Exploring the Proteomic Profile of Soybean Bran: Unlocking the Potential for Improving Protein Quality and Quantity
Abstract
1. Introduction
2. Results
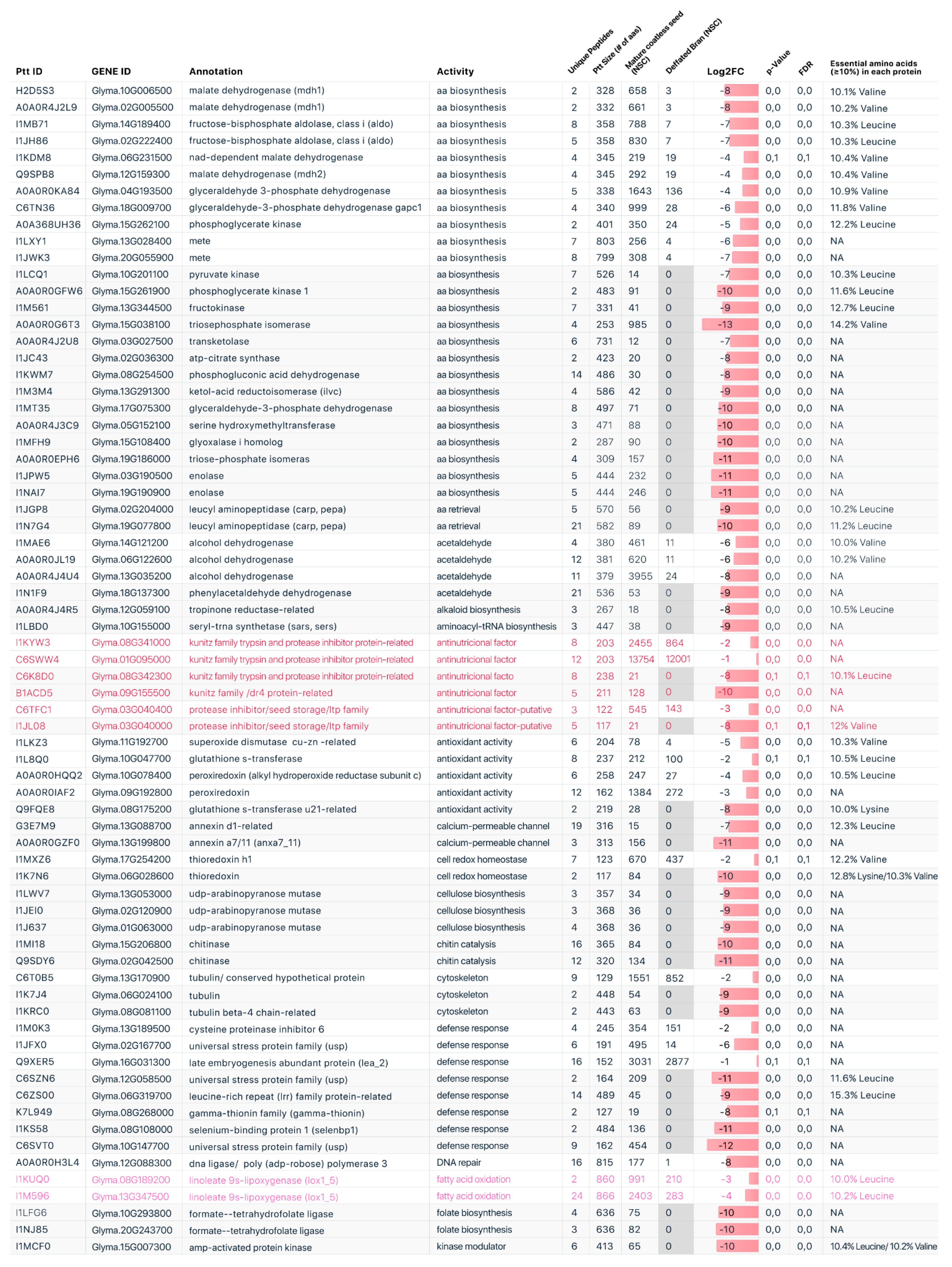
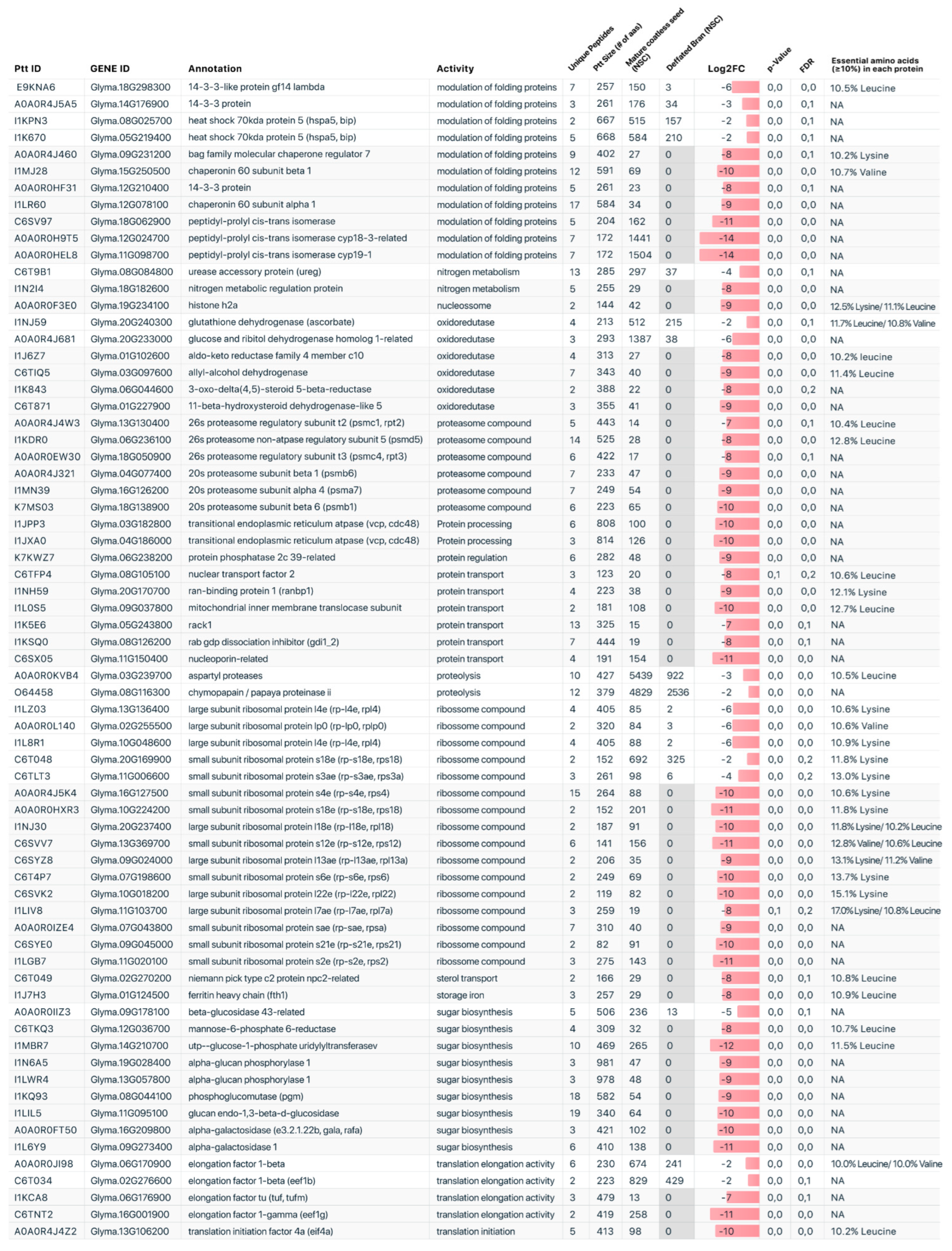
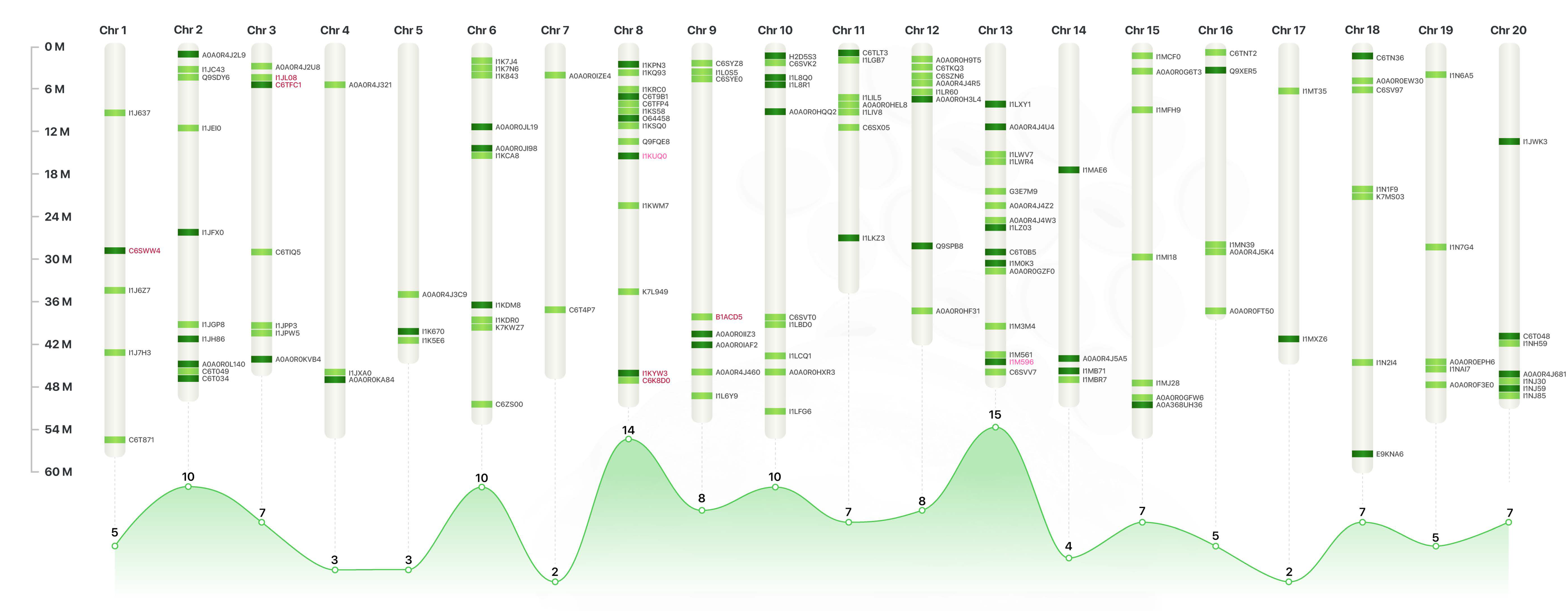
2.1. Comparative Proteomic Profile
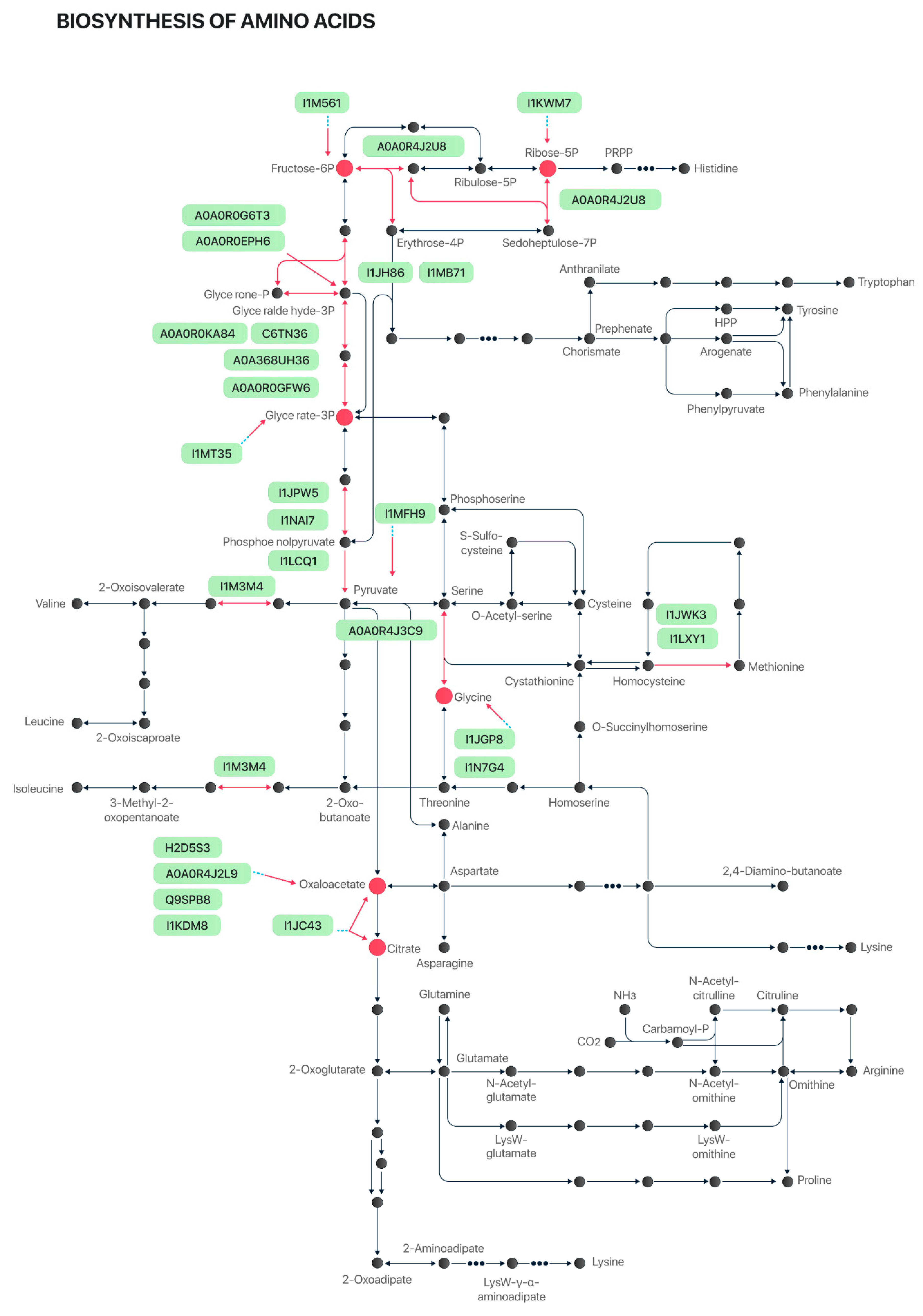
2.2. Protein Functional Categories
3. Discussion
4. Materials and Methods
4.1. Biological Material
4.2. LC–MS/MS Analysis
4.3. Identification of Differentially Expressed Proteins
4.4. Systems Biology
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pathan, M.S.; Sleper, D.A. Advances in Soybean Breeding. In Genetics and Genomics of Soybean; Stacey, G., Ed.; Springer: New York, NY, USA, 2008; pp. 113–133. [Google Scholar]
- Hartman, G.L.; West, E.D.; Herman, T.K. Crops that feed the World 2. Soybean—Worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 2011, 3, 5–17. [Google Scholar] [CrossRef]
- Pettersson, D.; Pontoppidan, K. Soybean Meal and The Potential for Upgrading Its Feeding Value by Enzyme Supplementation. In Soybean; Hany, A.E.-S., Ed.; IntechOpen: Rijeka, Italy, 2013; Chapter 13. [Google Scholar]
- Phommalath, S.; Teraishi, M.; Yoshikawa, T.; Saito, H.; Tsukiyama, T.; Nakazaki, T.; Tanisaka, T.; Okumoto, Y. Wide genetic variation in phenolic compound content of seed coats among black soybean cultivars. Breed. Sci. 2014, 64, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-H.; Kim, K.-S.; Ko, J.-M.; Choi, M.-S.; Kang, B.-K.; Kwon, S.-W.; Jun, T.-H. Quantitative trait locus analysis for soybean (Glycine max) seed protein and oil concentrations using selected breeding populations. Plant Breed. 2019, 138, 95–104. [Google Scholar] [CrossRef]
