An Integrative Transcriptomics and Proteomics Approach to Identify Putative Genes Underlying Fruit Ripening in Tomato near Isogenic Lines with Long Shelf Life
Abstract
1. Introduction
2. Results
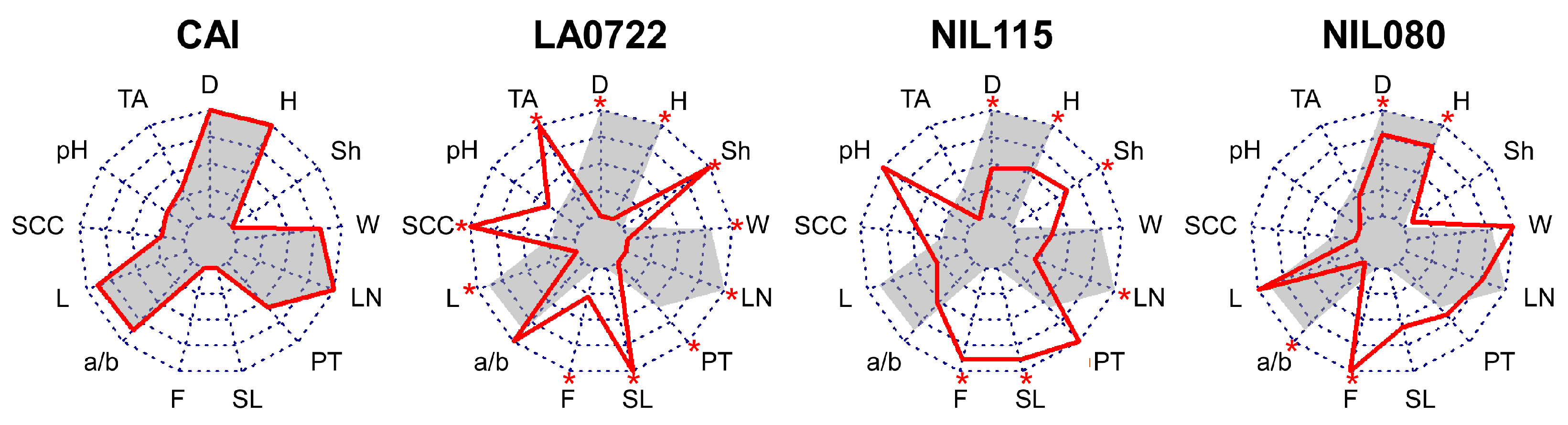
2.1. Phenotypic Analysis
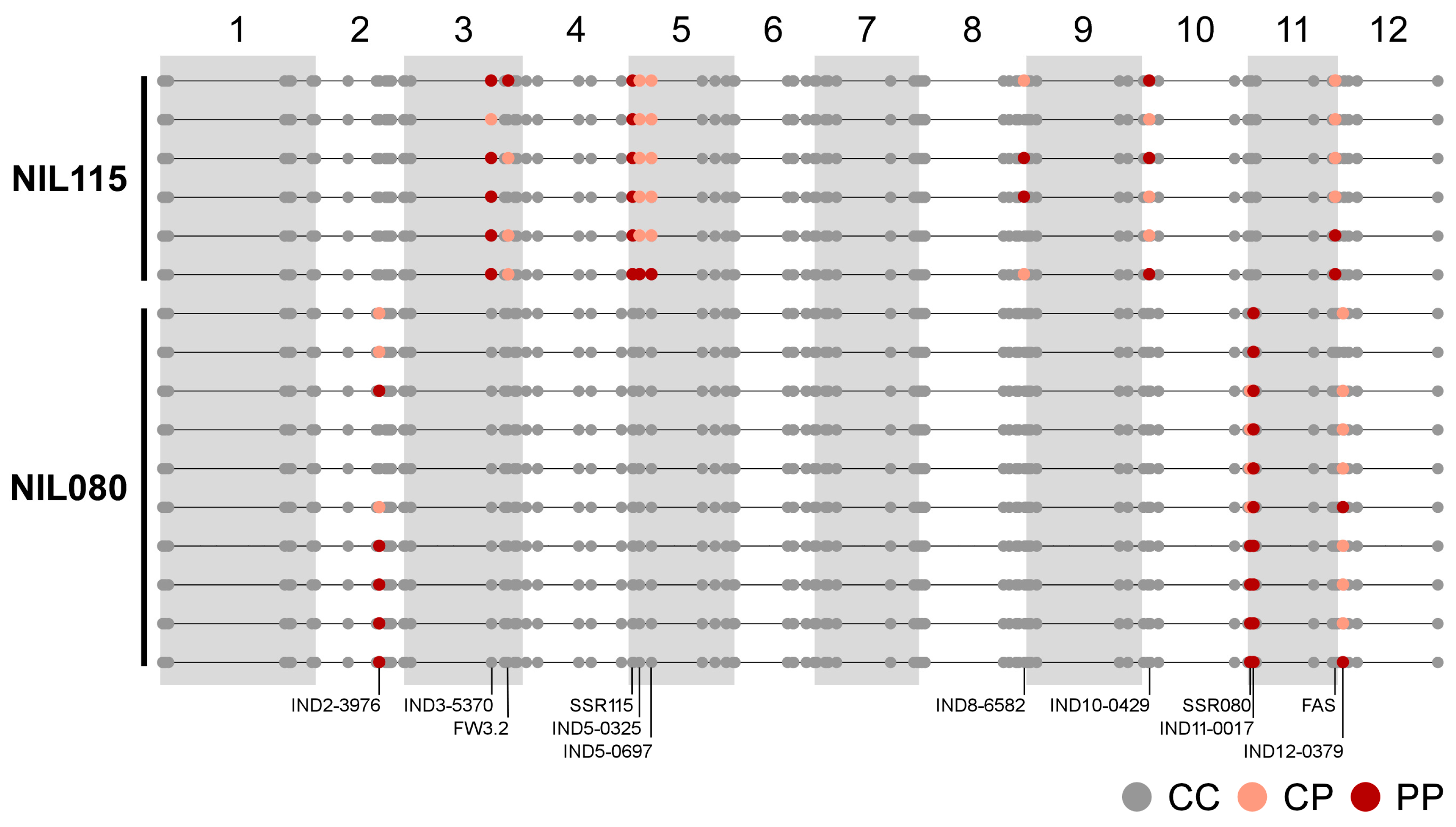
2.2. Molecular Characterization of the NILs
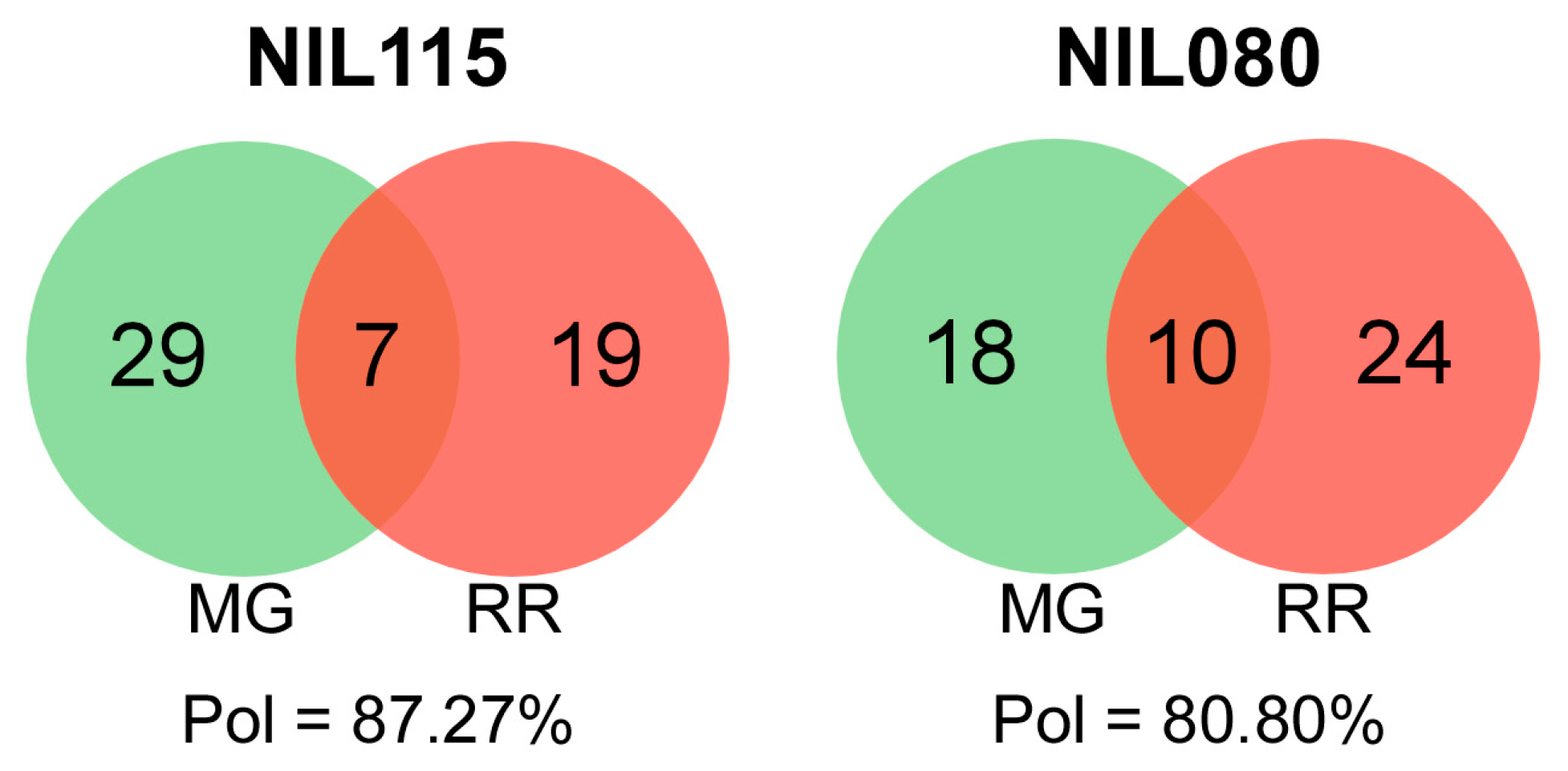
2.3. Ripening-Related Transcript Polymorphism
2.4. Differentially Expressed Ripening Proteins
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Phenotyping
4.3. Genomic Approach: Molecular Characterization
4.4. Transcript Profiling: cDNA-AFLP
4.5. Proteomic Approach: Label Free Quantification
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The Epigenome and Transcriptional Dynamics of Fruit Ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Seymour, G.B.; Chapman, N.H.; Chew, B.L.; Rose, J.K.C. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnol. J. 2013, 11, 269–278. [Google Scholar] [CrossRef]
- Gapper, N.E.; McQuinn, R.P.; Giovannoni, J.J. Molecular and genetic regulation of fruit ripening. Plant Mol. Biol. 2013, 82, 575–591. [Google Scholar] [CrossRef]
- Kumar, R.; Khurana, A.; Sharma, A.K. Role of plant hormones and their interplay in development and ripening of fleshy fruits. J. Exp. Bot. 2014, 65, 4561–4575. [Google Scholar] [CrossRef] [PubMed]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-Box Gene Necessary for Fruit Ripening at the Tomato Ripening-Inhibitor (Rin) Locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; da Rocha Tavano, E.C.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; de Maagd, R.A. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 2019, 9, 1696. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Samsulrizal, N.H.; Yan, C.; Allcock, N.S.; Craigon, J.; Blanco-Ulate, B.; Ortega-Salazar, I.; Marcus, S.E.; Bagheri, H.M.; Perez-Fons, L.; et al. Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 2019, 179, 544–557. [Google Scholar] [CrossRef]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef]
- Phan, T.D.; Bo, W.; West, G.; Lycett, G.W.; Tucker, G.A. Silencing of the major salt-dependent isoform of pectinesterase in tomato alters fruit softening. Plant Physiol. 2007, 144, 1960–1967. [Google Scholar] [CrossRef]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Vinaykumar, R.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; et al. Advances in omics approaches for abiotic stress tolerance in tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar] [CrossRef]
