Primary Metabolite Screening Shows Significant Differences between Embryogenic and Non-Embryogenic Callus of Tamarillo (Solanum betaceum Cav.)
Abstract
1. Introduction
2. Results
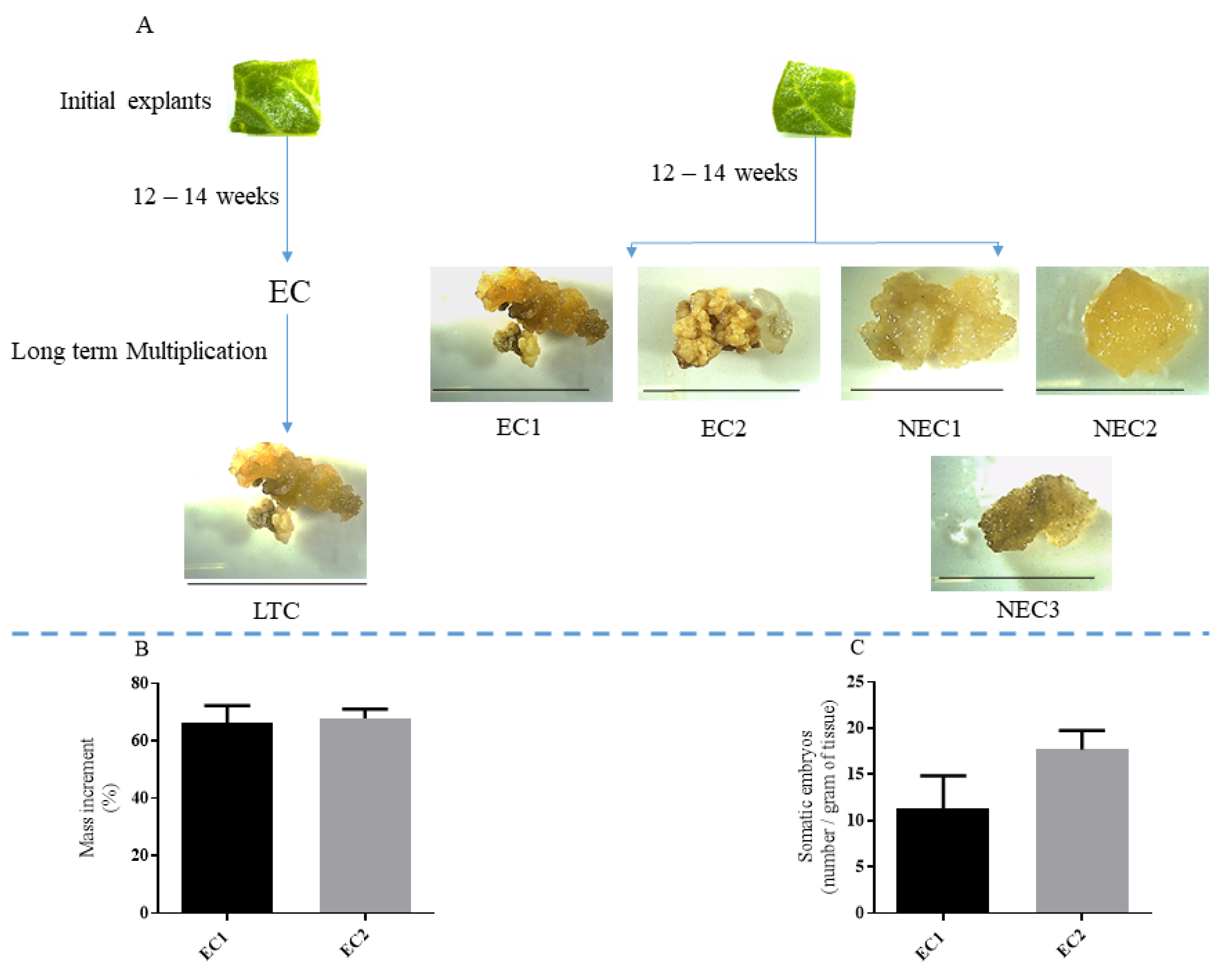
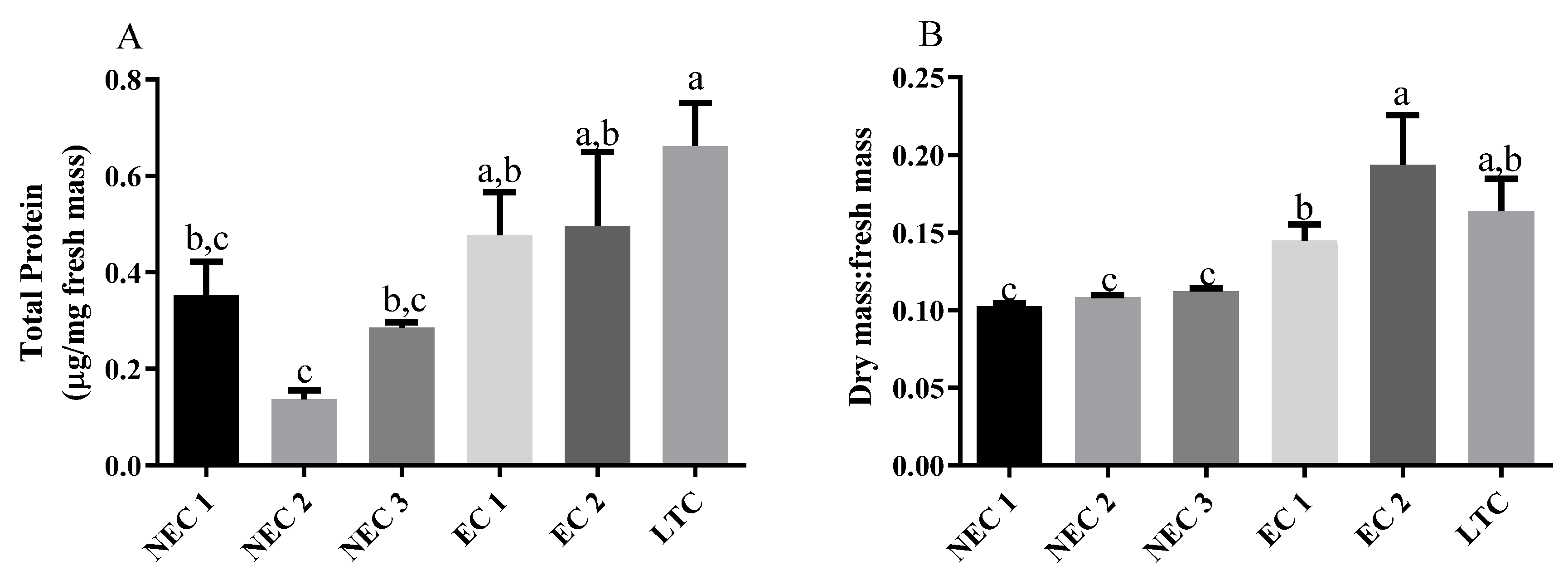
2.1. Callus Culture, Embryo Conversion and Protein Quantification
2.2. Metabolite Identification
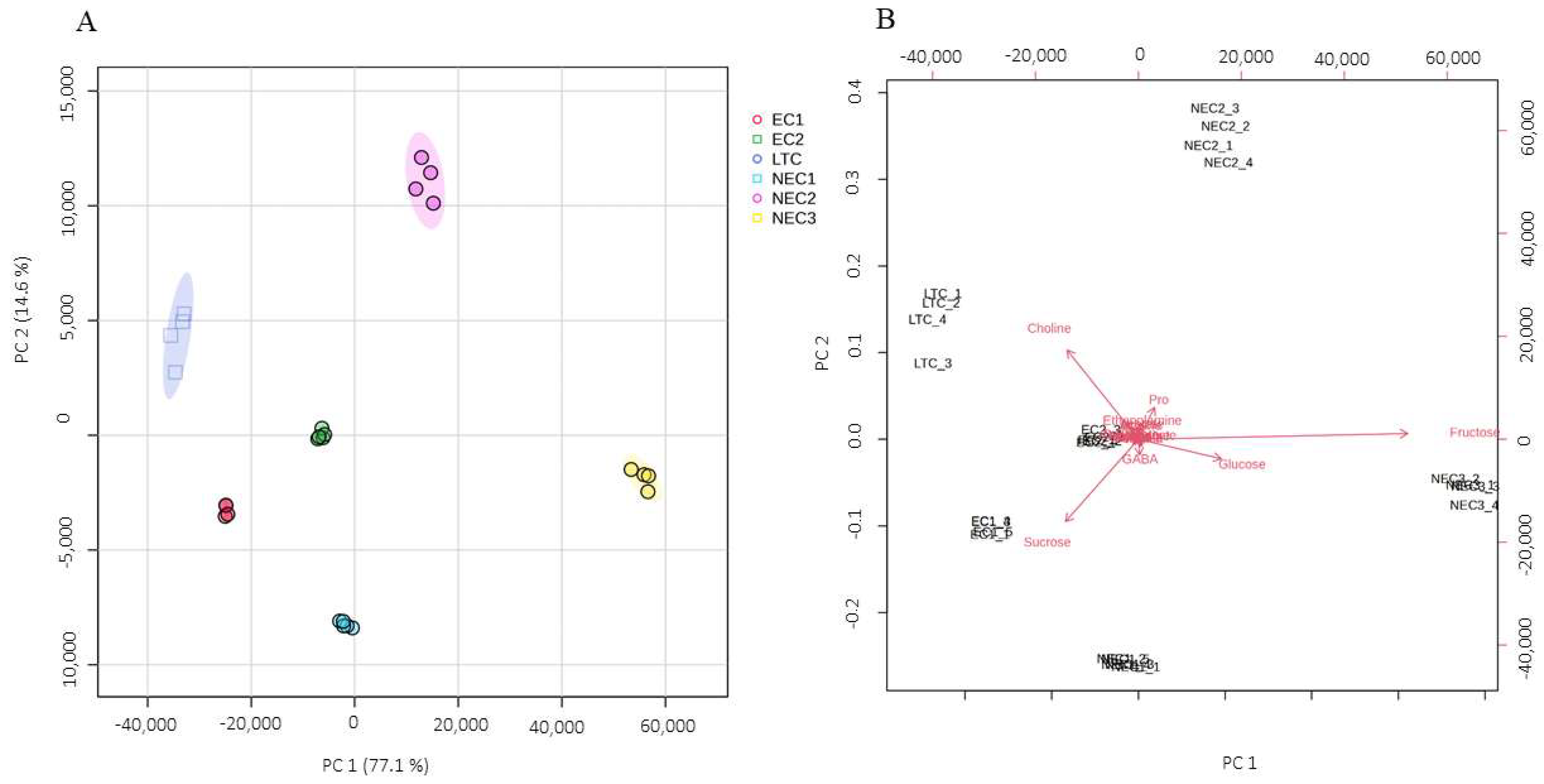
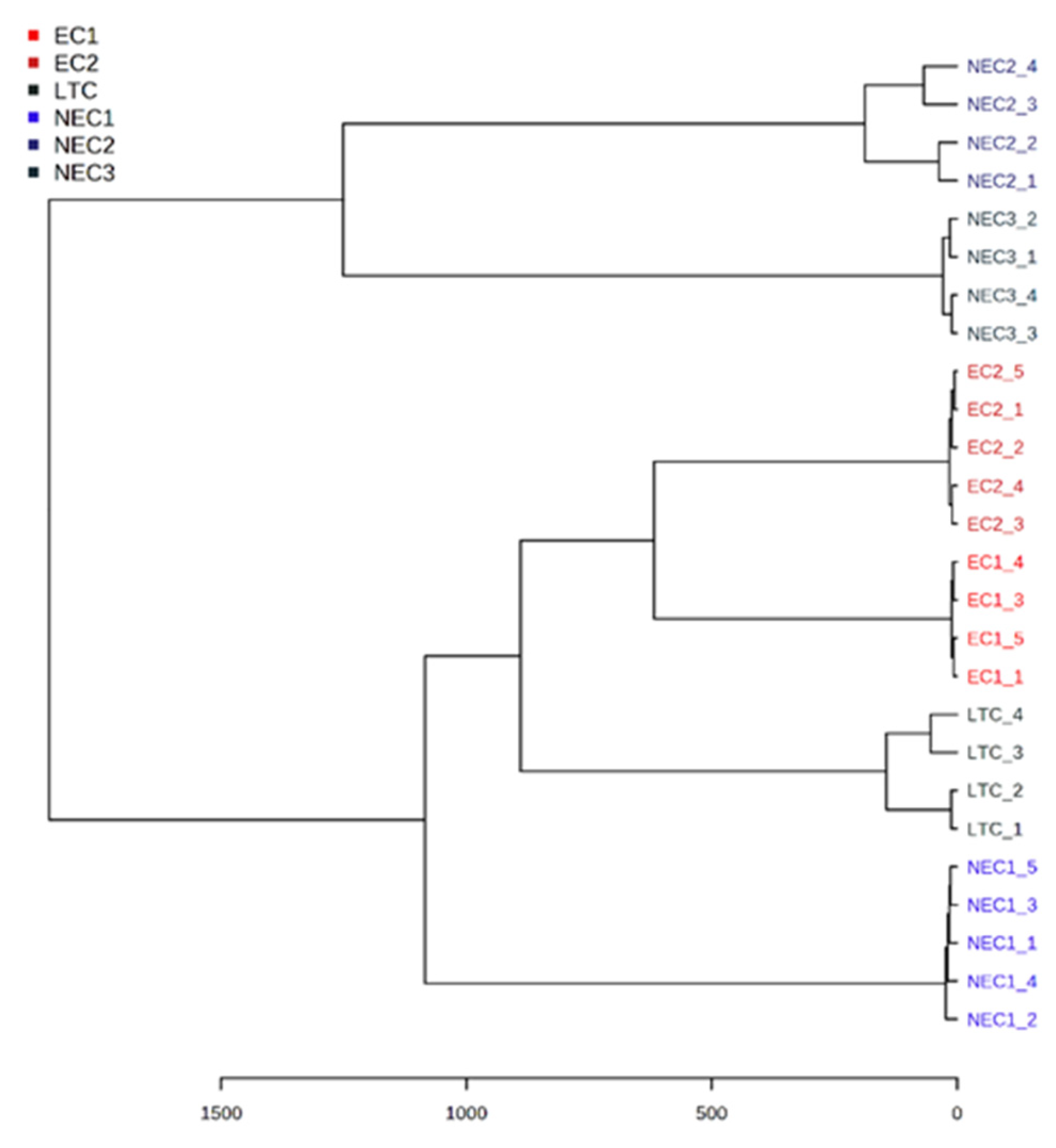
2.3. Chemometric Analysis
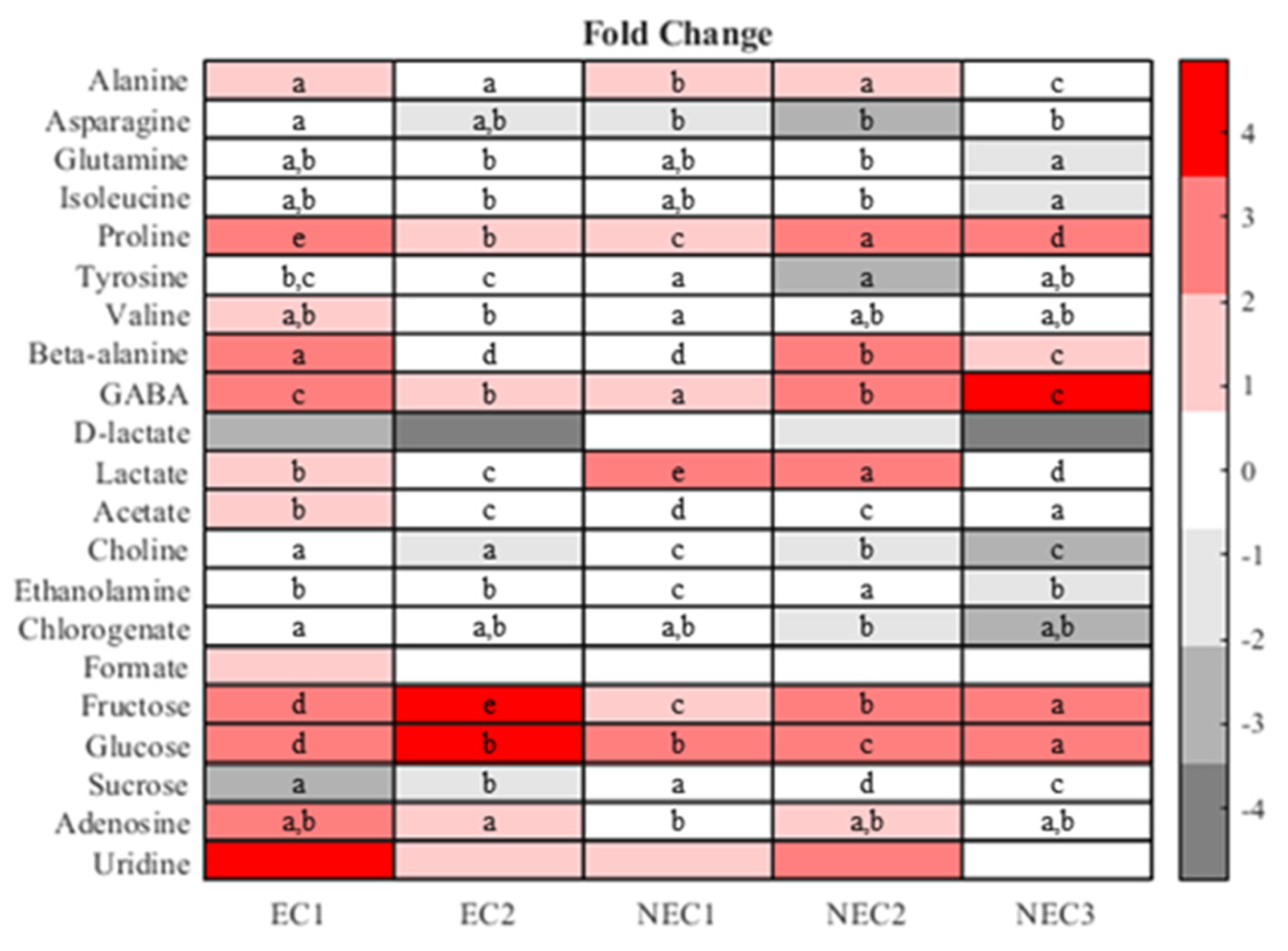
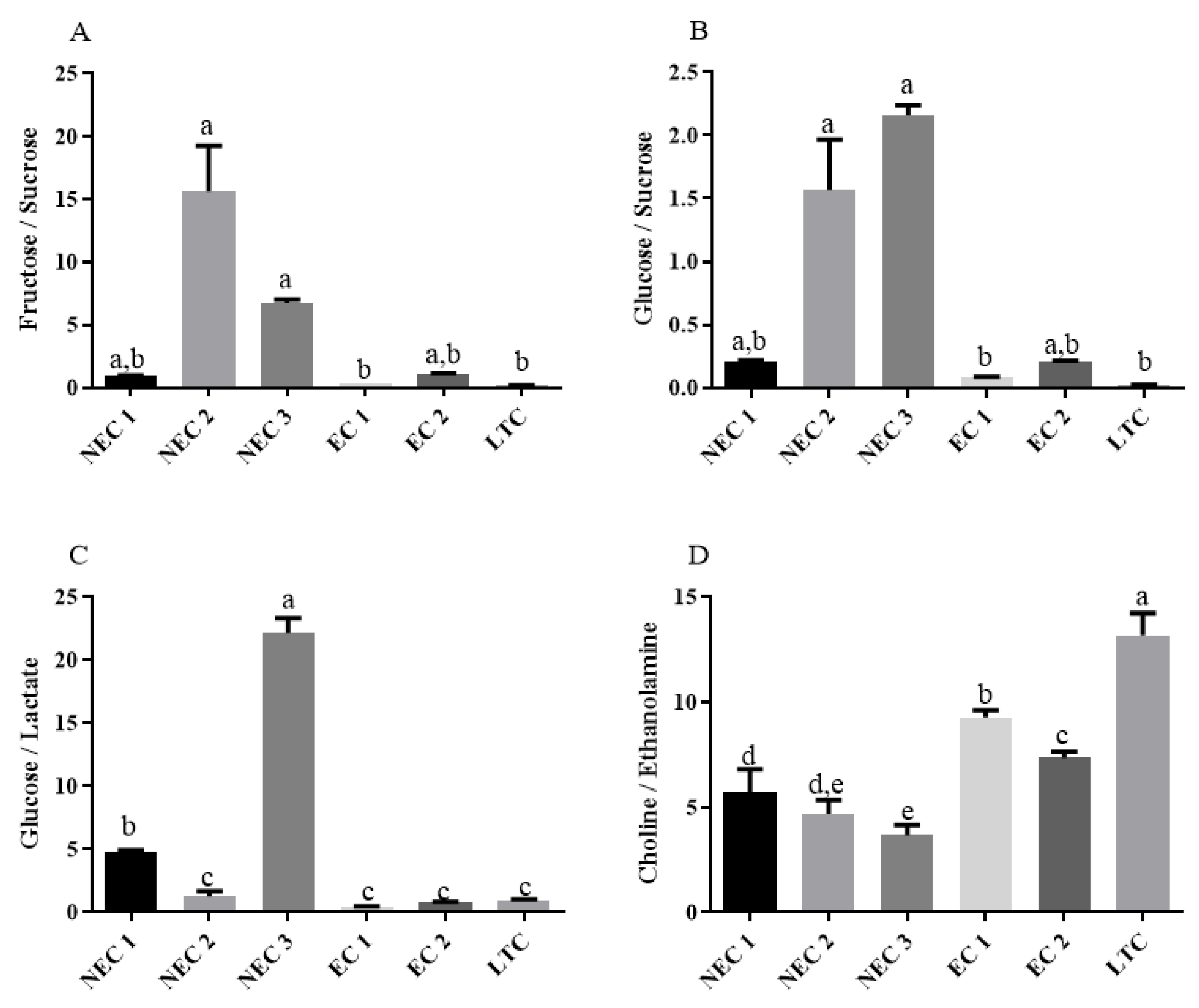
2.4. One-Way Analysis and Metabolite Ratios
3. Discussion
3.1. Somatic Embryogenesis and Metabolite Profile
3.2. Identified Metabolites and Possible Biosynthesis Pathways
3.3. Chemometric Analysis
3.4. One-Way Analysis and Metabolite Ratios
4. Materials and Methods
4.1. In Vitro Propagation and SE Induction
4.2. Metabolite Extraction
4.3. Protein Quantification
4.4. Nuclear Magnetic Resonance and Chemometric Analysis
4.5. One-Way Analysis and Metabolite Ratios
5. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kahia, J.; Sallah, P.K.; Diby, L.; Kouame, C.; Kirika, M.; Niyitegeka, S.; Asiimwe, T. A novel regeneration system for tamarillo (Cyphomandra betacea) via organogenesis from hypocotyl, leaf, and root explants. HortScience 2015, 50, 1375–1378. [Google Scholar] [CrossRef]
- Wang, S.; Zhu, F. Tamarillo (Solanum betaceum): Chemical composition, biological properties, and product innovation. Trends Food Sci. Technol. 2020, 95, 45–58. [Google Scholar] [CrossRef]
- Diep, T.T.; Rush, E.C.; Yoo, M.J.Y. Tamarillo (Solanum betaceum Cav.): A review of physicochemical and bioactive properties and potential applications. Food Rev. Int. 2022, 38, 1343–1367. [Google Scholar] [CrossRef]
- Prohens, J.; Nuez, F. The tamarillo (Cyphomandra betacea): A review of a promising small fruit crop. Small Fruits Rev. 2001, 1, 43–68. [Google Scholar] [CrossRef]
- Correia, S.; Canhoto, J. Biotechnology of tamarillo (Cyphomandra betacea): From in vitro cloning to genetic transformation. Sci. Hortic. 2012, 148, 161–168. [Google Scholar] [CrossRef]
- Guimarães, M.L.; Tomé, M.C.; Cruz, G.S. Cyphomandra betacea (Cav.) Sendtn. (Tamarillo)-Trees IV; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 1996; pp. 120–137. [Google Scholar]
- Correia, S.; Canhoto, J. Somatic embryogenesis of tamarillo (Solanum betaceum Cav.). In Step Wise Protocols for Somatic Embryogenesis on Important Woody Plants; Jain, S.M., Gupta, P., Eds.; Springer International Publishing: New York, NY, USA, 2018; Volume 2, pp. 171–179. [Google Scholar]
- Méndez-Hernández, H.A.; Ledezma-Rodriguez, M.; Avilez-Montalvo, R.N.; Juarez-Gomez, Y.L.; Skeete, A.; Avilez-Montalvo, J.; De-la-Pena, C.; Loyola-Vargas, V.M. Signaling overview of plant somatic embryogenesis. Front. Plant Sci. 2019, 10, 77. [Google Scholar] [CrossRef]
- Stasolla, C.; Yeung, E.C. Recent advances in conifer somatic embryogenesis: Improving somatic embryo quality. Plant Cell Tissue Organ Cult. 2003, 74, 15–35. [Google Scholar] [CrossRef]
- Rose, R.J.; Song, Y. Somatic Embryogenesis. In Encyclopedia of Applied Plant Sciences, 2nd ed.; Thomas, B., Murray, B.G., Murphy, D.J., Eds.; Academic Press: Oxford, UK, 2017; pp. 474–479. [Google Scholar]
- Salaün, C.; Lepiniec, L.; Dubreucq, B. Genetic and molecular control of somatic embryogenesis. Plants 2021, 10, 1467. [Google Scholar] [CrossRef]
