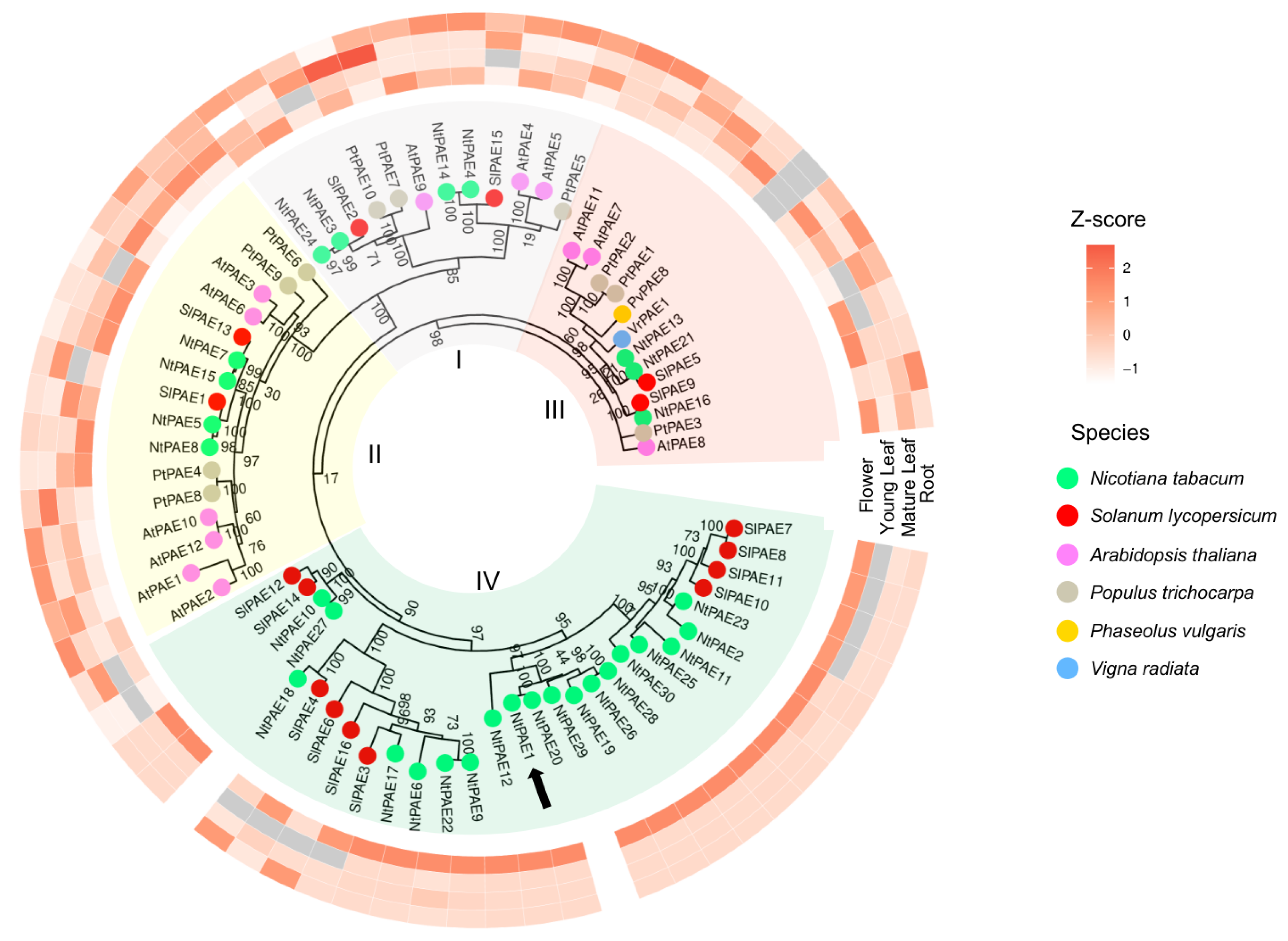
3.2. Genome-Wide Identification of PAE Encoding Genes in N. tabacum
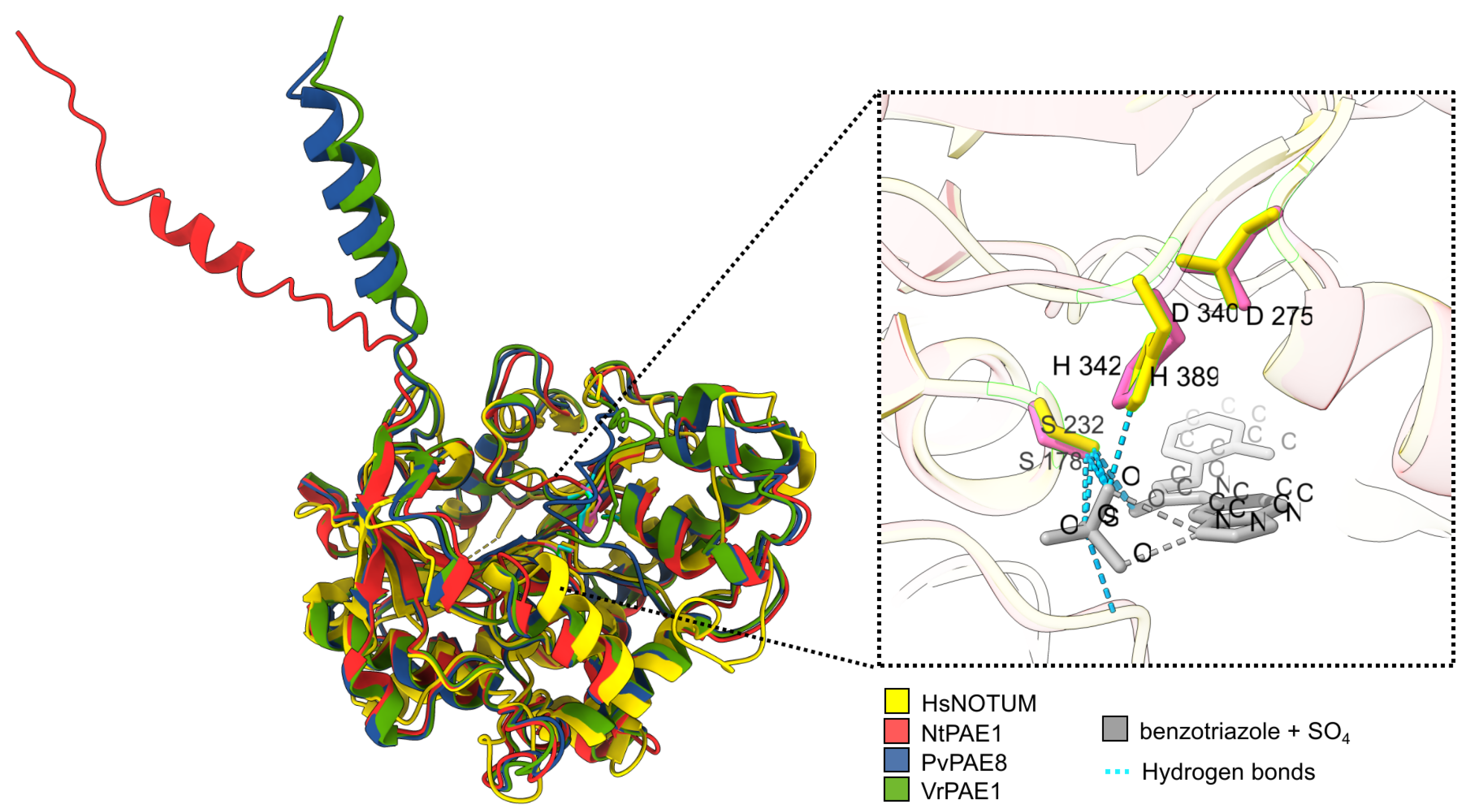
3.3. NtPAE1 Is Structurally Very Similar to Functionally Characterized PAEs and Other Carboxylesterases
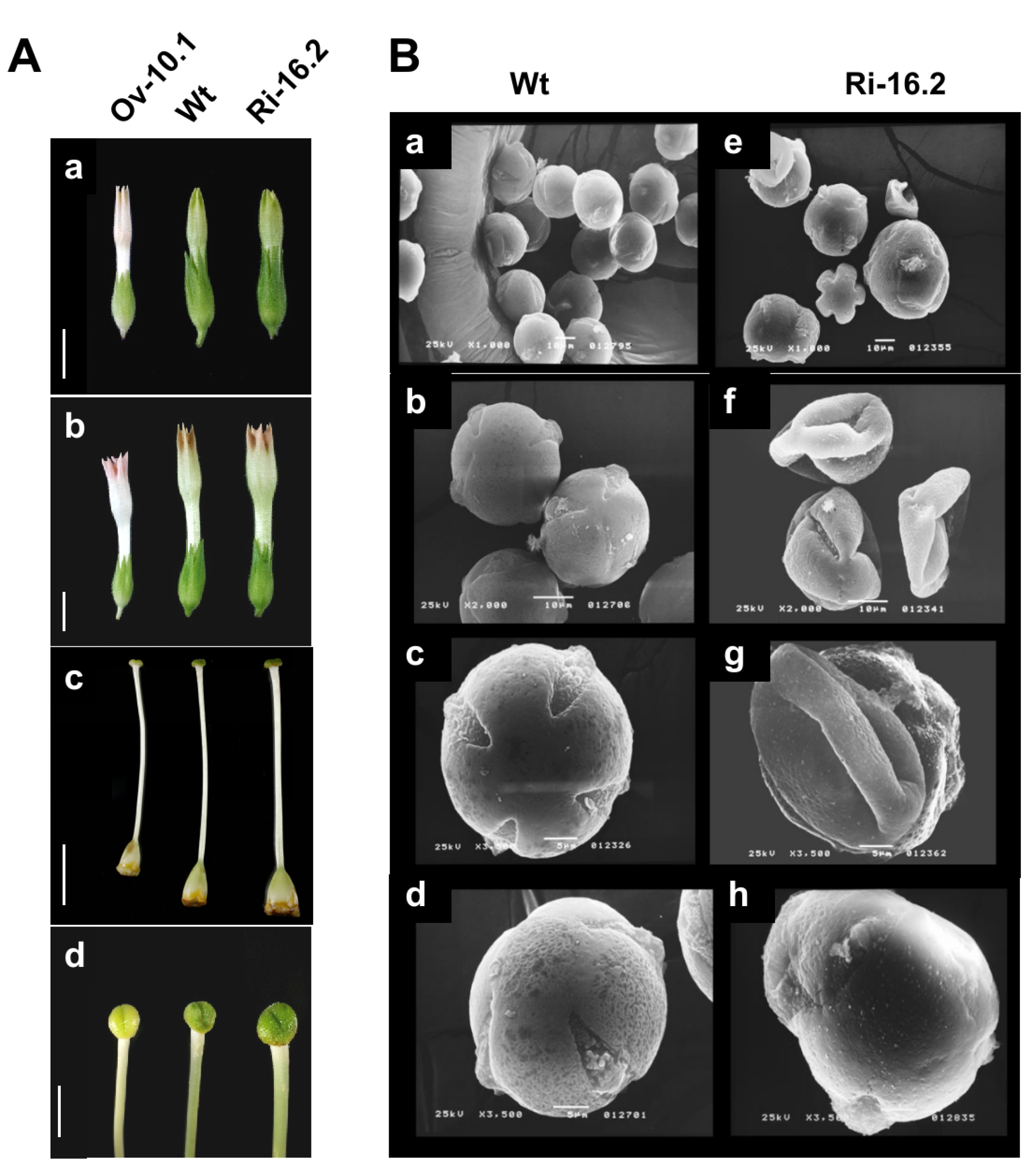
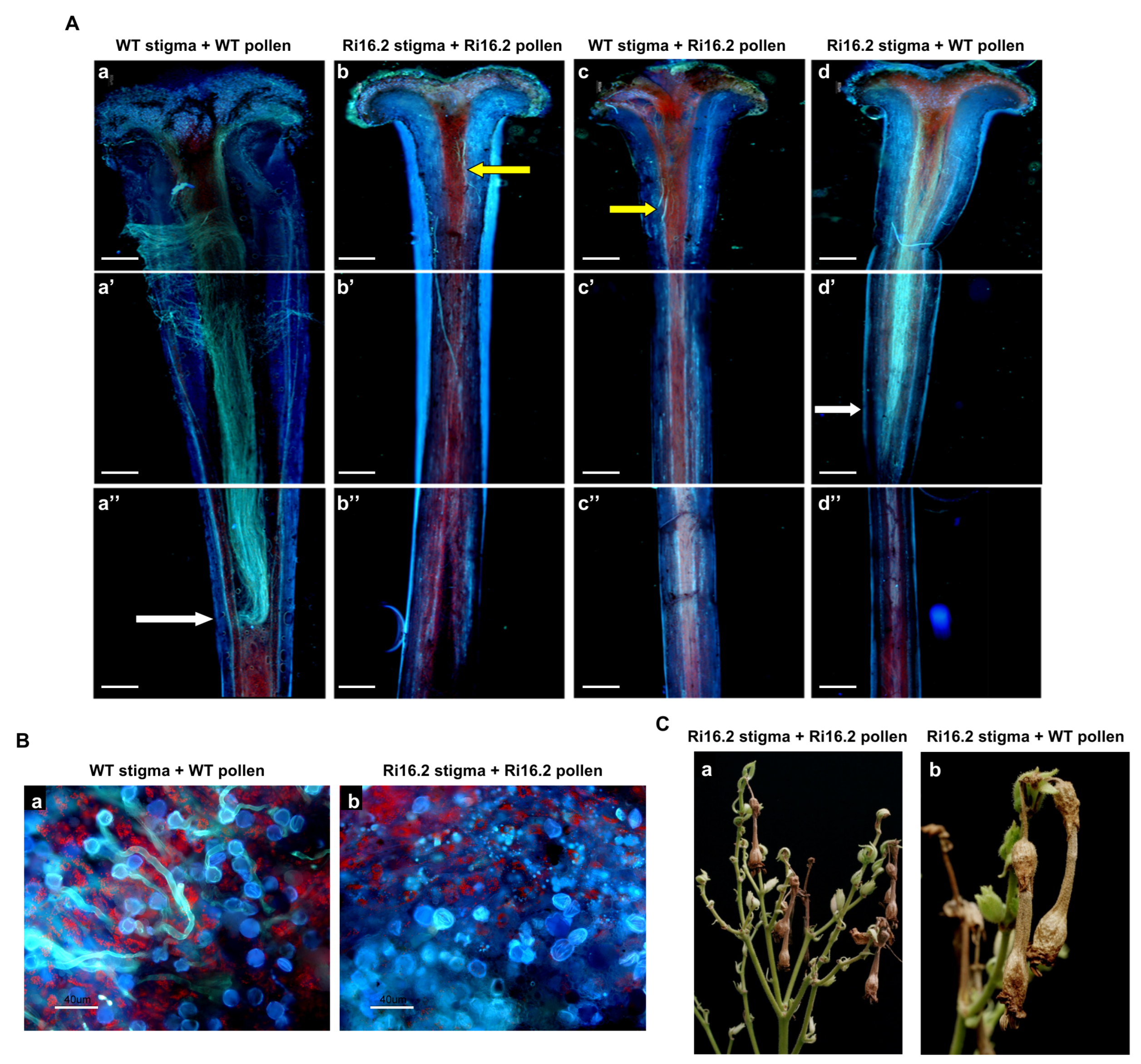
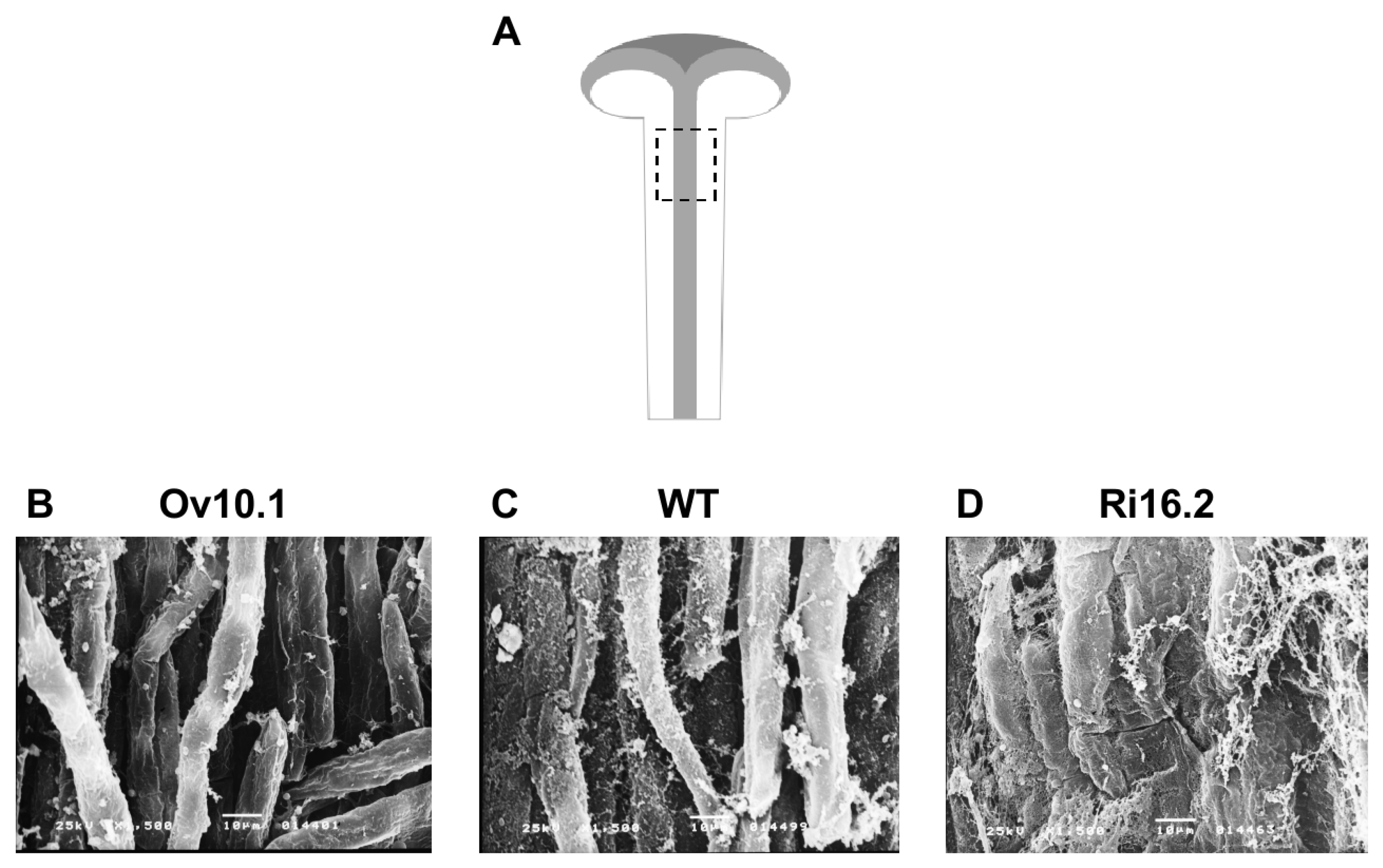
3.4. Altered Expression of the NtPAE1 Gene Affects Flower Development and Fertility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bell, J.; Hicks, G. Transmitting tissue in the pistil of tobacco: Light and electron microscopic observations. Planta 1976, 131, 187–200. [Google Scholar] [CrossRef]
- Goldman, M.H.; Goldberg, R.B.; Mariani, C. Female sterile tobacco plants are produced by stigma-specific cell ablation. EMBO J. 1994, 13, 2976. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; Kristen, U. Developmental Aspects of Ultrastructure, Histochemistry and Receptivity of the Stigma of Nicotiana sylvestris. Ann. Bot. 1987, 60, 427–437. [Google Scholar] [CrossRef]
- Wolters-Arts, M.; Lush, W.M.; Mariani, C. Lipids are required for directional pollen-tube growth. Nature 1998, 392, 818–821. [Google Scholar] [CrossRef]
- Linskens, H.; Kroh, M. Regulation of Pollen Tube Growth. Curr. Top. Dev. Biol. 1970, 5, 89–113. [Google Scholar] [CrossRef]
- Daher, F.B.; Braybrook, S.A. How to let go: Pectin and plant cell adhesion. Front. Plant Sci. 2015, 6, 523. [Google Scholar] [CrossRef]
- Shin, Y.; Chane, A.; Jung, M.; Lee, Y. Recent advances in understanding the roles of pectin as an active participant in plant signaling networks. Plants 2021, 10, 1712. [Google Scholar] [CrossRef]
- Louvet, R.; Cavel, E.; Gutierrez, L.; Guénin, S.; Roger, D.; Gillet, F.; Guerineau, F.; Pelloux, J. Comprehensive expression profiling of the pectin methylesterase gene family during silique development in Arabidopsis thaliana. Planta 2006, 224, 782–791. [Google Scholar] [CrossRef]
