VPBR: An Automatic and Low-Cost Vision-Based Biophysical Properties Recognition Pipeline for Pumpkin
Abstract
1. Introduction
2. Results
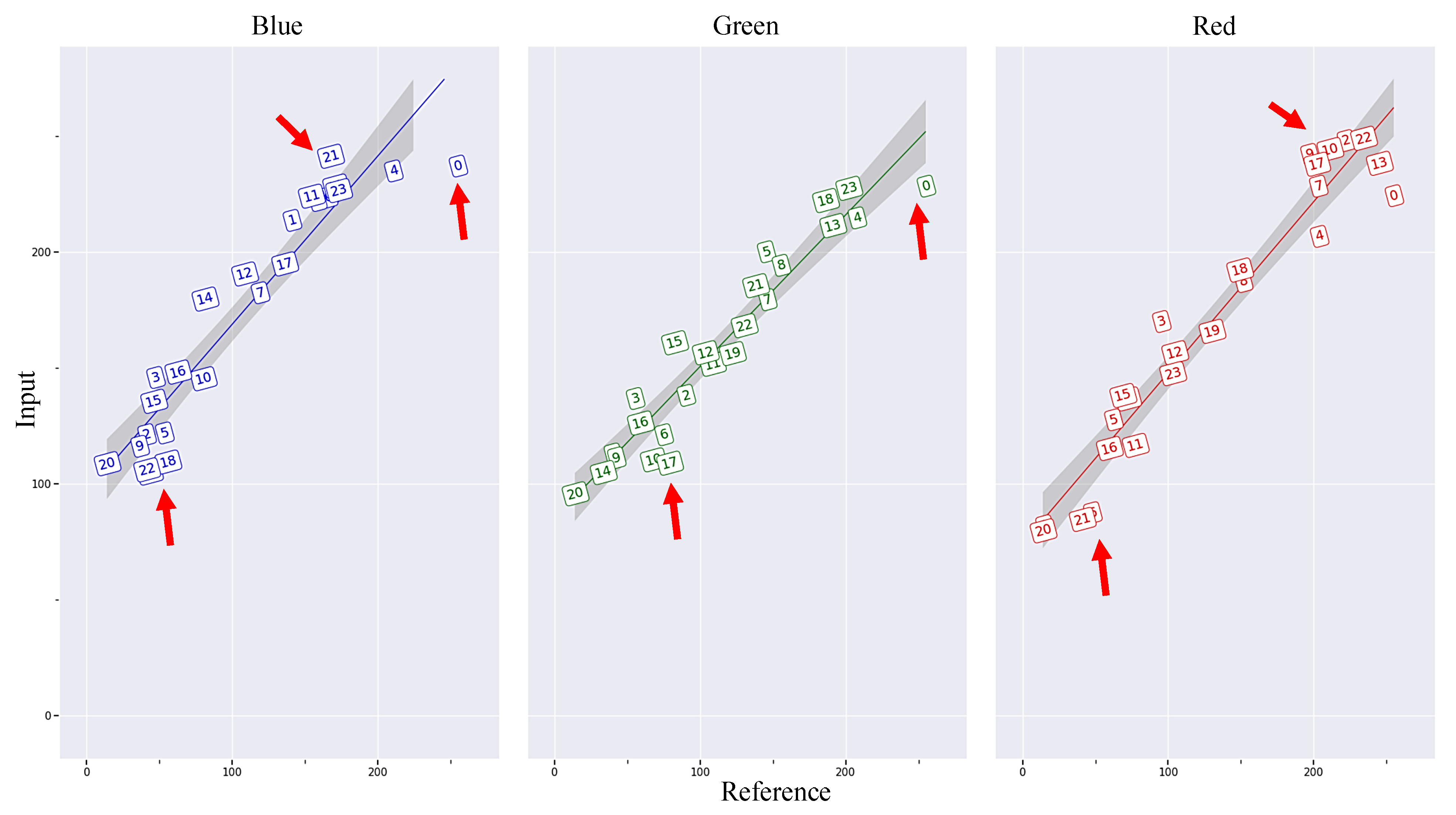
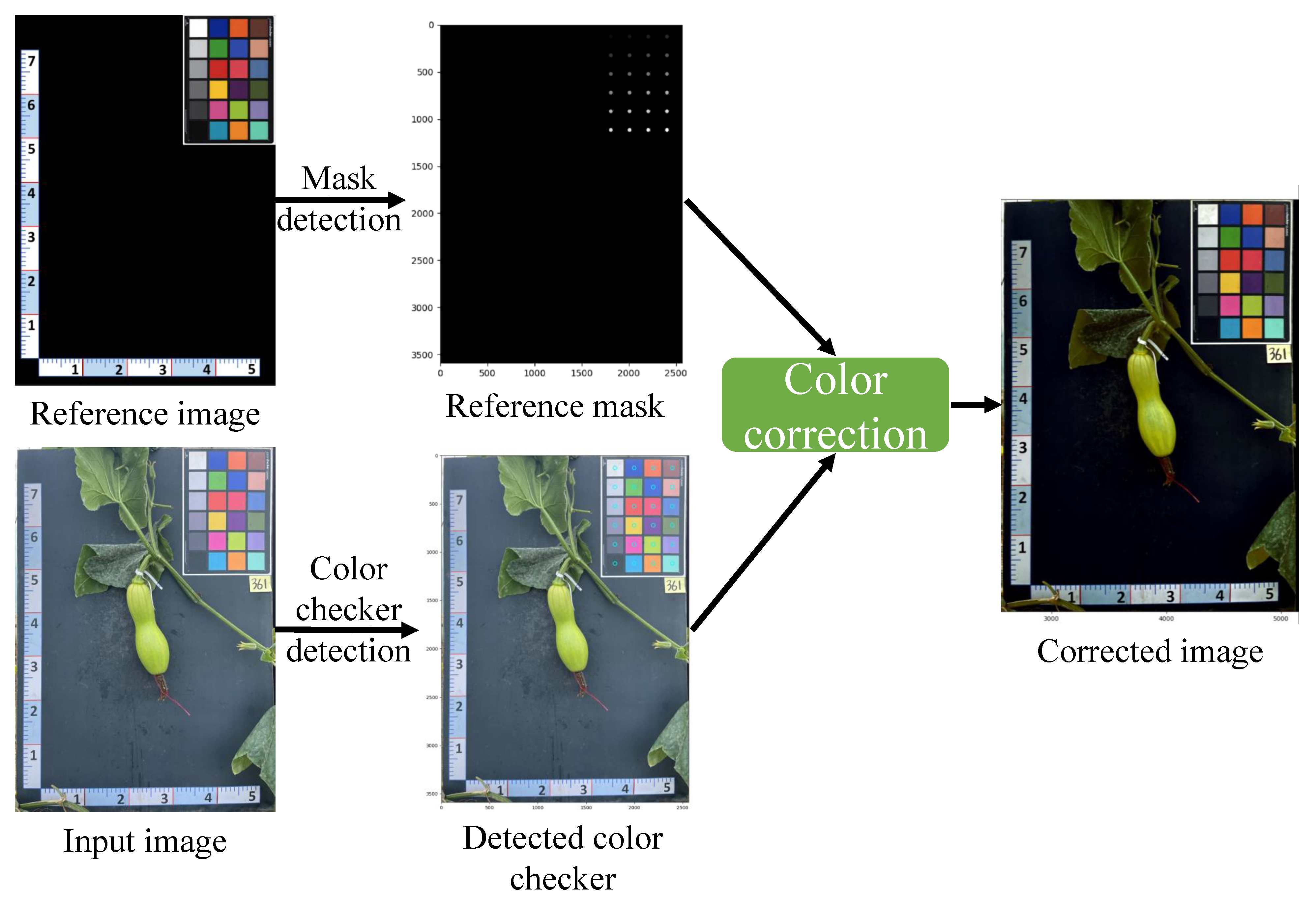
2.1. Color Calibration
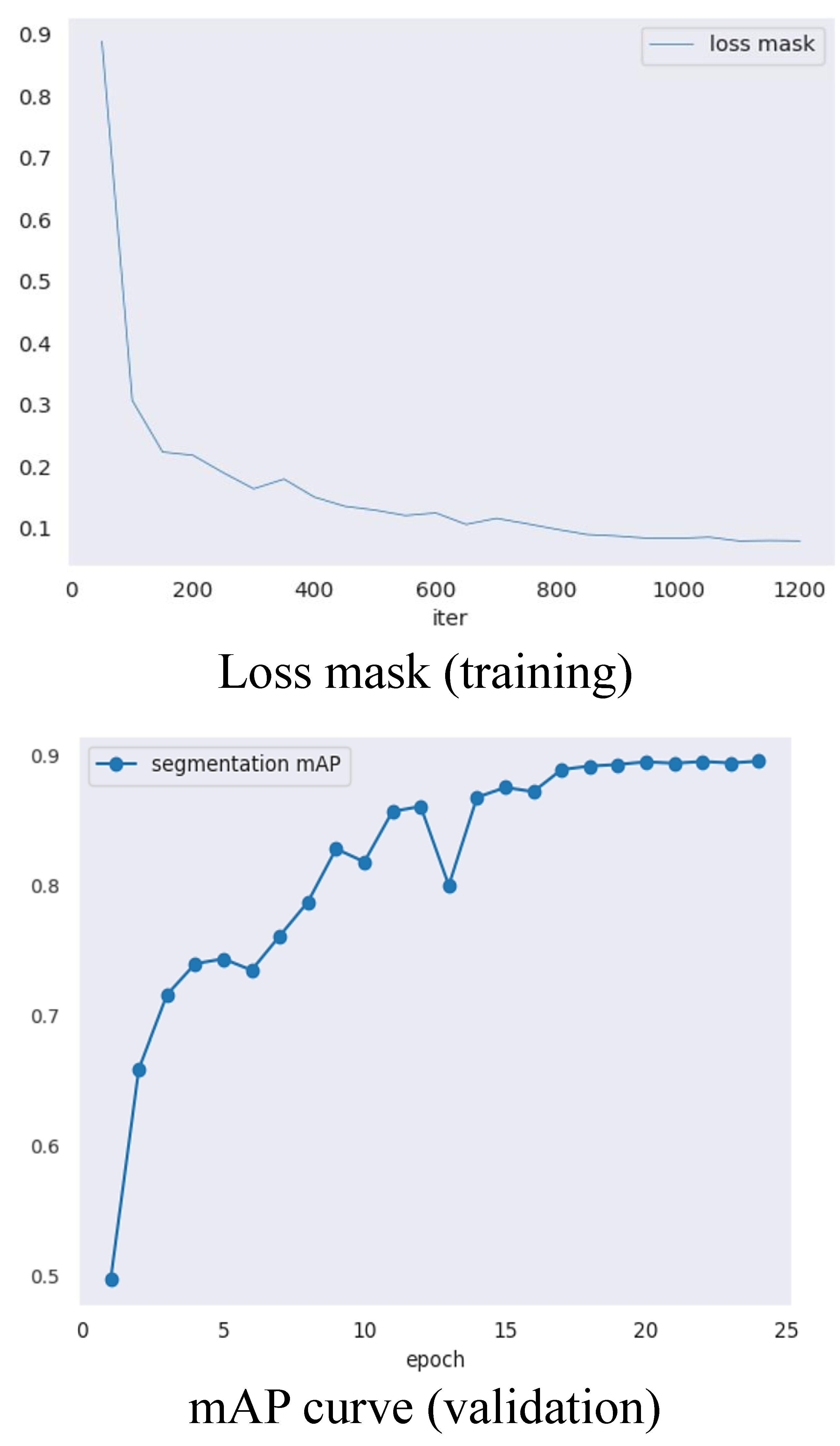
2.2. SOLOv2 Performance Evaluations