- Osorio, S.; Alba, R.; Damasceno, C.M.B.; Lopez-Casado, G.; Lohse, M.; Zanor, M.I.; Tohge, T.; Usadel, B.; Rose, J.K.C.; Fei, Z.; et al. Systems biology of tomato fruit development: Combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 2011, 157, 405–425. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Duan, W.; Chen, K.; Zhang, B. Transcriptome and methylome analysis reveals effects of ripening on and off the vine on flavor quality of tomato fruit. Postharvest Biol. Technol. 2020, 162, 111096. [Google Scholar] [CrossRef]
- Mata, C.I.; Hertog, M.L.A.T.M.; Van Raemdonck, G.; Baggerman, G.; Tran, D.; Nicolai, B.M. Omics analysis of the ethylene signal transduction in tomato as a function of storage temperature. Postharvest Biol. Technol. 2019, 155, 1–10. [Google Scholar] [CrossRef]
- Tang, H.; Zhang, X.; Gong, B.; Yan, Y.; Shi, Q. Proteomics and metabolomics analysis of tomato fruit at different maturity stages and under salt treatment. Food Chem. 2020, 311, 126009. [Google Scholar] [CrossRef] [PubMed]
- Goytia Bertero, V.; Pratta, G.R.; Arce, D.P. Independent transcriptomic and proteomic networks reveal common differentially expressed chaperone and interactor genes during tomato cv. Micro-Tom fruit ripening. Plant Gene 2021, 28, 100346. [Google Scholar] [CrossRef]
- Cigliano, R.A.; Aversano, R.; Di Matteo, A.; Palombieri, S.; Termolino, P.; Angelini, C.; Bostan, H.; Cammareri, M.; Consiglio, F.M.; Della Ragione, F.; et al. Multi-omics data integration provides insights into the post-harvest biology of a long shelf-life tomato landrace. Hortic. Res. 2022, 9, uhab042. [Google Scholar] [CrossRef]
- Choi, H.G.; Park, D.Y.; Kang, N.J. The Fruit Proteome Response to the Ripening Stages in Three Tomato Genotypes. Plants 2022, 11, 553. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Tian, Y.; Hu, Z.; Zhang, L.; Tang, B.; Wang, Y.; Li, J.; Chen, G. Novel translational and phosphorylation modification regulation mechanisms of tomato (Solanum lycopersicum) fruit ripening revealed by integrative proteomics and phosphoproteomics. Int. J. Mol. Sci. 2021, 22, 11782. [Google Scholar] [CrossRef]
- Cai, J.; Wang, P.; Tian, S.; Qin, G. Quantitative proteomic analysis reveals the involvement of mitochondrial proteins in tomato fruit ripening. Postharvest Biol. Technol. 2018, 145, 213–221. [Google Scholar] [CrossRef]