- Wang, M.; Yuan, D.; Gao, W.; Li, Y.; Tan, J.; Zhang, X. A Comparative Genome Analysis of PME and PMEI Families Reveals the Evolution of Pectin Metabolism in Plant Cell Walls. PLoS ONE 2013, 8, e72082. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, H.; Qin, X.; Chen, K.; Zhao, J.; Zhao, Y.; Yue, B. Genome-wide identification, phylogeny and expression analysis of the PME and PMEI gene families in maize. Sci. Rep. 2019, 9, 19918. [Google Scholar] [CrossRef]
- Pilling, J.; Willmitzer, L.; Bücking, H.; Fisahn, J. Inhibition of a ubiquitously expressed pectin methyl esterase in Solanum tuberosum L. affects plant growth, leaf growth polarity, and ion partitioning. Planta 2004, 219, 32–40. [Google Scholar] [CrossRef]
- Saffer, A.M. Expanding roles for pectins in plant development. J. Integr. Plant Biol. 2018, 60, 910–923. [Google Scholar] [CrossRef]
- Bethke, G.; Grundman, R.E.; Sreekanta, S.; Truman, W.; Katagiri, F.; Glazebrook, J. Arabidopsis PECTIN METHYLESTERASEs contribute to immunity against Pseudomonas syringae. Plant Physiol. 2014, 164, 1093–1107. [Google Scholar] [CrossRef]
- Del Corpo, D.; Fullone, M.R.; Miele, R.; Lafond, M.; Pontiggia, D.; Grisel, S.; Kieffer-Jaquinod, S.; Giardina, T.; Bellincampi, D.; Lionetti, V. AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea. Mol. Plant Pathol. 2020, 21, 1620–1633. [Google Scholar] [CrossRef]
- Huang, Y.C.; Wu, H.C.; Wang, Y.D.; Liu, C.H.; Lin, C.C.; Luo, D.L.; Jinn, T.L. PECTIN METHYLESTERASE34 Contributes to Heat Tolerance through Its Role in Promoting Stomatal Movement. Plant Physiol. 2017, 174, 748–763. [Google Scholar] [CrossRef]
- Wu, H.C.; Bulgakov, V.P.; Jinn, T.L. Pectin methylesterases: Cell wall remodeling proteins are required for plant response to heat stress. Front. Plant Sci. 2018, 871, 1612. [Google Scholar] [CrossRef]
- Yan, J.; He, H.; Fang, L.; Zhang, A. PECTIN METHYLESTERASE31 positively regulates salt stress tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2018, 496, 497–501. [Google Scholar] [CrossRef]