2.3. Biophysical Properties Measurement
3. Discussion
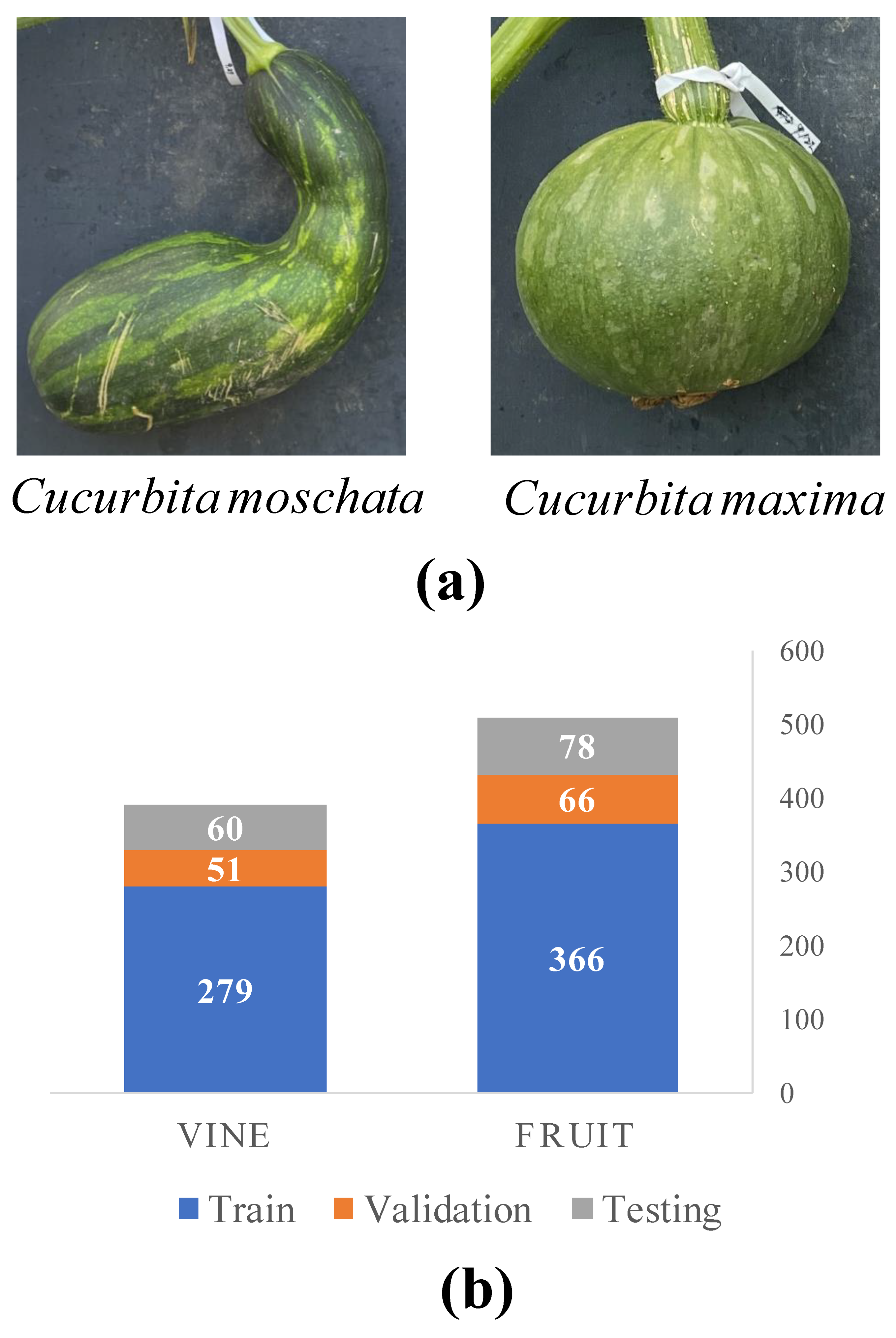
4. Materials
5. Methods
5.1. VPBR
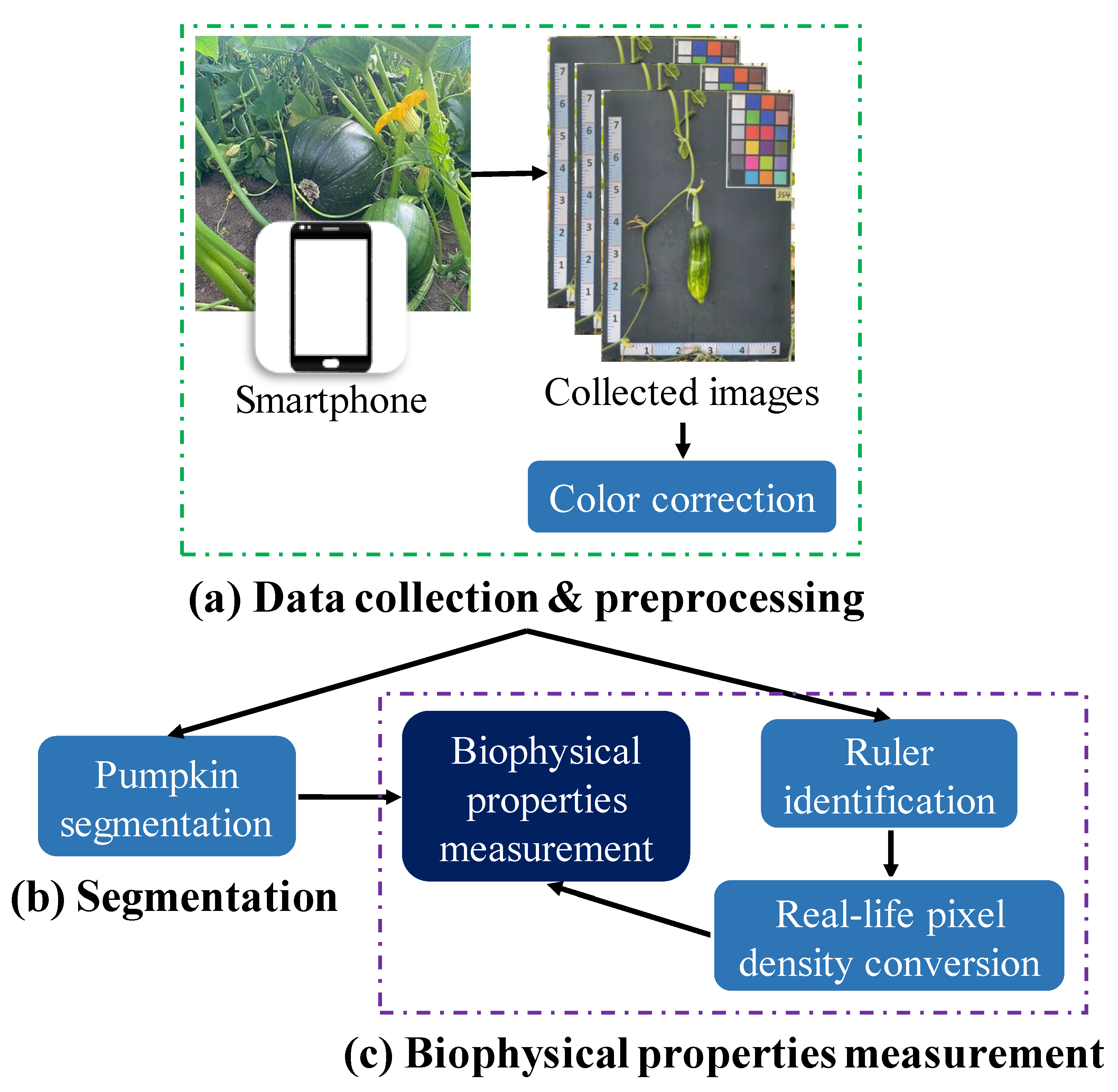
5.1.1. Overall Description
- Data collection and preprocessing (Figure 6a): The pumpkin component segmentation dataset was collected by a Samsung Galaxy S22 device on two pumpkin green houses in November 2022. The constantly changing light conditions of outdoor environments can lead to inconsistencies in the color of images taken at different times. To address this issue and ensure the quality of the dataset, color correction was performed before the training process.