- Fehér, A. Somatic embryogenesis—Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta-Gene Regul. Mech. 2015, 1849, 385–402. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X. Regulation of somatic embryogenesis in higher plants. CRC Crit. Rev. Plant Sci. 2010, 29, 36–57. [Google Scholar] [CrossRef]
- Awada, R.; Campa, C.; Gibault, E.; Déchamp, E.; Georget, F.; Lepelley, M.; Abdallah, C.; Erban, A.; Martinez-Seidel, F.; Kopka, J.; et al. Unravelling the metabolic and hormonal machinery during key steps of somatic embryogenesis: A case study in coffee. Int. J. Mol. Sci. 2019, 20, 4665. [Google Scholar] [CrossRef]
- Correia, S.; Canhoto, J. Somatic embryogenesis of tamarillo (Solanum betaceum Cav.). In Vegetative Propagation of Forest Trees; Jain, S., Gupta, P., Eds.; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Lopes, M.L.; Ferreira, M.R.; Carloto, J.M.; Cruz, G.S.; Canhoto, J.M. Somatic embryogenesis induction in Tamarillo (Cyphomandra betacea). In Somatic Embryogenesis in Woody Plants; Springer: Dordrecht, The Netherlands, 2000; pp. 433–455. [Google Scholar]
- Cordeiro, D.; Rito, M.; Borges, F.; Canhoto, J.; Correia, S. Selection and validation of reference genes for qPCR analysis of miRNAs and their targets during somatic embryogenesis in tamarillo (Solanum betaceum Cav.). Plant Cell Tissue Organ Cult. 2020, 143, 109–120. [Google Scholar] [CrossRef]
- Correia, S.; Vinhas, R.; Manadas, B.; Lourenço, A.S.; Veríssimo, P.; Canhoto, J.M. Comparative proteomic analysis of auxin induced embryogenic and nonembryogenic tissues of the solanaceous tree Cyphomandra betacea (Tamarillo). J. Proteome Res. 2012, 11, 1666–1675. [Google Scholar] [CrossRef]
- Wang, J.; Kambhampati, S.; Allen, D.K.; Chen, L.Q. Comparative metabolic analysis reveals a metabolic switch in mature, hydrated, and germinated pollen in Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 836665. [Google Scholar] [CrossRef]
- Gao, H.; Zhou, Q.; Yang, L.; Zhang, K.; Ma, Y.; Xu, Z.Q. Metabolomics analysis identifies metabolites associated with systemic acquired resistance in Arabidopsis. PeerJ 2020, 8, e10047. [Google Scholar] [CrossRef]
- Kumari, A.; Ray, K.; Sadhna, S.; Pandey, A.K.; Sreelakshmi, Y.; Sharma, R. Metabolomic homeostasis shifts after callus formation and shoot regeneration in tomato. PLoS ONE 2017, 12, 5. [Google Scholar] [CrossRef] [PubMed]
- Ge, F.; Hu, H.; Huang, X.; Zhang, Y.; Wang, Y.; Li, Z.; Zou, C.; Peng, H.; Li, L.; Gao, S.; et al. Metabolomic and proteomic analysis of maize embryonic callus induced from immature embryo. Sci. Rep. 2017, 7, 1004. [Google Scholar] [CrossRef]
- Sharma, K.; Sarma, S.; Bohra, A.; Mitra, A.; Sharma, N.K.; Kumar, A. Plant Metabolomics: An Emerging Technology for Crop Improvement; Çelik, Ö., Ed.; IntechOpen: Rijeka, Croatia, 2018; p. 4. [Google Scholar]
- Rebiai, A.; Seghir, B.; Hemmami, H.; Zeghoud, S.; Amor, I.; Kouadri, I.; Messaoudi, M.; Pasdaran, A.; Caruso, G.; Sharma, S.; et al. Quality assessment of medicinal plants via chemometric exploration of quantitative NMR aata: A review. Compounds 2022, 22, 163–181. [Google Scholar] [CrossRef]
- Sciubba, F.; Tomassini, A.; Giorgi, G.; Brasili, E.; Pasqua, G.; Capuani, G.; Aureli, W.; Miccheli, A. NMR-based metabolomic study of purple carrot optimal harvest time for utilization as a source of bioactive compounds. Appl. Sci. 2020, 10, 8493. [Google Scholar] [CrossRef]
- Correia, S.; Lopes, M.L.; Canhoto, J.M. Somatic embryogenesis induction system for cloning an adult Cyphomandra betacea (Cav.) Sendt. (tamarillo). Trees 2011, 25, 1009–1020. [Google Scholar] [CrossRef]
