Natural Variation in OASC Gene for Mitochondrial O-Acetylserine Thiollyase Affects Sulfate Levels in Arabidopsis
Abstract
1. Introduction
2. Results
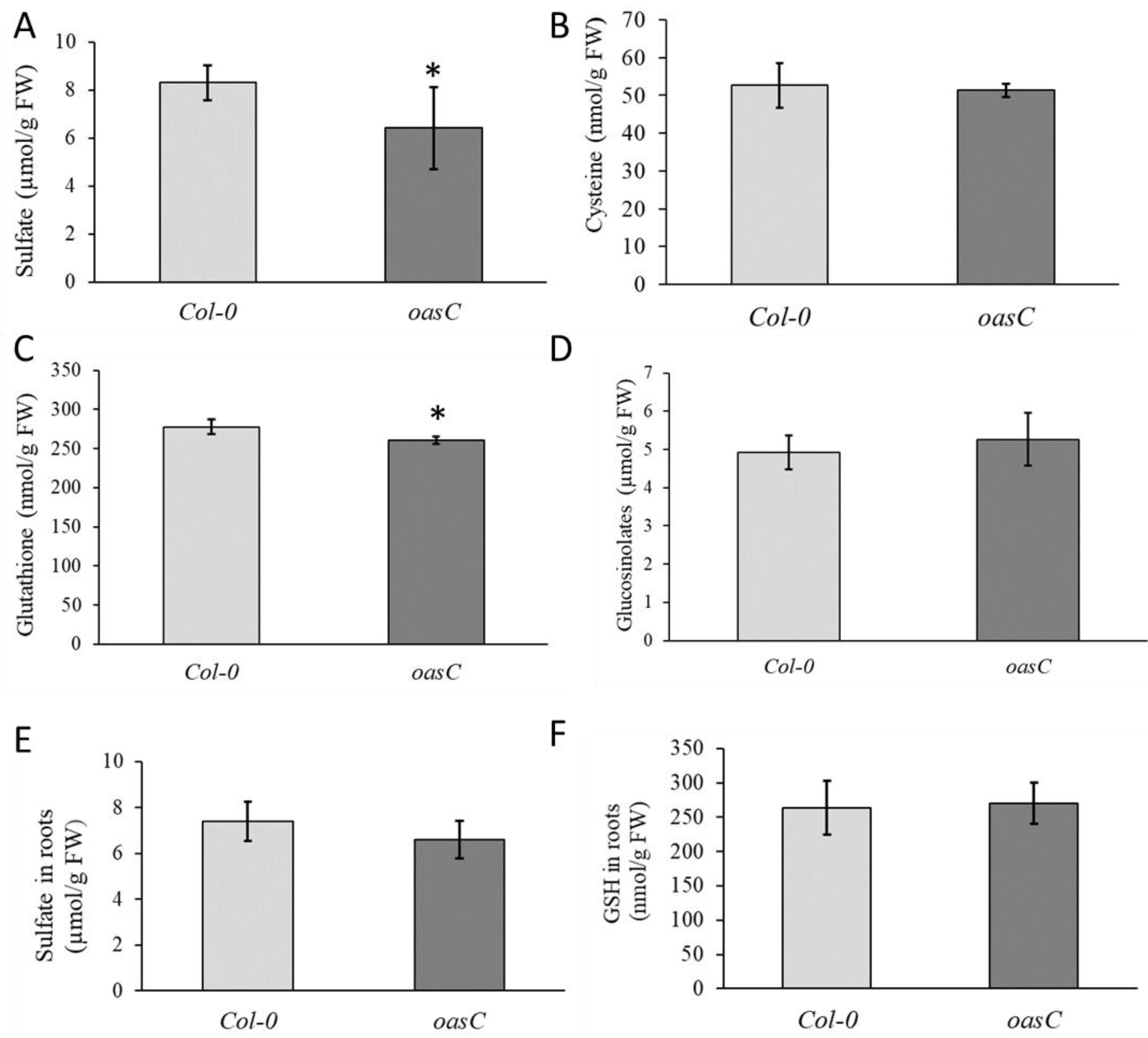