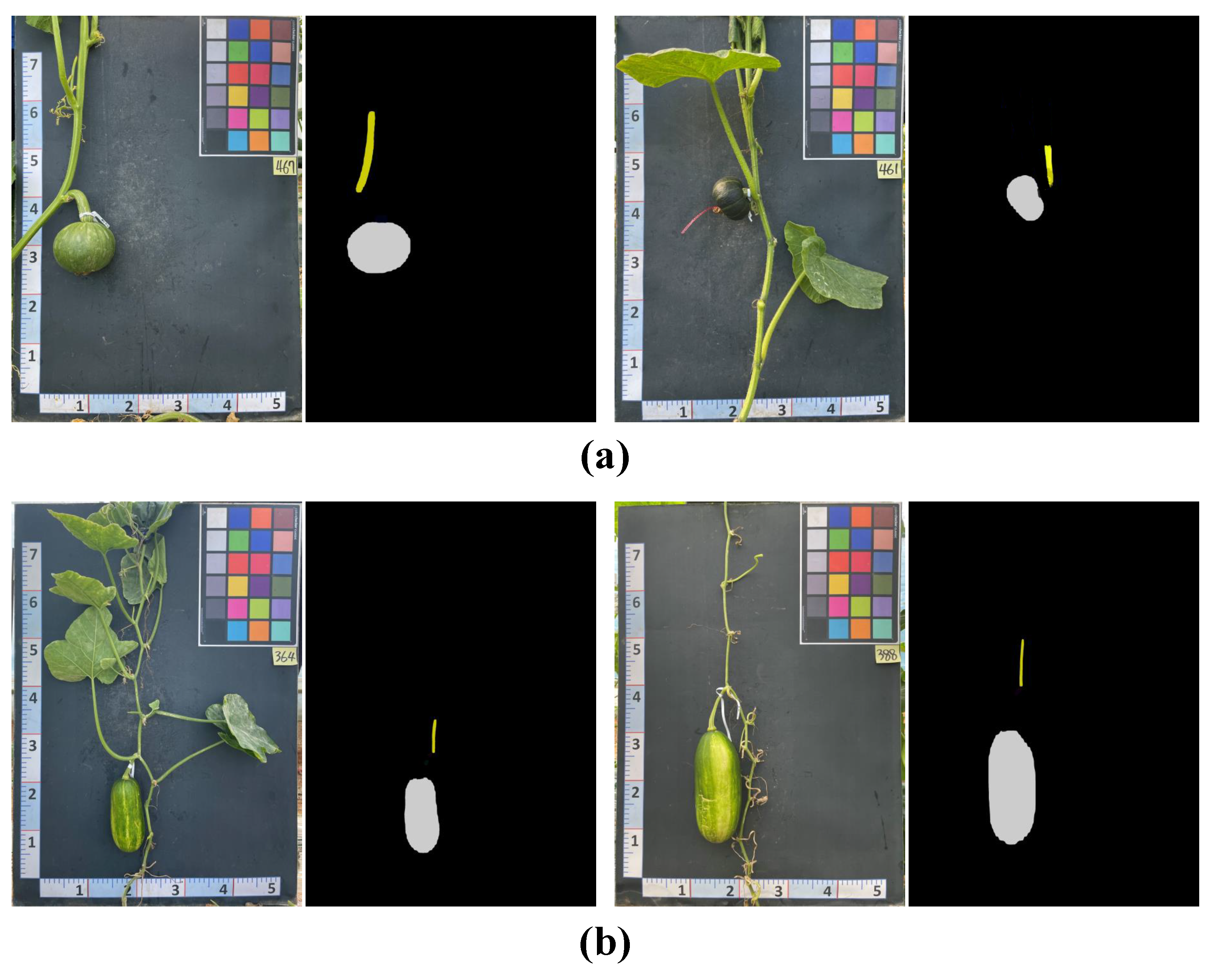
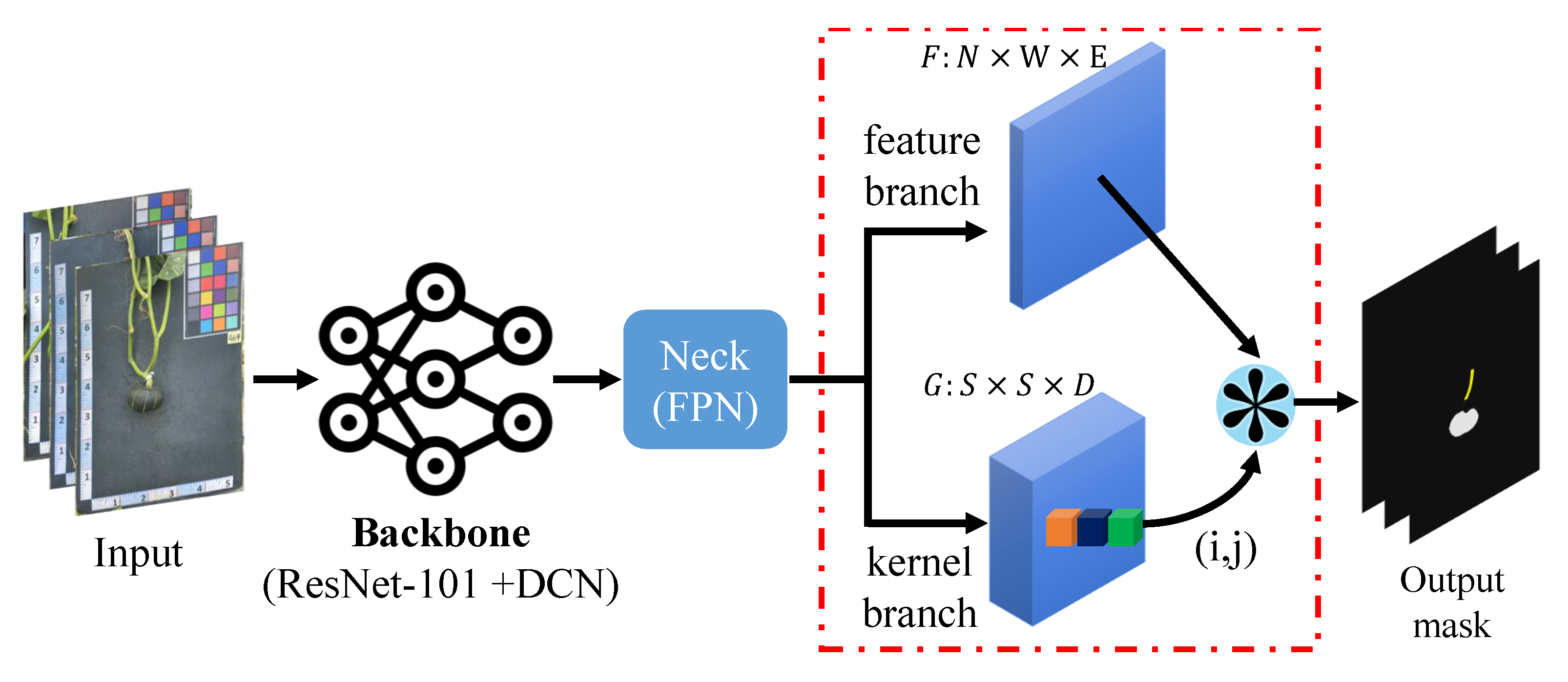
- Pumpkin segmentation (Figure 6b): SOLOv2 [29] is an extension of the mask region-based convolutional neural network (Mask R-CNN) architecture and enables the identification and localization of individual objects within an image. Unlike the traditional two-stage approach, SOLOv2 utilizes a single-stage network for object detection and segmentation, resulting in faster processing times while maintaining high accuracy. In this study, SOLOv2 is applied to segment the pumpkin’s components accurately.