- Lilian Hasegawa, F.; Rayane Nunes, L.; Mayla, D.C.M. Soybean Functional Proteins and the Synthetic Biology. In Soybean; Takuji, O., Yoshihiko, T., Norikuni, O., Takashi, S., Sayuri, T., Eds.; IntechOpen: Rijeka, Italy, 2022; Chapter 11. [Google Scholar]
- Sessa, D.J. Processing of soybean hulls to enhance the distribution and extraction of value-added proteins. J. Sci. Food Agric. 2004, 84, 75–82. [Google Scholar] [CrossRef]
- Tkaczewska, J.; Zając, M.; Jamróz, E.; Derbew, H. Utilising waste from soybean processing as raw materials for the production of preparations with antioxidant properties, serving as natural food preservatives—A pilot study. LWT 2022, 160, 113282. [Google Scholar] [CrossRef]
- Krishnan, H.B. Engineering Soybean for Enhanced Sulfur Amino Acid Content. Crop Sci. 2005, 45, 454–461. [Google Scholar] [CrossRef]
- Basson, A.R.; Ahmed, S.; Almutairi, R.; Seo, B.; Cominelli, F. Regulation of Intestinal Inflammation by Soybean and Soy-Derived Compounds. Foods 2021, 10, 774. [Google Scholar] [CrossRef] [PubMed]
- Erickson, D.R. Chapter 5—Overview of Modern Soybean Processing and Links Between Processes. In Practical Handbook of Soybean Processing and Utilization; Erickson, D.R., Ed.; AOCS Press: Urbana, IL, USA, 1995; pp. 56–64. [Google Scholar]
- Patil, G.; Mian, R.; Vuong, T.; Pantalone, V.; Song, Q.; Chen, P.; Shannon, G.J.; Carter, T.C.; Nguyen, H.T. Molecular mapping and genomics of soybean seed protein: A review and perspective for the future. TAG. Theor. Appl. Genet. Theor. Und Angew. Genet. 2017, 130, 1975–1991. [Google Scholar] [CrossRef]