- Leroux, C.; Bouton, S.; Kiefer-Meyer, M.C.; Fabrice, T.N.; Mareck, A.; Guénin, S.; Fournet, F.; Ringli, C.; Pelloux, J.; Driouich, A.; et al. PECTIN METHYLESTERASE48 Is Involved in Arabidopsis Pollen Grain Germination. Plant Physiol. 2015, 167, 367–380. [Google Scholar] [CrossRef]
- Bosch, M.; Cheung, A.Y.; Hepler, P.K. Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 2005, 138, 1334–1346. [Google Scholar] [CrossRef]
- Bosch, M.; Hepler, P.K. Silencing of the tobacco pollen pectin methylesterase NtPPME1 results in retarded in vivo pollen tube growth. Planta 2006, 223, 736–745. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Lin, S.; Yu, Y.; Huang, L.; Cao, J. The putative pectin methylesterase gene, BcMF23a, is required for microspore development and pollen tube growth in Brassica campestris. Plant Cell Rep. 2018, 37, 1003–1009. [Google Scholar] [CrossRef]
- Duan, Q.; Liu, M.C.J.; Kita, D.; Jordan, S.S.; Yeh, F.L.J.; Yvon, R.; Carpenter, H.; Federico, A.N.; Garcia-Valencia, L.E.; Eyles, S.J.; et al. FERONIA controls pectin- and nitric oxide-mediated male–female interaction. Nature 2020, 579, 561–566. [Google Scholar] [CrossRef]
- Lu, Y.; Lauter, A.N.M.; Makkena, S.; Scott, M.P.; Evans, M.M. Insights into the molecular control of cross-incompatibility in Zea mays. Plant Reprod. 2020, 33, 117–128. [Google Scholar] [CrossRef]
- Wen, B.; Zhang, F.; Wu, X.; Li, H. Characterization of the Tomato (Solanum lycopersicum) Pectin Methylesterases: Evolution, Activity of Isoforms and Expression During Fruit Ripening. Front. Plant Sci. 2020, 11, 238. [Google Scholar] [CrossRef]
- Jeong, H.Y.; Nguyen, H.P.; Eom, S.H.; Lee, C. Integrative analysis of pectin methylesterase (PME) and PME inhibitors in tomato (Solanum lycopersicum): Identification, tissue-specific expression, and biochemical characterization. Plant Physiol. Biochem. 2018, 132, 557–565. [Google Scholar] [CrossRef]