- Magnani, E.; Jiménez-Gómez, J.M.; Soubigou-Taconnat, L.; Lepiniec, L.; Fiume, E. Profiling the onset of somatic embryogenesis in Arabidopsis. BMC Genom. 2017, 18, 998. [Google Scholar] [CrossRef]
- Wen, L.; Li, W.; Parris, S.; West, M.; Lawson, J.; Smathers, M.; Li, Z.; Jones, D.; Jin, S.; Saski, C. Transcriptomic profiles of non-embryogenic and embryogenic callus cells in a highly regenerative upland cotton line (Gossypium hirsutum L.). BMC Dev. Biol. 2020, 20, 25. [Google Scholar] [CrossRef]
- Zhang, S.; Yan, S.; An, P.; Cao, Q.; Wang, C.; Wang, J.; Zhang, H.; Zhang, L. Embryogenic callus induction from immature zygotic embryos and genetic transformation of Larix kaempferi 3× Larix gmelinii 9. PLoS ONE 2021, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Välimäki, S.; Hazubska-Przybył, T.; Ratajczak, E.; Tikkinen, M.; Varis, S.; Aronen, T. Somatic embryo yield and quality from Norway Spruce embryogenic tissue proliferated in suspension culture. Front. Plant Sci. 2021, 12, 791549. [Google Scholar] [CrossRef] [PubMed]
- Correia, S.; Alhinho, A.; Casimiro, B.; Miguel, C.; Oliveira, M.; Veríssimo, P.; Canhoto, J. NEP-TC a rRNA methyltransferase involved on somatic embryogenesis of tamarillo (Solanum betaceum Cav.). Front. Plant Sci. 2019, 10, 438. [Google Scholar] [CrossRef]
- Alves, A.; Caeiro, A.; Correia, S.; Veríssimo, P.; Canhoto, J. Establishment and biochemical characterization of tamarillo (Solanum betaceum Cav.) embryogenic cell suspension cultures. In Vitro Cell. Dev. Biol.-Plant 2017, 53, 606–618. [Google Scholar] [CrossRef]
- Bartos, P.; Gomes, H.; Amaral, L.; Teixeira, J.; Scherwinski-Pereira, J. Biochemical events during somatic embryogenesis in Coffea arabica L. 3 Biotech 2018, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Trigiano, R.; Conger, B. Regulation of growth and somatic embryogenesis by proline and serine in suspension cultures of Dactylis glomerata. J. Plant Physiol. 1987, 130, 49–55. [Google Scholar] [CrossRef]
- Bahmankar, M.; Mortazavian, S.; Tohidfar, M.; Sadat Noori, S.; Izadi Darbandi, A.; Corrado, G.; Rao, R. Chemical compositions, somatic embryogenesis, and somaclonal variation inCumin. Biomed. Res. Int. 2017, 2017, 7283806. [Google Scholar] [CrossRef]
- Mahmud, I.; Thapaliya, M.; Boroujerdi, A.; Chowdhury, K. NMR-based metabolomics study of the biochemical relationship between sugarcane callus tissues and their respective nutrient culture media. Anal. Bioanal. Chem. 2014, 406, 5997–6005. [Google Scholar] [CrossRef]
- Paventi, G.; Pizzuto, R.; Chieppa, G.; Passarella, S. L-Lactate metabolism in potato tuber mitochondria. FEBS J. 2007, 274, 1459–1469. [Google Scholar] [CrossRef]
- Igamberdie, A. Review Origins and metabolism of formate in higher plants. Plant Physiol. Biochem. 1999, 37, 503–513. [Google Scholar] [CrossRef]
- Lin, M.; Oliver, D. The role of acetyl-coenzyme a synthetase in Arabidopsis. Plant Physiol. 2008, 147, 1822–1829. [Google Scholar] [CrossRef]
- Araújo, W.; Martins, A.; Fernie, A.; Tohge, T. 2-oxoglutarate: Linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 2014, 5, 552. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Zhao, D.; Liu, Q. Connections Between Amino Acid Metabolisms in Plants: Lysine as an Example. Front. Plant Sci. 2020, 11, 928. [Google Scholar] [CrossRef]
- Li, L.; Dou, N.; Zhang, H.; Wu, C. The versatile GABA in plants. Plant Signal. Behav. 2021, 16, 3. [Google Scholar] [CrossRef] [PubMed]
- Breitkreuz, K.; Allan, W.; Cauwenberghe, O.; Jakobs, C.; Talibi, D.; Andre, B.; Shelp, B. A Novel gamma-hydroxybutyrate dehydrogenase. J. Biol. Chem. 2003, 278, 41552–41556. [Google Scholar] [CrossRef] [PubMed]