- Patil, G.; Vuong, T.D.; Kale, S.; Valliyodan, B.; Deshmukh, R.; Zhu, C.; Wu, X.; Bai, Y.; Yungbluth, D.; Lu, F.; et al. Dissecting genomic hotspots underlying seed protein, oil, and sucrose content in an interspecific mapping population of soybean using high-density linkage mapping. Plant Biotechnol. J. 2018, 16, 1939–1953. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Z.; Hou, X.; Li, X. Genetic mapping and functional genomics of soybean seed protein. Mol. Breed. New Strateg. Plant Improv. 2023, 43, 29. [Google Scholar] [CrossRef]
- Roell, M.S.; Zurbriggen, M.D. The impact of synthetic biology for future agriculture and nutrition. Curr. Opin. Biotechnol. 2020, 61, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Min, C.W.; Hyeon, H.; Gupta, R.; Park, J.; Cheon, Y.E.; Lee, G.H.; Jang, J.W.; Ryu, H.W.; Lee, B.W.; Park, S.U.; et al. Integrated Proteomics and Metabolomics Analysis Highlights Correlative Metabolite-Protein Networks in Soybean Seeds Subjected to Warm-Water Soaking. J. Agric. Food Chem. 2020, 68, 8057–8067. [Google Scholar] [CrossRef] [PubMed]
- Islam, N.; Krishnan, H.B.; Natarajan, S. Proteomic Profiling of Fast Neutron-Induced Soybean Mutant Unveiled Pathways Associated with Increased Seed Protein Content. J. Proteome Res. 2020, 19, 3936–3944. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xu, W.; Li, L.; Dong, M.; Wan, Y.; He, X.; Huang, K.; Jin, W. iTRAQ-based quantitative tissue proteomic analysis of differentially expressed proteins (DEPs) in non-transgenic and transgenic soybean seeds. Sci. Rep. 2018, 8, 17681. [Google Scholar] [CrossRef]
- Kim, H.T.; Choi, U.K.; Ryu, H.S.; Lee, S.J.; Kwon, O.S. Mobilization of storage proteins in soybean seed (Glycine max L.) during germination and seedling growth. Biochim. Et Biophys. Acta 2011, 1814, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.J.; Gao, J.; Xu, D.; Thelen, J.J. Phosphoproteomic analysis of seed maturation in Arabidopsis, rapeseed, and soybean. Plant Physiol. 2012, 159, 517–528. [Google Scholar] [CrossRef]