- Sun, J.; Tian, Z.; Li, X.; Li, S.; Li, Z.; Wang, J.; Hu, Z.; Chen, H.; Guo, C.; Xie, M.; et al. Systematic analysis of the pectin methylesterase gene family in Nicotiana tabacum and reveal their multiple roles in plant development and abiotic stresses. Front. Plant Sci. 2022, 13, 998841. [Google Scholar] [CrossRef]
- Philippe, F.; Pelloux, J.; Rayon, C. Plant pectin acetylesterase structure and function: New insights from bioinformatic analysis. BMC Genom. 2017, 18, 456. [Google Scholar] [CrossRef]
- Liners, F.; Gaspar, T.; Van Cutsem, P. Acetyl- and methyl-esterification of pectins of friable and compact sugar-beet calli: Consequences for intercellular adhesion. Planta 1994, 192, 545–556. [Google Scholar] [CrossRef]
- Gou, J.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef]
- de Souza, A.; Hull, P.A.; Gille, S.; Pauly, M. Identification and functional characterization of the distinct plant pectin esterases PAE8 and PAE9 and their deletion mutants. Planta 2014, 240, 1123–1138. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, S.; Suo, J.; Chang, R.; Xu, X.; Xu, Z.; Yang, C.; Qu, C.; Liu, G. Bioinformatics analysis of PAE family in Populus trichocarpa and responsiveness to carbon and nitrogen treatment. 3 Biotech 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Roig-Oliver, M.; Rayon, C.; Roulard, R.; Fournet, F.; Bota, J.; Flexas, J. Reduced photosynthesis in Arabidopsis thaliana atpme17.2 and atpae11.1 mutants is associated to altered cell wall composition. Physiol. Plant. 2021, 172, 1439–1451. [Google Scholar] [CrossRef]
- Breton, C.; Bordenave, M.; Richard, L.; Pernollet, J.C.; Huet, J.C.; Pérez, S.; Goldberg, R. PCR cloning and expression analysis of a cDNA encoding a pectinacetylesterase from Vigna radiata L. FEBS Lett. 1996, 388, 139–142. [Google Scholar] [CrossRef]