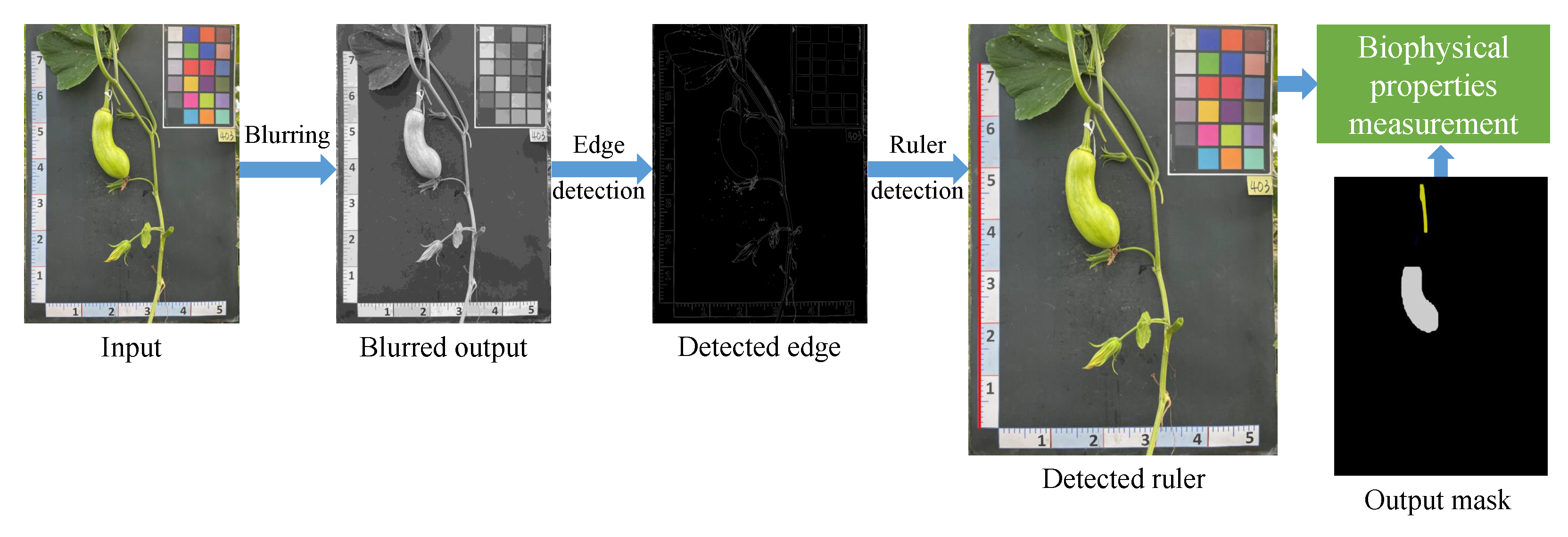
- Biophysical properties measurement (Figure 6c): This study proposes an automated pipeline that utilizes various CV techniques to measure the real-life values of diverse biophysical properties of pumpkins. The pipeline includes recognizing the ruler positioned near the pumpkin as a reference for measurement. This approach enables the efficient and accurate extraction of biophysical property measurements from the captured images.
5.1.2. Data Preprocessing
5.1.3. Pumpkin Segmentation
5.1.4. Biophysical Properties Measurement
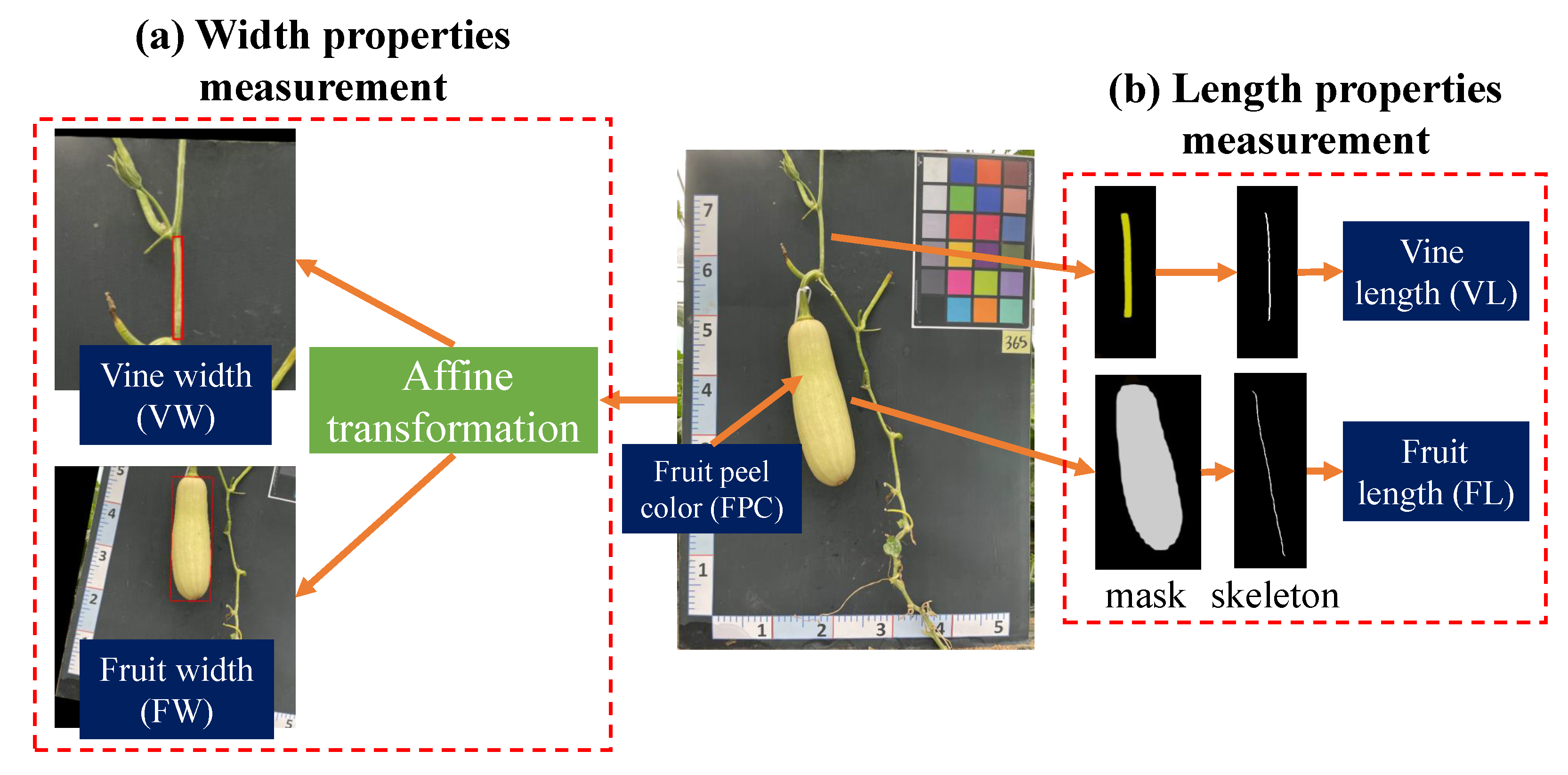
- Width properties measurement (Figure 9a): When dealing with objects that do not have symmetrical shapes, an affine transformation method provides an effective approach to accurately calculate their width, as demonstrated in Figure 9a [39]. Initially, an ellipse is utilized to fit the pumpkin’s fruit and vine components, as it offers a better approximation of the shape compared to a rectangular bounding box. The center coordinates, major and minor axis lengths, and rotation angle of the best-fit ellipse are then extracted, enabling the construction of an affine transformation matrix for rotation. By applying this transformation, the object is aligned with the image’s x and y axes, facilitating the measurement of width by determining the distance between the two farthest points within the transformed object.