- Yu, X.; Jin, H.; Fu, X.; Yang, Q.; Yuan, F. Quantitative proteomic analyses of two soybean low phytic acid mutants to identify the genes associated with seed field emergence. BMC Plant Biol. 2019, 19, 569. [Google Scholar] [CrossRef]
- Murad, A.M.; Rech, E.L. NanoUPLC-MSE proteomic data assessment of soybean seeds using the Uniprot database. BMC Biotechnol. 2012, 12, 82. [Google Scholar] [CrossRef]
- Barbosa, H.S.; Arruda, S.C.; Azevedo, R.A.; Arruda, M.A. New insights on proteomics of transgenic soybean seeds: Evaluation of differential expressions of enzymes and proteins. Anal. Bioanal. Chem. 2012, 402, 299–314. [Google Scholar] [CrossRef]
- Yin, Y.; Ren, Z.; Zhang, L.; Qin, L.; Chen, L.; Liu, L.; Jia, R.; Xue, K.; Liu, B.; Wang, X. In Situ Proteomic Analysis of Herbicide-Resistant Soybean and Hybrid Seeds via Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging. J. Agric. Food Chem. 2023, 71, 7140–7151. [Google Scholar] [CrossRef]
- Min, C.W.; Park, J.; Bae, J.W.; Agrawal, G.K.; Rakwal, R.; Kim, Y.; Yang, P.; Kim, S.T.; Gupta, R. In-Depth Investigation of Low-Abundance Proteins in Matured and Filling Stages Seeds of Glycine max Employing a Combination of Protamine Sulfate Precipitation and TMT-Based Quantitative Proteomic Analysis. Cells 2020, 9, 1517. [Google Scholar] [CrossRef]
- Soy Protein Ingredients Market To Reach USD 16.6 Billion By 2026, Reports and Data. Available online: https://www.globenewswire.com/news-release/2019/09/24/1920183/0/en/Soy-Protein-Ingredients-Market-To-Reach-USD-16-6-Billion-By-2026-Reports-And-Data.html (accessed on 22 May 2022).