- Orfila, C.; Degan, F.D.; Jørgensen, B.; Scheller, H.V.; Ray, P.M.; Ulvskov, P. Expression of mung bean pectin acetyl esterase in potato tubers: Effect on acetylation of cell wall polymers and tuber mechanical properties. Planta 2012, 236, 185–196. [Google Scholar] [CrossRef]
- Li, Q.; Fu, J.; Qin, X.; Yang, W.; Qi, J.; Li, Z.; Chen, S.; He, Y. Systematic analysis and functional validation of citrus pectin acetylesterases (CsPAES) reveals that CsPAE2 negatively regulates citrus bacterial canker development. Int. J. Mol. Sci. 2020, 21, 9429. [Google Scholar] [CrossRef]
- Palmer, J.P.; Pajak, A.; Robson, B.; Zhang, B.; Joshi, J.; Diapari, M.; Pauls, K.P.; Marsolais, F. Pectin acetylesterase 8 influences pectin acetylation in the seed coat, seed imbibition, and dormancy in common bean (Phaseolus vulgaris L.). Legume Sci. 2021, 4, e130. [Google Scholar] [CrossRef]
- Quiapim, A.C.; Brito, M.S.; Bernardes, L.A.; DaSilva, I.; Malavazi, I.; DePaoli, H.C.; Molfetta-Machado, J.B.; Giuliatti, S.; Goldman, G.H.; Goldman, M.H.S. Analysis of the Nicotiana tabacum stigma/style transcriptome reveals gene expression differences between wet and dry stigma species. Plant Physiol. 2009, 149, 1211–1230. [Google Scholar] [CrossRef]
- Koltunow, A.M.; Truettner, J.; Cox, K.H.; Wallroth, M.; Goldberg, R.B. Different Temporal and Spatial Gene Expression Patterns Occur during Anther Development. Plant Cell 1990, 2, 1201–1224. [Google Scholar] [CrossRef]
- Jackson, D.P. In-situ hybridisation in plants. In Molecular Plant Pathology: A Practical Approach, 1st ed.; Gurr, S.J., McPherson, M.J., Bowles, D.J., Eds.; Oxford University Press: Oxford, UK, 1992; Chapter 85; pp. 163–174. [Google Scholar]
- Brasileiro, A.C.M.; Carneiro, V.T.d.C. Manual de Transformação Genética de Plantas, 2nd ed.; Embrapa: Distrito Federal, Brazil, 2015; p. 453. [Google Scholar]
- Johansen, D.A. Plant Microtechnique, 1st ed.; McGraw-Hill Publishing Company, Ltd.: London, UK, 1940; p. 523. [Google Scholar]