- Ching, S.; Gidda, S.; Rochon, A.; Van Cauwenberghe, O.; Shelp, B.; Mullen, R. Glyoxylate reductase isoform 1 is localized in the cytosol and not peroxisomes in Plant Cells. J. Integr. Plant Biol. 2012, 54, 152–168. [Google Scholar] [CrossRef]
- Rontein, D.; Dieuaide-Noubhani, M.; Dufourc, E.; Raymond, P.; Rolin, D. The metabolic architecture of plant cells: Stability of central metabolism and flexibility of anabolic pathways during the growth cycle of tomato cells. J. Biol. Chem. 2002, 277, 43948–43960. [Google Scholar] [CrossRef]
- Kwon, Y.; Yu, S.; Lee, H.; Yim, J.; Zhu, J.; Lee, B. Arabidopsis serine decarboxylase mutants implicate the roles of ethanolamine in plant growth and development. Int. J. Mol. Sci. 2012, 13, 3176–3188. [Google Scholar] [CrossRef]
- Gibellini, F.; Smith, T. The Kennedy pathway—De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010, 62, 414–428. [Google Scholar] [CrossRef]
- Gramazio, P.; Prohens, J.; Plazas, M.; Andjar, I.; Herraiz, F.; Castillo, E.; Knapp, S.; Meyer, R.; Vilanova, S. Location of chlorogenic acid biosynthesis pathway and polyphenol oxidase genes in a new interspecific anchored linkage map of eggplant. BMC Plant Biol. 2014, 14, 350. [Google Scholar] [CrossRef]
- Schenck, C.; Maeda, H. Tyrosine biosynthesis, metabolism, and catabolism in plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef]
- Fraser, C.; Chapple, C. The phenylpropanoid pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef]
- Matsui, A.; Yin, Y.; Yamanaka, K.; Iwasaki, M.; Ashihara, H. Metabolic fate of nicotinamide in higher plants. Physiol. Plant 2007, 131, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Witte, C.; Herde, M. Nucleotide Metabolism in Plants 1. Plant Physiol. 2020, 182, 63–78. [Google Scholar] [CrossRef]
- Herrmann, K.; Weaver, L. The shikimate pathway. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 473–503. [Google Scholar] [CrossRef] [PubMed]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar]
- Del Coco, L.; Felline, S.; Girelli, C.; Angilè, F.; Magliozzi, L.; Almada, F.; D’Aniello, B.; Mollo, E.; Terlizzi, A.; Fanizzi, F. 1H NMR spectroscopy and MVA to evaluate the effects of caulerpin-based diet on Diplodus sargus lipid profiles. Mar. Drugs 2008, 16, 390. [Google Scholar] [CrossRef]
- Shiokawa, Y.; Date, Y.; Kikuchi, J. Application of kernel principal component analysis and computational machine learning to exploration of metabolites strongly associated with diet. Sci. Rep. 2018, 8, 3426. [Google Scholar] [CrossRef]
- Jollife, I.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. Royal Soc. 2016, 374, 2065. [Google Scholar] [CrossRef]
- Lou, H.; Kako, S. Role of high sugar concentrations in inducing somatic embryogenesis from cucumber cotyledons. Sci. Hortic. 1995, 64, 11–20. [Google Scholar] [CrossRef]
- Satoh, K.; Ooka, H.; Wakai, A.; Takahara, Y.; Yamamoto, K. Osmotic and non-osmotic induction of somatic embryogenesis by sucrose at high concentrations in Daucus carota L. Plant Biotechnol. 2000, 17, 155–158. [Google Scholar] [CrossRef][Green Version]
- Karami, O.; Deljou, A.; Esna-Ashari, M.; Ostad-Ahmadi, P. Effect of sucrose concentrations on somatic embryogenesis in carnation (Dianthus caryophyllus L.). Sci. Hortic. 2006, 110, 340–344. [Google Scholar] [CrossRef]
- Blanckaert, A.; Belingheri, L.; Vasseur, J.; Hilbert, J. Changes in lipid composition during somatic embryogenesis in leaves of Cichorium. Plant Sci. 2000, 157, 165–172. [Google Scholar] [CrossRef]