- Chen, Y.; Rofidal, V.; Hem, S.; Gil, J.; Nosarzewska, J.; Berger, N.; Demolombe, V.; Bouzayen, M.; Azhar, B.J.; Shakeel, S.N.; et al. Targeted Proteomics Allows Quantification of Ethylene Receptors and Reveals SlETR3 Accumulation in Never-Ripe Tomatoes. Front. Plant Sci. 2019, 10, 1054. [Google Scholar] [CrossRef] [PubMed]
- Belouah, I.; Bénard, C.; Denton, A.; Blein-Nicolas, M.; Balliau, T.; Teyssier, E.; Gallusci, P.; Bouchez, O.; Usadel, B.; Zivy, M.; et al. Transcriptomic and proteomic data in developing tomato fruit. Data Brief 2020, 28, 105015. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Nicolas, P.; Fernandez-Pozo, N.; Ma, Q.; Evanich, D.J.; Shi, Y.; Xu, Y.; Zheng, Y.; Snyder, S.I.; Martin, L.B.B.; et al. High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 2018, 9, 364. [Google Scholar] [CrossRef] [PubMed]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.; Francis, D.; Causse, M.; van der Knaap, E.; Cañizares, J. Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genom. 2015, 16, 257. [Google Scholar] [CrossRef]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Razifard, H.; Ramos, A.; Della Valle, A.L.; Bodary, C.; Goetz, E.; Manser, E.J.; Li, X.; Zhang, L.; Visa, S.; Tieman, D.; et al. Genomic evidence for complex domestication history of the cultivated tomato in Latin America. Mol. Biol. Evol. 2020, 37, 1118–1132. [Google Scholar] [CrossRef]
- Zorzoli, R.; Pratta, G.; Picardi, L. Variabilidad genética para la vida postcosecha y el peso de los frutos en tomate para familias F3 de un híbrido interespecífico. Pesqui. Agropecu. Bras. 2000, 35, 2423–2427. [Google Scholar] [CrossRef]
- Pratta, G.; Zorzoli, R.; Picardi, L.A. Diallel analysis of production traits among domestic, exotic and mutant germplasms of Lycopersicon. Genet. Mol. Res. 2003, 2, 206–213. [Google Scholar]
- Rodríguez, G.; Pratta, G.; Zorzoli, R.; Picardi, L. Recombinant lines obtained from an interspecific cross between Lycopersicon species selected by fruit weight and fruit shelf life. J. Am. Soc. Hortic. Sci. 2006, 131, 651–656. [Google Scholar] [CrossRef]
- Cambiaso, V.; Gimenez, M.; Pereira da Costa, J.; Vazquez, D.; Picardi, L.; Pratta, G.; Rodríguez, G. Selected genome regions for fruit weight and shelf life in tomato RILs discernible by markers based on genomic sequence information. Breed. Sci. 2019, 69, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, M.; Luciani, M.D.; Cambiaso, V.; Zorzoli, R.; Rodríguez, G.R.; Pereira da Costa, J.H. Tomato near isogenic lines to unravel the genetic diversity of S. pimpinellifolium LA0722 for fruit quality and shelf life breeding. Euphytica 2020, 216, 126. [Google Scholar] [CrossRef]