- Krishnan, H.B.; Jez, J.M. Review: The promise and limits for enhancing sulfur-containing amino acid content of soybean seed. Plant Sci. Int. J. Exp. Plant Biol. 2018, 272, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Shoveller, A.K.; Stoll, B.; Ball, R.O.; Burrin, D.G. Nutritional and functional importance of intestinal sulfur amino acid metabolism. J. Nutr. 2005, 135, 1609–1612. [Google Scholar] [CrossRef] [PubMed]
- Samuel, N.N.; Agnes, K.K.-N. Advances in Soybean and Soybean By-Products in Monogastric Nutrition and Health. In Soybean and Nutrition; Hany, E.-S., Ed.; IntechOpen: Rijeka, Italy, 2011; Chapter 7. [Google Scholar]
- Dilger, R.N.; Sands, J.S.; Ragland, D.; Adeola, O. Digestibility of nitrogen and amino acids in soybean meal with added soyhulls. J. Anim. Sci. 2004, 82, 715–724. [Google Scholar] [CrossRef]
- Opapeju, F.O.; Golian, A.; Nyachoti, C.M.; Campbell, L.D. Amino acid digestibility in dry extruded-expelled soybean meal fed to pigs and poultry1. J. Anim. Sci. 2006, 84, 1130–1137. [Google Scholar] [CrossRef]
- Ansia, I.; Stein, H.H.; Brøkner, C.; Hayes, C.A.; Drackley, J.K. Nutrient digestibility and endogenous protein losses in the foregut and small intestine of weaned dairy calves fed calf starters with conventional or enzyme-treated soybean meal. J. Dairy Sci. 2021, 104, 2979–2995. [Google Scholar] [CrossRef]
- Kertz, A.F.; Hill, T.M.; Quigley, J.D., 3rd; Heinrichs, A.J.; Linn, J.G.; Drackley, J.K. A 100-Year Review: Calf nutrition and management. J. Dairy Sci. 2017, 100, 10151–10172. [Google Scholar] [CrossRef]
- Maria John, K.M.; Khan, F.; Luthria, D.L.; Garrett, W.; Natarajan, S. Proteomic analysis of anti-nutritional factors (ANF’s) in soybean seeds as affected by environmental and genetic factors. Food Chem. 2017, 218, 321–329. [Google Scholar] [CrossRef]