- Ling, J.; Xia, Y.; Hu, J.; Zhu, T.; Wang, J.; Zhang, H.; Kong, L. Integrated lipidomic and transcriptomic analysis reveals phospholipid changes in somatic embryos of Picea asperata in response to partial desiccation. Int. J. Mol. Sci. 2022, 23, 6494. [Google Scholar] [CrossRef] [PubMed]
- Murtagh, F.; Contreras, P. Algorithms for hierarchical clustering: An overview. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 86–97. [Google Scholar] [CrossRef]
- Qiu, W.; Su, W.; Cai, Z.; Dong, L.; Li, C.; Xin, M.; Li, C.; Fang, W.; Liu, Y.; Wang, X.; et al. Combined analysis of transcriptome and metabolome reveals the potential mechanism of coloration and fruit quality in yellow and purple Passiflora edulis Sims. J. Agric. Food Chem. 2020, 68, 12096–12106. [Google Scholar] [CrossRef]
- Pires, R.; Cardoso, H.; Ribeiro, A.; Peixe, A.; Cordeiro, A. Somatic embryogenesis from mature embryos of Olea europaea L. cv. ‘Galega Vulgar’ and long-term management of Calli morphogenic capacity. Plants 2020, 9, 758. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Abdi, H. Partial least squares regression and projection on latent structure regression (PLS Regression). Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 97–106. [Google Scholar] [CrossRef]
- Castander-Olarieta, A.; Pereira, C.; Montalbán, I.; Mendes, V.; Correia, S.; Suárez-Álvarez, S.; Manadas, B.; Canhoto, J.; Moncaleán, P. Proteome-wide analysis of heat-stress in Pinus radiata somatic embryos reveals a combined response of sugar metabolism and translational regulation mechanisms. Front. Plant Sci. 2021, 12, 631239. [Google Scholar] [CrossRef]
- Das, P. Somatic embryogenesis in four tree legumes. Biotechnol. Res. Int. 2011, 2011, 737636. [Google Scholar] [CrossRef][Green Version]
- Hayat, S.; Hayat, Q.; Alyemeni, M.; Wani, A.; Pichtel, J.; Ahmad, A. Role of proline under changing environments: A review. Plant Signal Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, H.; Chen, S.; Ji, M.; Perl, A.; Kovacs, L.; Chen, S. Stress response proteins’ differential expression in embryogenic and non-embryogenic callus of Vitis vinifera L. cv. Cabernet Sauvignon—A proteomic approach. Plant Sci. 2009, 177, 103–113. [Google Scholar] [CrossRef]
- Caeiro, A.; Jarak, I.; Correia, S.; Carvalho, R.; Canhoto, J. Comparative analysis of primary metabolome and secondary metabolites in the somatic embryogenesis of tamarillo (Solanum betaceum Cav.). In Proceedings of the 5th International Conference of the IUFRO Unit 2.09.02 on Clonal Trees in the Bioeconomy Age: Opportunities and Challenges, Coimbra, Portugal, 10–15 September 2018; pp. 10–15. [Google Scholar]
- Saccenti, E.; Hoefsloot, H.; Smilde, A.; Westerhuis, J.; Hendriks, M. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2014, 10, 361–374. [Google Scholar] [CrossRef]
- Hur, M.; Campbell, A.; Almeida-de-Macedo, M.; Li, L.; Ransom, N.; Jose, A.; Crispin, M.; Nikolau, B.; Wurtele, E. A global approach to analysis and interpretation of metabolic data for plant natural product discovery. Nat. Prod. Rep. 2013, 30, 565–583. [Google Scholar] [CrossRef]
- Gieger, C.; Geistlinger, L.; Altmaier, E.; Hrabé de Angelis, M.; Kronenberg, F.; Meitinger, T.; Mewes, H.; Wichmann, H.; Weinberger, K.; Adamski, J.; et al. Genetics meets metabolomics: A genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008, 4, e1000282. [Google Scholar] [CrossRef]
- Iraqi, D.; Tremblay, F. Analysis of carbohydrate metabolism enzymes and cellular contents of sugars and proteins during spruce somatic embryogenesis suggests a regulatory role of exogenous sucrose in embryo development. J. Exp. Bot. 2001, 52, 2301–2311. [Google Scholar] [CrossRef]