- Liu, D.; Lu, J.; Li, H.; Wang, J.; Pei, Y. Characterization of the O-acetylserine(thiol)lyase gene family in Solanum lycopersicum L. Plant Mol. Biol. 2019, 99, 123–134. [Google Scholar] [CrossRef]
- Cao, J.; Tan, X. Comprehensive analysis of the chitinase family genes in tomato (Solanum Lycopersicum). Plants 2019, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Celik, I. In Silico Integrated Analysis of Genomic, Transcriptomic, and Proteomic Data Reveals QTL-Specific Genes for Bacterial Canker Resistance in Tomato (Solanum lycopersicum L.). Curr. Issues Mol. Biol. 2023, 45, 1387–1395. [Google Scholar] [CrossRef]
- Martel, C.; Vrebalov, J.; Tafelmeyer, P.; Giovannoni, J.J. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 2011, 157, 1568–1579. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. A critical evaluation of the role of ethylene and MADS transcription factors in the network controlling fleshy fruit ripening. New Phytol. 2019, 221, 1724–1741. [Google Scholar] [CrossRef]
- Gao, Y.; Wei, W.; Fan, Z.; Zhao, X.; Zhang, Y.; Jing, Y.; Zhu, B.; Zhu, H.; Shan, W.; Chen, J.; et al. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J. Exp. Bot. 2020, 71, 3560–3574. [Google Scholar] [CrossRef]
- Uluisik, S.; Chapman, N.H.; Smith, R.; Poole, M.; Adams, G.; Gillis, R.B.; Besong, T.M.D.; Sheldon, J.; Stiegelmeyer, S.; Perez, L.; et al. Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 2016, 34, 950–952. [Google Scholar] [CrossRef]
- Barry, C.S.; Giovannoni, J.J. Ethylene and fruit ripening. J. Plant Growth Regul. 2007, 26, 143–159. [Google Scholar] [CrossRef]
- Iftikhar, H.; Naveed, N.; Virk, N.; Bhatti, M.F.; Song, F. In silico analysis reveals widespread presence of three gene families, MAPK, MAPKK and MAPKKK, of the MAPK cascade from crop plants of Solanaceae in comparison to the distantly-related syntenic species from Rubiaceae, coffee. PeerJ 2017, 5, e3255. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, X.; Wang, Y.; Wang, W.; Tian, S.; Qin, G. Redox proteomic analysis reveals the involvement of oxidative post-translational modification in tomato fruit ripening. Postharvest Biol. Technol. 2021, 178, 111556. [Google Scholar] [CrossRef]