- Hawes, C.; Satiat-Jeunemaitre, B. Plant Cell Biology: A Practical Approach; OUP Oxford: Oxford, UK, 2001; Volume 250. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- O’Rourke, J.A.; Iniguez, L.P.; Fu, F.; Bucciarelli, B.; Miller, S.S.; Jackson, S.A.; McClean, P.E.; Li, J.; Dai, X.; Zhao, P.X.; et al. An RNA-Seq based gene expression atlas of the common bean. BMC Genom. 2014, 15, 866. [Google Scholar] [CrossRef]
- Sierro, N.; Battey, J.N.; Ouadi, S.; Bakaher, N.; Bovet, L.; Willig, A.; Goepfert, S.; Peitsch, M.C.; Ivanov, N.V. The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 2014, 5, 3833. [Google Scholar] [CrossRef]
- Yu, G.; Smith, D.K.; Zhu, H.; Guan, Y.; Lam, T.T.Y. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017, 8, 28–36. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. lDDT: A local superposition-free score for comparing protein structures and models using distance difference tests. Bioinformatics 2013, 29, 2722. [Google Scholar] [CrossRef]
- Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Huang, C.C.; Ferrin, T.E. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinform. 2006, 7, 339. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Armenteros, J.J.A.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, D.; Yuchi, J.; He, F.; Jiang, Y.; Cai, S.; Li, J.; Xu, D. MusiteDeep: A deep-learning based webserver for protein post-translational modification site prediction and visualization. Nucleic Acids Res. 2020, 48, W140–W146. [Google Scholar] [CrossRef] [PubMed]
- Brito, M.S.; Bertolino, L.T.; Cossalter, V.; Quiapim, A.C.; DePaoli, H.C.; Goldman, G.H.; Teixeira, S.P.; Goldman, M.H. Pollination triggers female gametophyte development in immature Nicotiana tabacum flowers. Front. Plant Sci. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Edwards, K.D.; Fernandez-Pozo, N.; Drake-Stowe, K.; Humphry, M.; Evans, A.D.; Bombarely, A.; Allen, F.; Hurst, R.; White, B.; Kernodle, S.P.; et al. A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genom. 2017, 18, 448. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Leitch, I.J.; Hanson, L.; Lim, K.Y.; Kovarik, A.; Chase, M.W.; Clarkson, J.J.; Leitch, A.R. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae). Ann. Bot. 2008, 101, 805–814. [Google Scholar] [CrossRef]