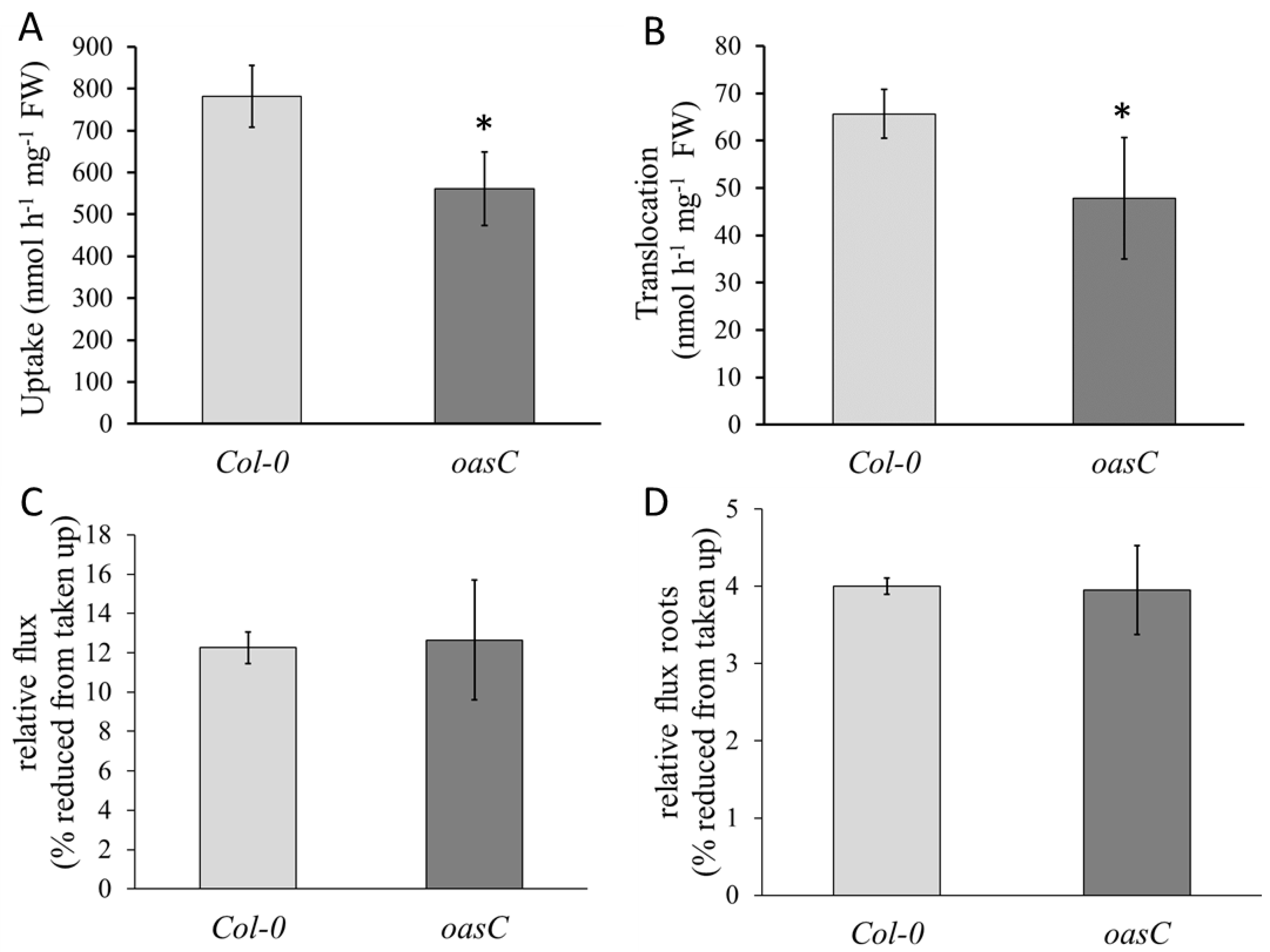
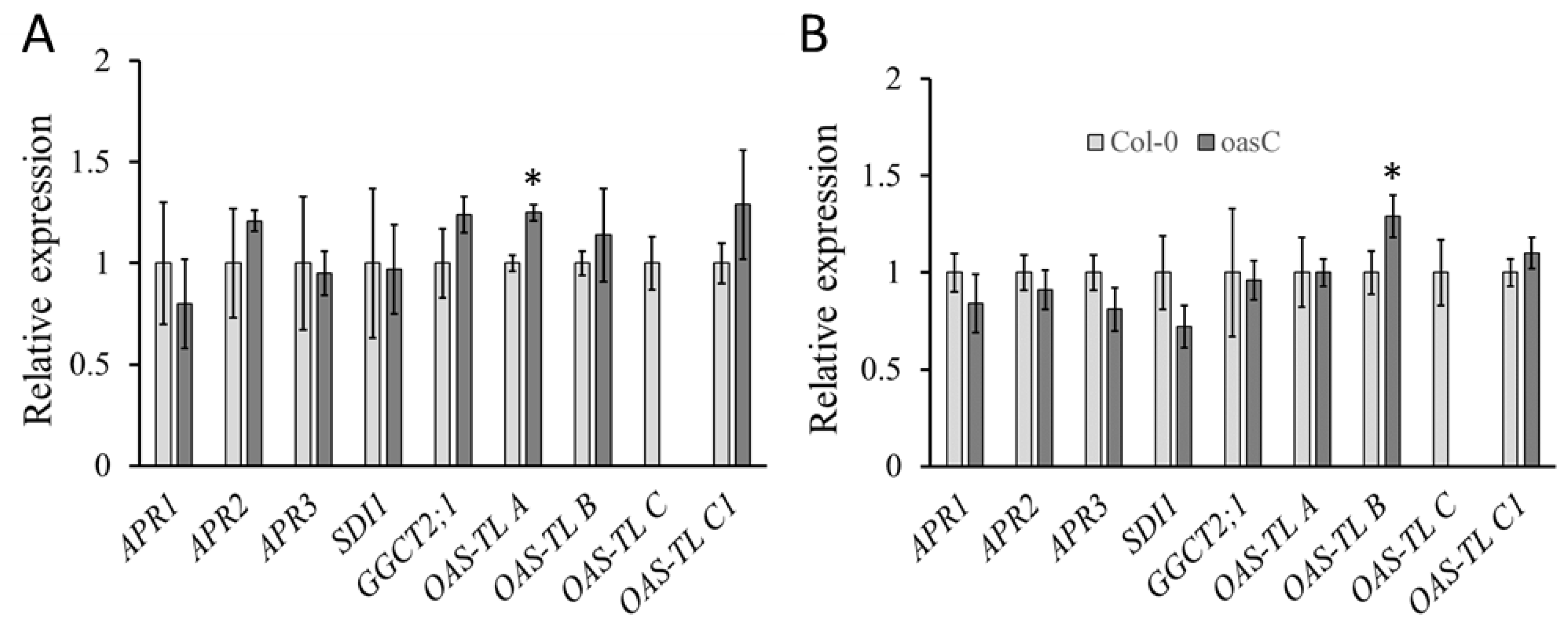
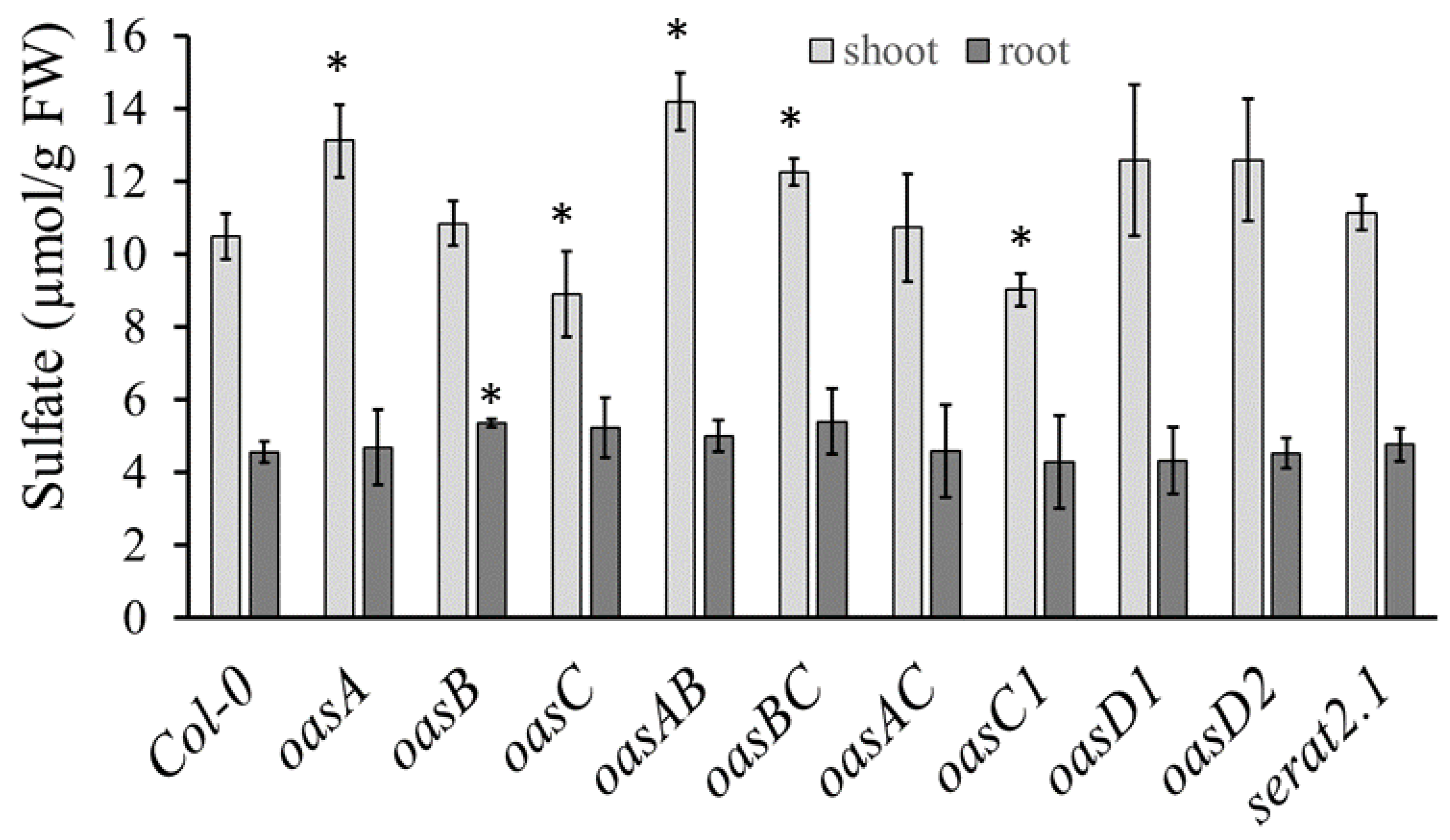