- Length properties measurement (Figure 9b): In order to precisely compute the length property, this study proposes the use of a skeletonization algorithm applied to the segmented mask. This approach offers a more accurate and precise measurement of the length, taking into account any irregularities or asymmetries that may be present in the shape of the fruit. Additionally, the skeleton, being a simplified representation of the fruit’s shape, enables more efficient data processing, facilitating the analysis of large datasets of fruit images.The skeletonization process involves iteratively thinning the object or shape until a one-pixel-wide skeleton is obtained. This simplified representation captures the essential features and structure of the object, providing a streamlined depiction of the object [40]. One commonly used method for generating the skeleton is the medial axis transform, which calculates the centerline of the object and produces a skeleton that represents its main axis of symmetry [41]. Figure 9a displays the skeleton output obtained from the medial axis skeletonization method applied to the input vine and fruit masks. The resulting image is binary, with pixels on the skeleton assigned a value of 1, while all other pixels are assigned a value of 0.Previous research has shown that the object length can be determined using the following formula once the object skeleton is extracted [8]:where the finite length of L is denoted by , and C represents the geometric calibration. Initially, C was introduced as a calibration factor to account for pixel displacements in the mask outputs. However, since the dataset used in this study exhibited no geometric distortion, the parameter C was set to 1. This allowed for the direct summation of the total pixels along the skeleton to calculate L.
- Color estimation: The estimation of the pumpkin’s FPC was based on the standard color variations observed in pumpkins. According to findings of Kaur et al. [1], the color of the pumpkin fruit peel can exhibit variations based on factors such as the pumpkin variety and maturity stage. Typically, pumpkin fruit peel is characterized by an orange hue, which can range from a pale, light orange to a deep, rich shade. Additionally, certain pumpkin varieties may feature green, yellow, or white stripes or patches on their peel. As the pumpkin undergoes ripening, the peel color tends to deepen and become more vibrant. Hence, this study focused on three main FPC categories: orange, green, and light green.Figure 10 visually presents the process of classifying pumpkin’s FPC using the hue, saturation, and value (HSV) color space. The HSV color space is favored over RGB for color detection tasks due to its ability to separate color information from brightness or luminance information, thereby providing a more intuitive representation [42]. In this process, specific color ranges within the HSV color space were defined, enabling the creation of binary masks for each color range. These binary masks were then employed to determine the FPC by identifying the color range with the highest pixel count.
5.2. Experimental Settings
5.2.1. Hardware and Software Platform
5.2.2. Optimizer, Loss Function and Hyperparameters
5.2.3. Evaluation Metrics
6. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RGB | Red–green–blue color channel |
| RCNN | Region-based convolutional neural network |
| UAV | Unmanned aerial vehicle |
| ISSR | Inter-simple sequence repeat |
| FPN | Feature pyramid network |
| FL | Fruit length |
| FW | Fruit width |
| VL | Vine length |
| VW | Vine width |
| FPC | Fruit peel color |
| HSV | Hue, saturation, and value |
| CNN | Convolutional neural network |
| DCN | Deformable convolutional networks |
References
- Kaur, S.; Panghal, A.; Garg, M.; Mann, S.; Khatkar, S.K.; Sharma, P.; Chhikara, N. Functional and nutraceutical properties of pumpkin–A review. Nutr. Food Sci. 2020, 50, 384–401. [Google Scholar] [CrossRef]
- Hussain, A.; Kausar, T.; Sehar, S.; Sarwar, A.; Ashraf, A.H.; Jamil, M.A.; Noreen, S.; Rafique, A.; Iftikhar, K.; Aslam, J.; et al. Utilization of pumpkin, pumpkin powders, extracts, isolates, purified bioactives and pumpkin based functional food products: A key strategy to improve health in current post COVID 19 period: An updated review. Appl. Food Res. 2022, 2, 100241. [Google Scholar] [CrossRef]
- Lee, G.S.; Han, G.P. Characteristics of sponge cake prepared by the addition of sweet pumpkin powder. Korean J. Food Preserv. 2018, 25, 507–515. [Google Scholar] [CrossRef]
- Yunli, W.; Yangyang, W.; Wenlong, X.; Chaojie, W.; Chongshi, C.; Shuping, Q. Genetic diversity of pumpkin based on morphological and SSR markers. Pak. J. Bot. 2020, 52, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Mishra, D.; Yadav, G.; Dwivedi, D. Genetic diversity assessment for morphological, yield and biochemical traits in genotypes of pumpkin. J. Pharmacogn. Phytochem. 2017, 6, 14–18. [Google Scholar]
- Nankar, A.N.; Todorova, V.; Tringovska, I.; Pasev, G.; Radeva-Ivanova, V.; Ivanova, V.; Kostova, D. A step towards Balkan Capsicum annuum L. core collection: Phenotypic and biochemical characterization of 180 accessions for agronomic, fruit quality, and virus resistance traits. PLoS ONE 2020, 15, e0237741. [Google Scholar] [CrossRef]
- Öztürk, H.İ.; Dönderalp, V.; Bulut, H.; Korkut, R. Morphological and molecular characterization of some pumpkin (Cucurbita pepo L.) genotypes collected from Erzincan province of Turkey. Sci. Rep. 2022, 12, 6814. [Google Scholar] [CrossRef]
- Dang, L.M.; Wang, H.; Li, Y.; Park, Y.; Oh, C.; Nguyen, T.N.; Moon, H. Automatic tunnel lining crack evaluation and measurement using deep learning. Tunn. Undergr. Space Technol. 2022, 124, 104472. [Google Scholar] [CrossRef]
- Dang, L.M.; Lee, S.; Li, Y.; Oh, C.; Nguyen, T.N.; Song, H.K.; Moon, H. Daily and seasonal heat usage patterns analysis in heat networks. Sci. Rep. 2022, 12, 9165. [Google Scholar] [CrossRef]