- Lipavská, H.; Konrádová, H. Somatic embryogenesis in conifers: The role of carbohydrate metabolism. Vitr. Cell. Dev. Biol.-Plant 2004, 40, 23–30. [Google Scholar] [CrossRef]
- Ayre, B. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol. Plant 2001, 4, 377–394. [Google Scholar] [CrossRef]
- Ma, S.; Li, Y.; Li, X.; Sui, X.; Zhang, Z. Phloem Unloading Strategies and Mechanisms in Crop Fruits. J. Plant Growth Regul. 2019, 38, 494–500. [Google Scholar] [CrossRef]
- Ruan, Y. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Loboda, T.; Sung, S.; Black, C. Sucrose Synthase in Wild Tomato, Lycopersicon chmielewskii, and Tomato Fruit Sink Strength. Plant Physiol. 1992, 98, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Zanor, M.; Osorio, S.; Nunes-Nesi, A.; Carrari, F.; Lohse, M.; Usadel, B.; Kühn, C.; Bleiss, W.; Giavalisco, P.; Willmitzer, L.; et al. RNA interference of LIN5 in tomato confirms its role in controlling brix content, uncovers the influence of sugars on the levels of fruit hormones, and demonstrates the importance of sucrose cleavage for normal fruit development and fertility. Plant Physiol. 2009, 150, 1204–1218. [Google Scholar] [CrossRef]
- Carvalho, R.; Rodrigues, T.; Zhao, P.; Jeffrey, F.; Malloy, C.; Sherry, A. A 13C isotopomer kinetic analysis of cardiac metabolism: Influence of altered cytosolic redox and [Ca2+]. Am. J. Physiol.-Heart Circ. Physiol. 2004, 287, H889–H895. [Google Scholar] [CrossRef]
- Chung, Y.; Leach, M.; Eykyn, T. Magnetic resonance spectroscopy to study Ggycolytic metabolism during autophagy. Methods Enzymol. 2017, 588, 133–153. [Google Scholar]
- Kendziorek, M.; Paszkowski, A.; Zagdańska, B. Differential regulation of alanine aminotransferase homologues by abiotic stresses in wheat (Triticum aestivum L.) seedlings. Plant Cell Rep. 2012, 31, 1105–1117. [Google Scholar] [CrossRef]
- Rocha, M.; Sodek, L.; Licausi, F.; Hameed, M.; Dornelas, M.; Van Dongen, J. Analysis of alanine aminotransferase in various organs of soybean (Glycine max) and in dependence of different nitrogen fertilisers during hypoxic stress. Amino Acids 2010, 39, 1043–1053. [Google Scholar] [CrossRef]
- Parthasarathy, A.; Cross, P.; Dobson, R.; Adams, L.; Savka, M.; Hudson, A. A Three-Ring circus: Metabolism of the three proteogenic aromatic amino acids and their role in the health of plants and animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Xie, T.; Jin, J.; Chei, W.; Yue, J.; Du, C.; Sun, J.; Chen, L.; Jiang, Z.; Shi, S. GABA negatively regulates adventitious root development in poplar. J. Exp. Bot. 2020, 4, 1459–1474. [Google Scholar] [CrossRef]
- Pei, L.; Zhao, Y.; Shi, X.; Chen, R.; Yan, J.; Li, X.; Jiang, Z.; Wang, J.; Shi, S. The role of γ-aminobutyric acid (GABA) in the occurrence of adventitious roots and somatic embryos in woody plants. Plants 2022, 11, 3512. [Google Scholar] [CrossRef]
- Zimmermann, S.; Benstein, R.; Flores-Tornero, M.; Blau, S.; Anoman, A.; Rosa-Téllez, S.; Gerlich, S.; Salem, M.; Alseekh, S.; Kopriva, S.; et al. The phosphorylated pathway of serine biosynthesis links plant growth with nitrogen metabolism. Plant Physiol. 2021, 186, 1487–1506. [Google Scholar] [CrossRef]
- Singh, K.; van der Hooft, J.; van Wees, S.; Medema, M. Integrative omics approaches for biosynthetic pathway discovery in plants. Nat. Prod. Rep. 2022, 39, 1876–1896. [Google Scholar] [CrossRef]