- Freire, M.O.; Van Dyke, T.E. Natural resolution of inflammation. Periodontology 2000 2013, 63, 149–164. [Google Scholar] [CrossRef]
- Krishna, A.; Singh, G.; Kumar, D.; Agrawal, K. Physico-chemical characteristics of some new varieties of soybean. J. Food Sci. Technol. 2003, 40, 490–492. [Google Scholar]
- Shi, Y.; Mandal, R.; Singh, A.; Pratap Singh, A. Legume lipoxygenase: Strategies for application in food industry. Legume Sci. 2020, 2, e44. [Google Scholar] [CrossRef]
- Yang, A.; Smyth, H.; Chaliha, M.; James, A. Sensory quality of soymilk and tofu from soybeans lacking lipoxygenases. Food Sci. Nutr. 2016, 4, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.D.C.; Fuganti-Pagliarini, R.; Barbosa, D.A.; Kafer, J.M.; Marin, S.R.R.; Mertz-Henning, L.M.; Nepomuceno, A.L.; Rech, E. Screening de proteínas bioativas com potencial antioxidante em proteoma de soja cv. BRS 537. In Proceedings of the IX Congresso de Soja 2022 Mercosoja, Foz do Iguaçu, Brazil, 19 May 2022. [Google Scholar]
- Embrapa. Soja-BRS 537. Available online: https://www.embrapa.br/en/busca-de-solucoes-tecnologicas/-/produto-servico/8340/soja-brs-537 (accessed on 29 May 2022).
- Yu, Y.; Pieper, R. Using Proteomics to Identify Inflammation During Urinary Tract Infection. Methods Mol. Biol. 2019, 2021, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Platt, M.P.; Fu, H.; Gui, Y.; Wang, Y.; Gonzalez-Juarbe, N.; Zhou, D.; Yu, Y. Global Proteome and Phosphoproteome Characterization of Sepsis-induced Kidney Injury. Mol. Cell. Proteom. MCP 2020, 19, 2030–2047. [Google Scholar] [CrossRef] [PubMed]