- Atkinson, B.N.; Steadman, D.; Zhao, Y.; Sipthorp, J.; Vecchia, L.; Ruza, R.R.; Jeganathan, F.; Lines, G.; Frew, S.; Monaghan, A.; et al. Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen. Med. Chem. Commun. 2019, 10, 1361–1369. [Google Scholar] [CrossRef]
- Hiscock, S.J.; Hoedemaekers, K.; Friedman, W.E.; Dickinson, H.G. The Stigma Surface and Pollen-Stigma Interactions in Senecio squalidus L. (Asteraceae) following Cross (Compatible) and Self (Incompatible) Pollinations. Int. J. Plant Sci. 2002, 163, 1–16. [Google Scholar] [CrossRef]
- Lenartowska, M.; Rodríguez-García, M.I.; Bednarska, E. Immunocytochemical localization of esterified and unesterified pectins in unpollinated and pollinated styles of Petunia hybrida hort. Planta 2001, 213, 182–191. [Google Scholar] [CrossRef]
- Bednarska, E.; Lenartowska, M.; Niekraś, L. Localization of pectins and Ca2+ ions in unpollinated and pollinated wet (Petunia hybrida Hort.) and dry (Haemanthus albiflos L.) stigma. Folia Histochem. Cytobiol. 2005, 43, 249–259. [Google Scholar]
- Broz, A.K.; Bedinger, P.A. Pollen-Pistil Interactions as Reproductive Barriers. Annu. Rev. Plant Biol. 2021, 72, 615–639. [Google Scholar] [CrossRef]
- Cascallares, M.; Setzes, N.; Marchetti, F.; López, G.A.; Distéfano, A.M.; Cainzos, M.; Zabaleta, E.; Pagnussat, G.C. A Complex Journey: Cell Wall Remodeling, Interactions, and Integrity During Pollen Tube Growth. Front. Plant Sci. 2020, 11, 599247. [Google Scholar] [CrossRef]
- Guo, J.; Yang, Z. Exocytosis and endocytosis: Coordinating and fine-tuning the polar tip growth domain in pollen tubes. J. Exp. Bot. 2020, 71, 2428–2438. [Google Scholar] [CrossRef]