- Resurreccion, E.; Fong, K. The Integration of Metabolomics with Other Omics: Insights into Understanding Prostate Cancer. Metabolites 2022, 12, 488. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bio Aassays with tobacco tissue cultures. Physiol. Plant 1962, 15, 474–497. [Google Scholar] [CrossRef]
- Berben, L.; Sereika, S.; Engberg, S. Effect size estimation: Methods and examples. Int. J. Nurs. Stud. 2012, 49, 1039–1047. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Amberg, A.; Barrett, D.; Beale, M.; Beger, R.; Daykin, C.; Fan, T.; Fiehn, O.; Goodacre, R.; Griffin, J.; Hankemeier, T.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar]
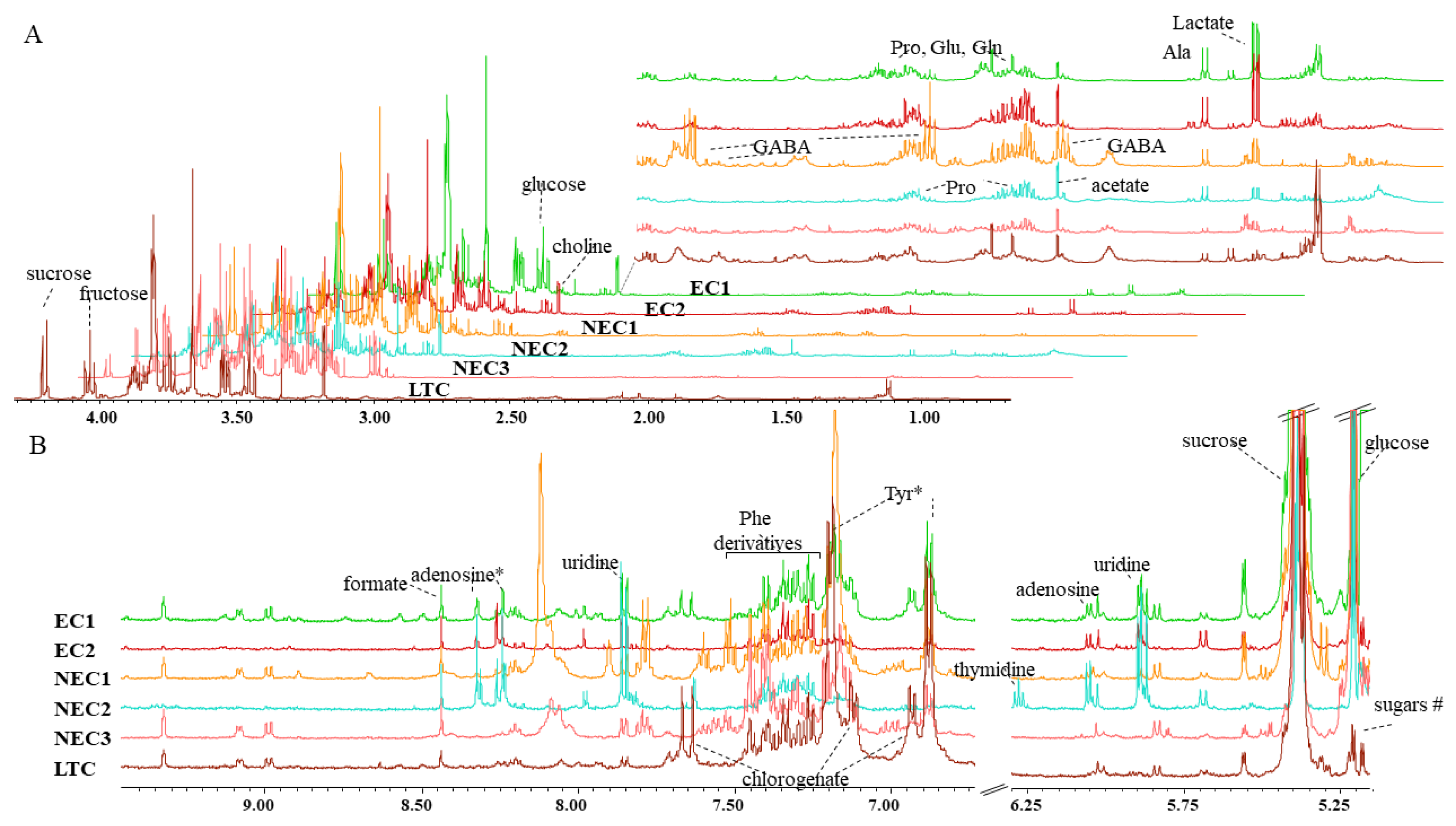
| Compound 1 | Assignment | δ 1H ppm (Multiplicity) | Cell Type | |
|---|---|---|---|---|
| 1 | Acetate | αCH3 | 1.9 (s) | all |
| 2 | Adenosine/inosine | C1’H ribose | 6.05 (d) | NEC2, EC1, EC2 |
| C8H ring | 8.24 (s) | |||
| C2H ring | 8.32 (s) | |||
| 3 | Alanine | βCH3 | 1.46 (d) | all |
| 4 | Asparagine | βCH | 2.84 (dd) | |
| β’CH | 2.94 (dd) | |||
| 5 | Aspartate * | βCH | 2.66 (dd) | all |
| β’CH | 2.79 (dd) | |||
| 6 | β-alanine | αCH2 | 2.54 (t) | NEC1-3, EC1, EC2 |
| βCH2 | 3.14 (t) | |||
| 7 | Choline | CH3 | 3.18 (s) | all |
| CH2 (N) | 3.49 (m) | |||
| CH2 (OH) | 4.04 (m) | |||
| 8 | Chlorogenic acid | C3H | 2.03 (m) | EC1, LTC |
| C5H | 2.17 (m) | |||
| C16H | 6.40 (d) | |||
| C21H | 6.93 (d) | |||
| C22H | 7.12 (dd) | |||
| C18H | 7.18 (d) | |||
| C15H | 7.65 (d) | |||
| 9 | Ethanolamine | C2H2 | 3.12 (m) | all |
| C3H2 | 3.81 (m) | |||
| 10 | Fructose | C1H | 3.55 (m) | all |
| C1H, C6H | 3.68 (m) | |||
| C1H, C2H, C5H | 3.77 (m) | |||
| C4H | 3.88 (m) | |||
| C5H | 3.99 (m) | |||
| C6H | 4.02 (m) | |||
| C3H, C4H | 4.09 (m) | |||
| 11 | Formate | CH | 8.43 (s) | all |
| 12 | γ-aminobutyric acid (GABA) | C2H | 1.88 (m) | all |
| C3H | 2.28 (t) | |||
| C4H | 2.99 (dd) | |||
| 13 | α-glucose | C4H | 3.38 | all |
| C2H | 3.53 | |||
| C3H | 3.76 | |||
| C5H, C6H | 3.82 | |||
| C1H | 5.21 (d) | |||
| β-glucose | C2H | 3.22 | ||
| C4H | 3.38 | |||
| C5H, C3H | 3.45 | |||
| C6H | 3.72 | |||
| C6’H | 3.87 | |||
| C1H | 4.62 (d) | |||
| 14 | Glutamate * | βCH | 2.05 (m) | all |
| β’CH | 2.12 (m) | |||
| γCH2 | 2.35 (m) | |||
| 15 | Glutamine | βCH2 | 2.14 (m) | all |
| γCH2 | 2.45 (m) | |||
| 16 | 4-hydroxybutyrate * | C3H | 1.8 (m) | all |
| C2H | 2.21 (dd) | |||
| 17 | Isoleucine | δCH3 | 0.94 (t) | all |
| β’CH3 | 1.01 (d) | |||
| 18 | Lactate | βCH3 | 1.31 (d) | all |
| αCH | 4.11 (q) | |||
| 19 | D-lactate | βCH3 | 1.38 (d) | all |
| 20 | Nicotinamide 2,* | 8.98 (d) | NEC1-3, EC1 | |
| 9.08 (d) | ||||
| 9.32 (s) | ||||
| 21 | Phenylalanine * | C2H, C6H ring | 7.31 (m) | all |
| C4H ring | 7.36 (m) | |||
| C3H, C5H ring | 7.40 (m) | |||
| 22 | Proline | γCH2 | 1.97 (m) | all |
| β’CH | 2.05 (m) | |||
| βCH | 2.33 (m) | |||
| δCH2 | 3.31 (m) | |||
| αCH | 4.11 (m) | |||
| 23 | Sucrose | C4H | 3.45 (m) | all |
| C2H | 3.54 (m) | |||
| C’1H2 | 3.67 (m) | |||
| C3H | 3.74 (m) | |||
| C’6H2, C6H2 | 3.80 (m) | |||
| C’5H, C5H | 3.87 (m) | |||
| C’4H | 4.03 (m) | |||
| C’3H | 4.19 (d) | |||
| C1H | 5.39 (d) | |||
| 24 | Thymidine * | C1’H ribose | 6.28 (t) | NEC2 |
| C6H ring | 7.63 (d) | |||
| 25 | Tyrosine 2 | C3H, C5H ring | 6.88 (d) | all |
| C2H, C6H ring | 7.17 (d) | |||
| 26 | Uridine | C1’H ribose, C5H ring | 5.89 (dd) | NEC2, EC1, EC2 |
| C6H ring | 7.85 (d) | |||
| 27 | Valine | γCH3 | 0.99 (d) | all |
| γ’CH3 | 1.04 (d) |
| Model | Q2 | R2 | Component Number | Important Features |
|---|---|---|---|---|