- Slugina, M.A.; Shchennikova, A.; Kochieva, E.Z. The expression pattern of the Pho1a genes encoding plastidic starch phosphorylase correlates with the degradation of starch during fruit ripening in green-fruited and red-fruited tomato species. Funct. Plant Biol. 2019, 46, 1146–1157. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, J.; Wang, B.; Huang, S.; Hu, J.; Yang, T.; Asmutola, P.; Lan, H.; Qinghui, Y. Identification of the Carbohydrate and Organic Acid Metabolism Genes Responsible for Brix in Tomato Fruit by Transcriptome and Metabolome Analysis. Front. Genet. 2021, 12, 714942. [Google Scholar] [CrossRef]
- Leszczuk, A.; Chylińska, M.; Zięba, E.; Skrzypek, T.; Szczuka, E.; Zdunek, A. Structural network of arabinogalactan proteins (AGPs) and pectins in apple fruit during ripening and senescence processes. Plant Sci. 2018, 275, 36–48. [Google Scholar] [CrossRef]
- Leszczuk, A.; Kalaitzis, P.; Blazakis, K.N.; Zdunek, A. The role of arabinogalactan proteins (AGPs) in fruit ripening—A review. Hortic. Res. 2020, 7, 176. [Google Scholar] [CrossRef]
- Fulop, D.; Ranjan, A.; Ofner, I.; Covington, M.F.; Chitwood, D.H.; West, D.; Ichihashi, Y.; Headland, L.; Zamir, D.; Maloof, J.N.; et al. A new advanced backcross tomato population enables high resolution leaf QTL mapping and gene identification. G3 Genes Genomes Genet. 2016, 6, 3169–3184. [Google Scholar] [CrossRef]
- Ofner, I.; Lashbrooke, J.; Pleban, T.; Aharoni, A.; Zamir, D. Solanum pennellii backcross inbred lines (BILs) link small genomic bins with tomato traits. Plant J. 2016, 87, 151–160. [Google Scholar] [CrossRef]
- Xu, J.; Feng, Q.; Liu, Q.; Tang, S.; Gu, M.; Han, B.; Liang, G. Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genom. 2010, 11, 656. [Google Scholar] [CrossRef]
- Schmalenbach, I.; March, T.J.; Pillen, K.; Bringezu, T.; Waugh, R. High-resolution genotyping of wild barley introgression lines and fine-mapping of the threshability locus thresh-1 using the illumina goldengate assay. G3 Genes Genomes Genet. 2011, 1, 187–196. [Google Scholar] [CrossRef]
- Barrantes, W.; Fernández-del-Carmen, A.; López-Casado, G.; González-Sánchez, M.Á.; Fernández-Muñoz, R.; Granell, A.; Monforte, A.J. Highly efficient genomics-assisted development of a library of introgression lines of Solanum pimpinellifolium. Mol. Breed. 2014, 34, 1817–1831. [Google Scholar] [CrossRef]