- Wittstruck, L.; Kühling, I.; Trautz, D.; Kohlbrecher, M.; Jarmer, T. UAV-based RGB imagery for Hokkaido pumpkin (Cucurbita max.) detection and yield estimation. Sensors 2020, 21, 118. [Google Scholar] [CrossRef]
- Dang, L.M.; Hassan, S.I.; Suhyeon, I.; kumar Sangaiah, A.; Mehmood, I.; Rho, S.; Seo, S.; Moon, H. UAV based wilt detection system via convolutional neural networks. Sustain. Comput. Inform. Syst. 2020, 28, 100250. [Google Scholar] [CrossRef]
- Ropelewska, E.; Popińska, W.; Sabanci, K.; Aslan, M.F. Flesh of pumpkin from ecological farming as part of fruit suitable for non-destructive cultivar classification using computer vision. Eur. Food Res. Technol. 2022, 248, 893–898. [Google Scholar] [CrossRef]
- Longchamps, L.; Tisseyre, B.; Taylor, J.; Sagoo, L.; Momin, A.; Fountas, S.; Manfrini, L.; Ampatzidis, Y.; Schueller, J.K.; Khosla, R. Yield sensing technologies for perennial and annual horticultural crops: A review. Precis. Agric. 2022, 23, 2407–2448. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Dang, L.M.; Sadeghi-Niaraki, A.; Moon, H. Crop pest recognition in natural scenes using convolutional neural networks. Comput. Electron. Agric. 2020, 169, 105174. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Lee, S.; Nguyen, P.C.; Nguyen-Xuan, H.; Lee, J. Geometrically nonlinear postbuckling behavior of imperfect FG-CNTRC shells under axial compression using isogeometric analysis. Eur. J. Mech. A Solids 2020, 84, 104066. [Google Scholar] [CrossRef]
- Li, Z.; Guo, R.; Li, M.; Chen, Y.; Li, G. A review of computer vision technologies for plant phenotyping. Comput. Electron. Agric. 2020, 176, 105672. [Google Scholar] [CrossRef]
- Mao, Y.; Merkle, C.; Allebach, J. A color image analysis tool to help users choose a makeup foundation color. Electron. Imaging 2022, 34, 373-1–373-6. [Google Scholar] [CrossRef]
- Chen, H.; Sun, K.; Tian, Z.; Shen, C.; Huang, Y.; Yan, Y. BlendMask: Top-down meets bottom-up for instance segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 8573–8581. [Google Scholar]
- Chen, K.; Pang, J.; Wang, J.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Shi, J.; Ouyang, W.; et al. Hybrid task cascade for instance segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 4974–4983. [Google Scholar]
- Huang, Z.; Huang, L.; Gong, Y.; Huang, C.; Wang, X. Mask scoring R-CNN. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 6409–6418. [Google Scholar]
- Yu, Y.; Zhang, K.; Yang, L.; Zhang, D. Fruit detection for strawberry harvesting robot in non-structural environment based on Mask-RCNN. Comput. Electron. Agric. 2019, 163, 104846. [Google Scholar] [CrossRef]
- Falk, K.G.; Jubery, T.Z.; O’Rourke, J.A.; Singh, A.; Sarkar, S.; Ganapathysubramanian, B.; Singh, A.K. Soybean root system architecture trait study through genotypic, phenotypic, and shape-based clusters. Plant Phenom. 2020, 2020, 1925495. [Google Scholar] [CrossRef]
- Sunoj, S.; Igathinathane, C.; Saliendra, N.; Hendrickson, J.; Archer, D. Color calibration of digital images for agriculture and other applications. ISPRS J. Photogramm. Remote Sens. 2018, 146, 221–234. [Google Scholar] [CrossRef]
- Dang, L.M.; Min, K.; Nguyen, T.N.; Park, H.Y.; Lee, O.N.; Song, H.K.; Moon, H. Vision-Based White Radish Phenotypic Trait Measurement with Smartphone Imagery. Agronomy 2023, 13, 1630. [Google Scholar] [CrossRef]
- Nguyen, T.K.; Dang, L.M.; Song, H.K.; Moon, H.; Lee, S.J.; Lim, J.H. Wild Chrysanthemums Core Collection: Studies on Leaf Identification. Horticulturae 2022, 8, 839. [Google Scholar] [CrossRef]
- Neupane, C.; Koirala, A.; Walsh, K.B. In-orchard sizing of mango fruit: 1. Comparison of machine vision based methods for on-the-go estimation. Horticulturae 2022, 8, 1223. [Google Scholar] [CrossRef]
- Gsmarena. Samsung Galaxy S22 5G. 2022. Available online: https://www.gsmarena.com/samsung_galaxy_s22_5g-11253.php (accessed on 7 July 2023).
- Xrite. Xrite Colorchecker. 2022. Available online: https://www.xrite.com (accessed on 7 July 2023).
- Wang, H.; Li, Y.; Dang, L.M.; Moon, H. An efficient attention module for instance segmentation network in pest monitoring. Comput. Electron. Agric. 2022, 195, 106853. [Google Scholar] [CrossRef]
- Zhou, Y.; Gao, K.; Guo, Y.; Dou, Z.; Cheng, H.; Chen, Z. Color correction method for digital camera based on variable-exponent polynomial regression. In Communications, Signal Processing, and Systems: Proceedings of the 2018 CSPS Volume II: Signal Processing, Dalian, China, 14–16 July 2018; Springer: Singapore, 2019; pp. 111–118. [Google Scholar]
- Wang, X.; Zhang, R.; Kong, T.; Li, L.; Shen, C. Solov2: Dynamic and fast instance segmentation. Adv. Neural Inf. Process. Syst. 2020, 33, 17721–17732. [Google Scholar]
- Minh, D.; Wang, H.X.; Li, Y.F.; Nguyen, T.N. Explainable artificial intelligence: A comprehensive review. Artif. Intell. Rev. 2022, 55, 3503–3568. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Lee, J.; Dinh-Tien, L.; Minh Dang, L. Deep learned one-iteration nonlinear solver for solid mechanics. Int. J. Numer. Methods Eng. 2022, 123, 1841–1860. [Google Scholar] [CrossRef]
- Sun, X.; Fang, W.; Gao, C.; Fu, L.; Majeed, Y.; Liu, X.; Gao, F.; Yang, R.; Li, R. Remote estimation of grafted apple tree trunk diameter in modern orchard with RGB and point cloud based on SOLOv2. Comput. Electron. Agric. 2022, 199, 107209. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common objects in context. In Proceedings of the Computer Vision—ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014, Proceedings, Part V; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- For the Protection of New Varieties of Plants. The International Union for the Protection of New Varieties of Plants. 2022. Available online: https://www.upov.int/portal/index.html.en (accessed on 7 July 2023).