| General (all calli) | 0.990 | 0.995 | 4 | Fructose Sucrose Glucose |
| EC1 vs. EC2 | 0.999 | 0.999 | 2 | Fructose Pro Sucrose Choline |
| NEC1 vs. NEC2 | 0.996 | 0.998 | 1 | Sucrose Choline Fructose Pro GABA |
| EC1-2 vs. LTC | 0.945 | 0.999 | 6 | Choline Lactate Ala Sucrose Gln Fructose |
| NEC1-3 vs. LTC | 0.997 | 0.999 | 5 | Fructose Glucose Sucrose Choline |
| EC1 | EC2 | ||||
|---|---|---|---|---|---|
| Metabolite | log2(F) | −log10(p) | Metabolite | log2(F) | −log10(p) |
| Lactate | 3.0157 | 8.9975 | Lactate | 3.0793 | 8.8111 |
| Glucose | 1.9727 | 4.5264 | Pro | 2.9962 | 8.1141 |
| Ala | 1.9164 | 5.2626 | Glucose | 2.854 | 6.7433 |
| GABA | 1.8693 | 8.539 | Fructose | 2.6606 | 8.3922 |
| Fructose | 1.3182 | 4.4695 | β-Ala | 2.3036 | 8.811 |
| Uridine | 1.2561 | 2.4695 | Uridine | 2.2245 | 5.4269 |
| Pro | 1.1759 | 2.5534 | GABA | 2.181 | 8.1141 |
| Asn | −1.1302 | 1.853 | Ala | 1.9012 | 6.6857 |
| Adenosine | 1.8386 | 2.5225 | |||
| Choline | −1.0833 | 9.1291 | |||
| D-lactate | −1.5989 | 7.4762 | |||
| Chlorogenate | −2.0409 | 1.3749 | |||
| Asn | −2.262 | 2.9808 | |||
| Tyr | −2.8503 | 1.4842 | |||
| NEC1 | NEC2 | ||||
| Metabolite | log2(F) | −log10(p) | Metabolite | log2(F) | -log10(p) |
| GABA | 4.0647 | 8.1504 | Uridine | 4.8861 | 3.8556 |
| Glucose | 3.3213 | 7.6686 | Adenosine | 4.3811 | 4.0685 |
| Fructose | 2.8625 | 7.9148 | Fructose | 3.4559 | 7.8242 |
| Pro | 2.2208 | 5.8718 | Pro | 3.3086 | 3.0872 |
| β-ala | 1.4617 | 5.9125 | Glucose | 2.7991 | 4.6198 |
| Ile | −1.0624 | 1.4783 | β-Ala | 2.5844 | 3.4663 |
| Ethanolamine | −1.0945 | 4.0848 | GABA | 2.4035 | 5.9496 |
| Gln | −1.8525 | 1.6637 | Lactate | 2.3676 | 3.7499 |
| Choline | −2.3282 | 10.34 | Ala | 2.114 | 3.8435 |
| Chlorogenate | −2.6901 | 3.8669 | Formate | 1.8583 | 2.1599 |
| D-lactate | −4.9126 | 8.3448 | Val | 1.6334 | 1.5457 |
| Acetate | 1.4829 | 3.24 | |||
| Ile | 1.0863 | 1.0379 | |||
| D-lactate | −1.1509 | 4.6059 | |||
| Sucrose | −3.1713 | 5.4015 | |||
| NEC3 | |||||
| Metabolite | log2(F) | −log10(p) | |||
| Glucose | 5.3571 | 10.719 | |||
| Fructose | 4.3449 | 9.8747 | |||
| GABA | 1.9708 | 6.9013 | |||
| Pro | 1.508 | 4.0053 | |||
| Ile | 1.3825 | ||||
| Sucrose | −1.9774 | 4.164 | |||
| Choline | −2.3462 | 9.1183 | |||
| Asn | −2.4002 | 2.8545 | |||
| Chlorogenate | −4.3551 | 1.3008 | |||
| D-lactate | 7.8344 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caeiro, A.; Jarak, I.; Correia, S.; Canhoto, J.; Carvalho, R. Primary Metabolite Screening Shows Significant Differences between Embryogenic and Non-Embryogenic Callus of Tamarillo (Solanum betaceum Cav.). Plants 2023, 12, 2869. https://doi.org/10.3390/plants12152869
Caeiro A, Jarak I, Correia S, Canhoto J, Carvalho R. Primary Metabolite Screening Shows Significant Differences between Embryogenic and Non-Embryogenic Callus of Tamarillo (Solanum betaceum Cav.). Plants. 2023; 12(15):2869. https://doi.org/10.3390/plants12152869
Chicago/Turabian StyleCaeiro, André, Ivana Jarak, Sandra Correia, Jorge Canhoto, and Rui Carvalho. 2023. "Primary Metabolite Screening Shows Significant Differences between Embryogenic and Non-Embryogenic Callus of Tamarillo (Solanum betaceum Cav.)" Plants 12, no. 15: 2869. https://doi.org/10.3390/plants12152869
APA StyleCaeiro, A., Jarak, I., Correia, S., Canhoto, J., & Carvalho, R. (2023). Primary Metabolite Screening Shows Significant Differences between Embryogenic and Non-Embryogenic Callus of Tamarillo (Solanum betaceum Cav.). Plants, 12(15), 2869. https://doi.org/10.3390/plants12152869