- Giovannoni, J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Bequette, C.J.; Xu, K.; Blakley, I.C.; Fu, Z.Q.; Stratmann, J.W.; Loraine, A.E. RNA-seq links the transcription factors AINTEGUMENTA and AINTEGUMENTA-LIKE6 to cell wall remodeling and plant defense pathways. Plant Physiol. 2016, 171, 2069–2084. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, C.; Ma, Q.; Shekasteband, R.; Stewart, K.S.; Hutton, S.F.; Scott, J.W.; Fei, Z.; Ling, K.S. Comprehensive transcriptome analysis and functional characterization of PR-5 for its involvement in tomato Sw-7 resistance to tomato spotted wilt tospovirus. Sci. Rep. 2019, 9, 7673. [Google Scholar] [CrossRef]
- Karlova, R.; Rosin, F.M.; Busscher-Lange, J.; Parapunova, V.; Do, P.T.; Fernie, A.R.; Fraser, P.D.; Baxter, C.; Angenent, G.C.; de Maagd, R.A. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 2011, 23, 923–941. [Google Scholar] [CrossRef]
- Liu, M.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.E.P.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.P.; Bouzayen, M.; et al. Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef]
- Shi, L.; Liu, Q.; Qiao, Q.; Zhu, Y.; Huang, W.; Wang, X.; Ren, Z. Exploring the effects of pectate and pectate lyase on the fruit softening and transcription profiling of Solanum lycopersicum. Food Control 2022, 133, 108636. [Google Scholar] [CrossRef]
- Zhou, T.; Li, R.; Yu, Q.; Wang, J.; Pan, J.; Lai, T. Proteomic Changes in Response to Colorless nonripening Mutation during Tomato Fruit Ripening. Plants 2022, 11, 3570. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene response factors in plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [CrossRef]
- Popescu, S.C.; Popescu, G.V.; Bachan, S.; Zhang, Z.; Gerstein, M.; Snyder, M.; Dinesh-Kumar, S.P. MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 2009, 23, 80–92. [Google Scholar] [CrossRef]
- Gu, Y.Q.; Yang, C.; Thara, V.K.; Zhou, J.; Martin, G.B. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 2000, 12, 771–785. [Google Scholar] [CrossRef] [PubMed]
- Cheong, Y.H.; Moon, B.C.; Kim, J.K.; Kim, C.Y.; Kim, M.C.; Kim, I.H.; Park, C.Y.; Kim, J.C.; Park, B.O.; Koo, S.C.; et al. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003, 132, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- De Boer, K.; Tilleman, S.; Pauwels, L.; Vanden Bossche, R.; De Sutter, V.; Vanderhaeghen, R.; Hilson, P.; Hamill, J.D.; Goossens, A. APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 2011, 66, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.S.; Chen, M.; Li, L.C.; Ma, Y.Z. Functions and Application of the AP2/ERF Transcription Factor Family in Crop Improvement. J. Integr. Plant Biol. 2011, 53, 570–585. [Google Scholar] [CrossRef]