- Searle, B.C. Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics 2010, 10, 1265–1269. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Bern, M.W.; Kil, Y.J. Two-dimensional target decoy strategy for shotgun proteomics. J. Proteome Res. 2011, 10, 5296–5301. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molinari, M.D.C.; Fuganti-Pagliarini, R.; Yu, Y.; Florentino, L.H.; Mertz-Henning, L.M.; Lima, R.N.; Bittencourt, D.M.d.C.; Freire, M.O.; Rech, E. Exploring the Proteomic Profile of Soybean Bran: Unlocking the Potential for Improving Protein Quality and Quantity. Plants 2023, 12, 2704. https://doi.org/10.3390/plants12142704
Molinari MDC, Fuganti-Pagliarini R, Yu Y, Florentino LH, Mertz-Henning LM, Lima RN, Bittencourt DMdC, Freire MO, Rech E. Exploring the Proteomic Profile of Soybean Bran: Unlocking the Potential for Improving Protein Quality and Quantity. Plants. 2023; 12(14):2704. https://doi.org/10.3390/plants12142704
Chicago/Turabian StyleMolinari, Mayla Daiane Corre, Renata Fuganti-Pagliarini, Yanbao Yu, Lilian Hasegawa Florentino, Liliane Marcia Mertz-Henning, Rayane Nunes Lima, Daniela Matias de Carvalho Bittencourt, Marcelo Oliveira Freire, and Elibio Rech. 2023. "Exploring the Proteomic Profile of Soybean Bran: Unlocking the Potential for Improving Protein Quality and Quantity" Plants 12, no. 14: 2704. https://doi.org/10.3390/plants12142704
APA StyleMolinari, M. D. C., Fuganti-Pagliarini, R., Yu, Y., Florentino, L. H., Mertz-Henning, L. M., Lima, R. N., Bittencourt, D. M. d. C., Freire, M. O., & Rech, E. (2023). Exploring the Proteomic Profile of Soybean Bran: Unlocking the Potential for Improving Protein Quality and Quantity. Plants, 12(14), 2704. https://doi.org/10.3390/plants12142704