- Giełczyk, A.; Marciniak, A.; Tarczewska, M.; Lutowski, Z. Pre-processing methods in chest X-ray image classification. PLoS ONE 2022, 17, e0265949. [Google Scholar] [CrossRef]
- Xue, X.; Zhang, K.; Tan, K.C.; Feng, L.; Wang, J.; Chen, G.; Zhao, X.; Zhang, L.; Yao, J. Affine transformation-enhanced multifactorial optimization for heterogeneous problems. IEEE Trans. Cybern. 2020, 52, 6217–6231. [Google Scholar] [CrossRef]
- Saha, P.K.; Borgefors, G.; di Baja, G.S. A survey on skeletonization algorithms and their applications. Pattern Recognit. Lett. 2016, 76, 3–12. [Google Scholar] [CrossRef]
- Mayer, J.; Wartzack, S. A concept towards automated reconstruction of topology optimized structures using medial axis skeletons. In Proceedings of the Munich Symposium on Lightweight Design 2020: Tagungsband zum Münchner Leichtbauseminar 2020; Springer: Berlin/Heidelberg, Germany, 2021; pp. 28–35. [Google Scholar]
- Ajmal, A.; Hollitt, C.; Frean, M.; Al-Sahaf, H. A comparison of RGB and HSV colour spaces for visual attention models. In Proceedings of the 2018 International Conference on Image and Vision Computing New Zealand (IVCNZ), Auckland, New Zealand, 19–21 November 2018; pp. 1–6. [Google Scholar]
- MMDetection. 2022. Available online: https://mmdetection.readthedocs.io/en/latest/ (accessed on 7 July 2023).
- PlantCV. 2022. Available online: https://plantcv.readthedocs.io/en/stable/ (accessed on 7 July 2023).
- Shafiq, M.; Gu, Z. Deep residual learning for image recognition: A survey. Appl. Sci. 2022, 12, 8972. [Google Scholar] [CrossRef]
- Hodson, T.O. Root-mean-square error (RMSE) or mean absolute error (MAE): When to use them or not. Geosci. Model Dev. 2022, 15, 5481–5487. [Google Scholar] [CrossRef]

| Model | Mask AP (%) | Inference Speed (FPS) |
|---|---|---|
| BlendMask [18] | 85.9 | 11 |
| HTC [19] | 85.6 | 6 |
| MS R-CNN [20] | 82.1 | 10.7 |
| Mask-RCNN [21] | 84.8 | 11.2 |
| SOLOv2 | 88 | 19.4 |
| Sample | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | ||
| GT | FL (mm) | 6.8 | 27.8 | 14.1 | 8.4 | 9.5 | 22.6 | 25.9 | 6.2 | 6.9 | 6.3 |
| FW (mm) | 10.2 | 9.6 | 12.2 | 12.9 | 12.1 | 9.2 | 8.1 | 8.6 | 11.1 | 9.7 | |
| VL (mm) | 12.2 | 10.3 | 16.2 | 12.8 | 10.7 | 10 | 8.9 | 14.8 | 10.6 | 6.5 | |
| VW (mm) | 1.4 | 0.9 | 1.2 | 1.64 | 1.8 | 0.67 | 1 | 1.1 | 1.4 | 1.6 | |
| FPC | G | OR | LG | G | G | LG | LG | G | G | G | |
| Pre | FL (mm) | 6.8 | 28.1 | 14.1 | 8.3 | 9.2 | 23 | 26.1 | 6.5 | 7 | 6.8 |
| FW (mm) | 9.9 | 9.1 | 12.3 | 13.1 | 12 | 9.1 | 8 | 8.9 | 11.5 | 9.5 | |
| VL (mm) | 12 | 10.1 | 15.9 | 12.6 | 10.9 | 9.8 | 8.4 | 14.2 | 10 | 6.3 | |
| VW (mm) | 1.4 | 0.85 | 1.3 | 1.63 | 1.8 | 0.68 | 1 | 1.2 | 1.3 | 1.6 | |
| FPC | G | OR | LG | G | G | LG | LG | G | G | G | |
| Accuracy (%) | 99.7 | 98.6 | 100 | 82.9 | 100 | 93.7 | 96.1 | 97.4 | 99 | 96.1 | |
| MAE | 0.12 | 0.26 | 0.12 | 0.15 | 0.15 | 0.2 | 0.2 | 0.32 | 0.3 | 0.22 | |
| MAPE (%) | 1.14 | 4.4 | 2.75 | 2.6 | 1.46 | 1.58 | 1.9 | 1.9 | 4.46 | 3.2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, L.M.; Nadeem, M.; Nguyen, T.N.; Park, H.Y.; Lee, O.N.; Song, H.-K.; Moon, H. VPBR: An Automatic and Low-Cost Vision-Based Biophysical Properties Recognition Pipeline for Pumpkin. Plants 2023, 12, 2647. https://doi.org/10.3390/plants12142647
Dang LM, Nadeem M, Nguyen TN, Park HY, Lee ON, Song H-K, Moon H. VPBR: An Automatic and Low-Cost Vision-Based Biophysical Properties Recognition Pipeline for Pumpkin. Plants. 2023; 12(14):2647. https://doi.org/10.3390/plants12142647
Chicago/Turabian StyleDang, L. Minh, Muhammad Nadeem, Tan N. Nguyen, Han Yong Park, O New Lee, Hyoung-Kyu Song, and Hyeonjoon Moon. 2023. "VPBR: An Automatic and Low-Cost Vision-Based Biophysical Properties Recognition Pipeline for Pumpkin" Plants 12, no. 14: 2647. https://doi.org/10.3390/plants12142647
APA StyleDang, L. M., Nadeem, M., Nguyen, T. N., Park, H. Y., Lee, O. N., Song, H.-K., & Moon, H. (2023). VPBR: An Automatic and Low-Cost Vision-Based Biophysical Properties Recognition Pipeline for Pumpkin. Plants, 12(14), 2647. https://doi.org/10.3390/plants12142647