- Pereira da Costa, J.; Rodríguez, G.; Pratta, G.; Picardi, L.; Zorzoli, R. QTL detection for fruit shelf life and quality traits across segregating populations of tomato. Sci. Hort. 2013, 156, 47–53. [Google Scholar] [CrossRef]
- Eriksson, E.M.; Bovy, A.; Manning, K.; Harrison, L.; Andrews, J.; De Silva, J.; Tucker, G.A.; Seymour, G.B. Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening. Plant Physiol. 2004, 136, 4184–4197. [Google Scholar] [CrossRef]
- Kok, E.J.; Lehesranta, S.J.; Van Dijk, J.P.; Helsdingen, J.R.; Dijksma, W.T.P.; Van Hoef, A.M.A.; Koistinen, K.M.; Kärenlampi, S.O.; Kuiper, H.A.; Keijer, J. Changes in gene and protein expression during tomato ripening—Consequences for the safety assessment of new crop plant varieties. Food Sci. Technol. Int. 2008, 14, 503–518. [Google Scholar] [CrossRef]
- Cambiaso, V.; Pratta, G.R.; Pereira da Costa, J.H.; Zorzoli, R.; Francis, D.M.; Rodríguez, G.R. Whole genome re-sequencing analysis of two tomato genotypes for polymorphism insight in cloned genes and a genetic map construction. Sci. Hort. 2019, 247, 58–66. [Google Scholar] [CrossRef]
- Pereira da Costa, J.H.; Rodríguez, G.R.; Picardi, L.A.; Zorzoli, R.; Pratta, G.R. Genome-wide expression analysis at three fruit ripening stages for tomato genotypes differing in fruit shelf life. Sci. Hort. 2018, 229, 125–131. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Vuylsteke, M.; Peleman, J.; van Eijk, M. AFLP-based transcript profiling (cDNA-AFLP) for genome-wide expression analysis. Nat. Protoc. 2007, 2, 1399–1413. [Google Scholar] [CrossRef]
- Wu, X.; Xiong, E.; Wang, W.; Scali, M.; Cresti, M. Universal sample preparation method integrating trichloroacetic acid/acetone precipitation with phenol extraction for crop proteomic analysis. Nat. Protoc. 2014, 9, 362–374. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Bai, J.; Bandla, C.; García-Seisdedos, D.; Hewapathirana, S.; Kamatchinathan, S.; Kundu, D.J.; Prakash, A.; Frericks-Zipper, A.; Eisenacher, M.; et al. The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022, 50, D543–D552. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. AgriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]

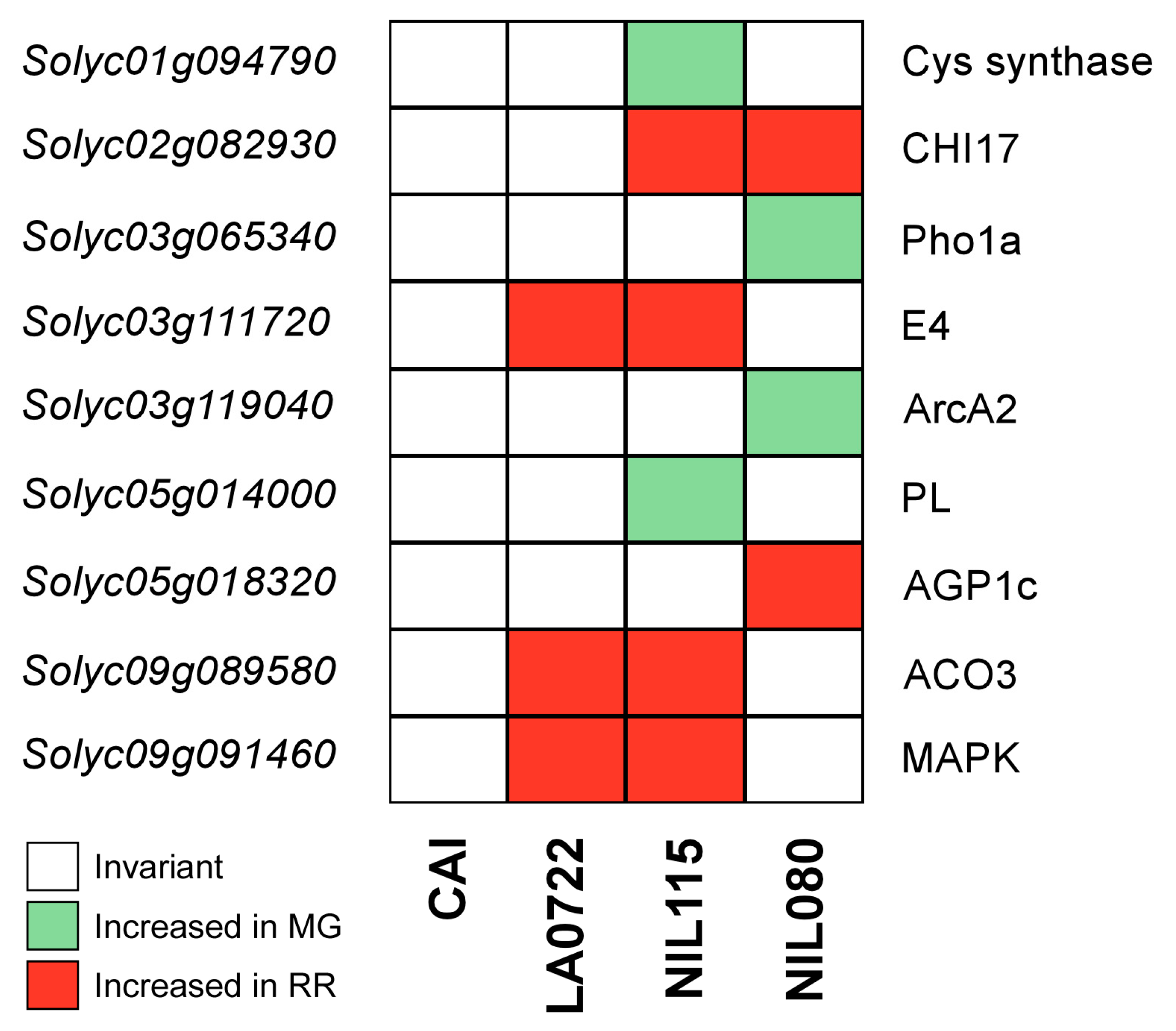
| NIL | Solyc ID | UniProt ID | Protein Name | Function | Increased Abundance | References |
|---|---|---|---|---|---|---|
| NIL115 | Solyc01g094790 | A0A3Q7F5F8 | Cys synthase | Cysteine synthase | MG | Liu et al., 2019 [33] |
| Solyc02g082930 | Q05540 | CHI17 | Acidic endochitinase | RR | Cao and Tan, 2019 [34] Celik et al., 2023 [35] | |