- Scholz, P.; Anstatt, J.; Krawczyk, H.E.; Ischebeck, T. Signalling pinpointed to the tip: The complex regulatory network that allows pollen tube growth. Plants 2020, 9, 1098. [Google Scholar] [CrossRef]
- Tonnabel, J.; David, P.; Janicke, T.; Lehner, A.; Mollet, J.C.; Pannell, J.R.; Dufay, M. The Scope for Postmating Sexual Selection in Plants. Trends Ecol. Evol. 2021, 36, 556–567. [Google Scholar] [CrossRef]
- Rejón, J.D.; Zienkiewicz, A.; Rodríguez-García, M.I.; Castro, A.J. Profiling and functional classification of esterases in olive (Olea europaea) pollen during germination. Ann. Bot. 2012, 110, 1035–1045. [Google Scholar] [CrossRef]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lubini, G.; Ferreira, P.B.; Quiapim, A.C.; Brito, M.S.; Cossalter, V.; Pranchevicius, M.C.S.; Goldman, M.H.S. Silencing of a Pectin Acetylesterase (PAE) Gene Highly Expressed in Tobacco Pistils Negatively Affects Pollen Tube Growth. Plants 2023, 12, 329. https://doi.org/10.3390/plants12020329
Lubini G, Ferreira PB, Quiapim AC, Brito MS, Cossalter V, Pranchevicius MCS, Goldman MHS. Silencing of a Pectin Acetylesterase (PAE) Gene Highly Expressed in Tobacco Pistils Negatively Affects Pollen Tube Growth. Plants. 2023; 12(2):329. https://doi.org/10.3390/plants12020329
Chicago/Turabian StyleLubini, Greice, Pedro Boscariol Ferreira, Andréa Carla Quiapim, Michael Santos Brito, Viviane Cossalter, Maria Cristina S. Pranchevicius, and Maria Helena S. Goldman. 2023. "Silencing of a Pectin Acetylesterase (PAE) Gene Highly Expressed in Tobacco Pistils Negatively Affects Pollen Tube Growth" Plants 12, no. 2: 329. https://doi.org/10.3390/plants12020329
APA StyleLubini, G., Ferreira, P. B., Quiapim, A. C., Brito, M. S., Cossalter, V., Pranchevicius, M. C. S., & Goldman, M. H. S. (2023). Silencing of a Pectin Acetylesterase (PAE) Gene Highly Expressed in Tobacco Pistils Negatively Affects Pollen Tube Growth. Plants, 12(2), 329. https://doi.org/10.3390/plants12020329