| Solyc03g111720 | P54153 | E4 | Peptide methionine sulfoxide reductase | RR | Martel et al., 2011 [36] Li et al., 2019 [37] Gao et al., 2020 [38] | |
| Solyc05g014000 | A0A3Q7GFD0 | PL | Pectate lyase | MG | Seymour et al., 2013 [39] | |
| Solyc09g089580 | P10967 | ACO3 | ACC oxidase | RR | Barry and Giovannoni, 2007 [40] Martel et al., 2011 [36] Li et al., 2019 [37] Gao et al., 2020 [38] | |
| Solyc09g091460 | A0A3Q7I9A3 | MAPK | Protein kinase of the Raf-like subfamily | RR | Iftikhar et al. 2017 [41] | |
| NIL080 | Solyc02g082930 | Q05540 | CHI17 | Acidic endochitinase | RR | Cao and Tan, 2019 [34] Celik et al., 2023 [35] |
| Solyc03g119040 | A0A3Q7GJ89 | ArcA2 | Redox-sensitive guanine nucleotide-binding protein | MG | Wang et al., 2021 [42] | |
| Solyc03g065340 | A0A3Q7G8G0 | Pho1a | Alpha-1,4 glucan phosphorylase | MG | Slugina et al., 2019 [43] Li et al., 2021 [44] | |
| Solyc05g018320 | A0A3Q7GJJ9 | AGP1c | Arabinogalactan | RR | Leszczuk et al., 2018 [45] Leszczuk et al., 2020 [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Giacomo, M.; Vega, T.A.; Cambiaso, V.; Picardi, L.A.; Rodríguez, G.R.; Pereira da Costa, J.H. An Integrative Transcriptomics and Proteomics Approach to Identify Putative Genes Underlying Fruit Ripening in Tomato near Isogenic Lines with Long Shelf Life. Plants 2023, 12, 2812. https://doi.org/10.3390/plants12152812
Di Giacomo M, Vega TA, Cambiaso V, Picardi LA, Rodríguez GR, Pereira da Costa JH. An Integrative Transcriptomics and Proteomics Approach to Identify Putative Genes Underlying Fruit Ripening in Tomato near Isogenic Lines with Long Shelf Life. Plants. 2023; 12(15):2812. https://doi.org/10.3390/plants12152812
Chicago/Turabian StyleDi Giacomo, Melisa, Tatiana Alejandra Vega, Vladimir Cambiaso, Liliana Amelia Picardi, Gustavo Rubén Rodríguez, and Javier Hernán Pereira da Costa. 2023. "An Integrative Transcriptomics and Proteomics Approach to Identify Putative Genes Underlying Fruit Ripening in Tomato near Isogenic Lines with Long Shelf Life" Plants 12, no. 15: 2812. https://doi.org/10.3390/plants12152812
APA StyleDi Giacomo, M., Vega, T. A., Cambiaso, V., Picardi, L. A., Rodríguez, G. R., & Pereira da Costa, J. H. (2023). An Integrative Transcriptomics and Proteomics Approach to Identify Putative Genes Underlying Fruit Ripening in Tomato near Isogenic Lines with Long Shelf Life. Plants, 12(15), 2812. https://doi.org/10.3390/plants12152812