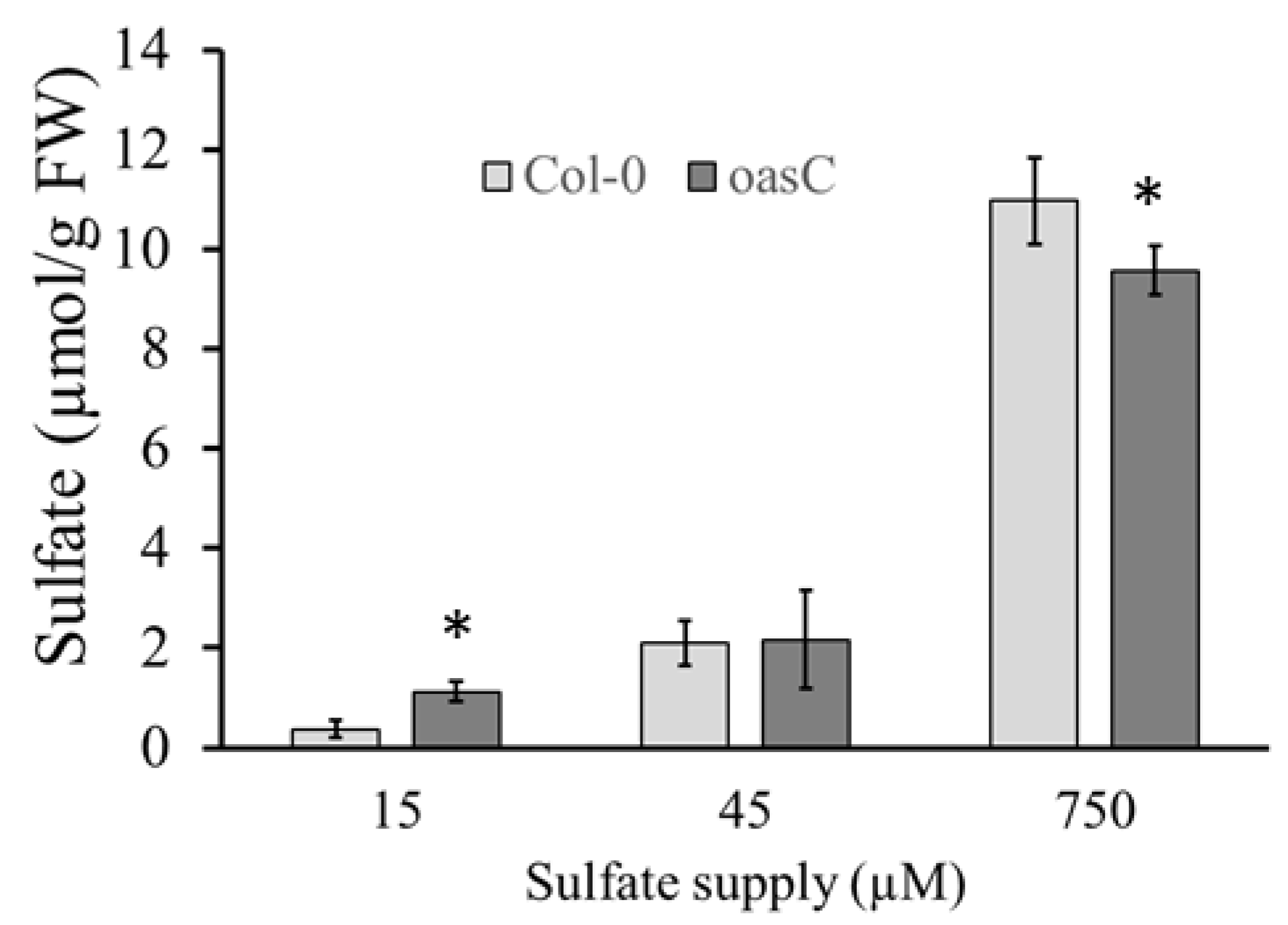
2.1. Characterization of the oasC Mutant
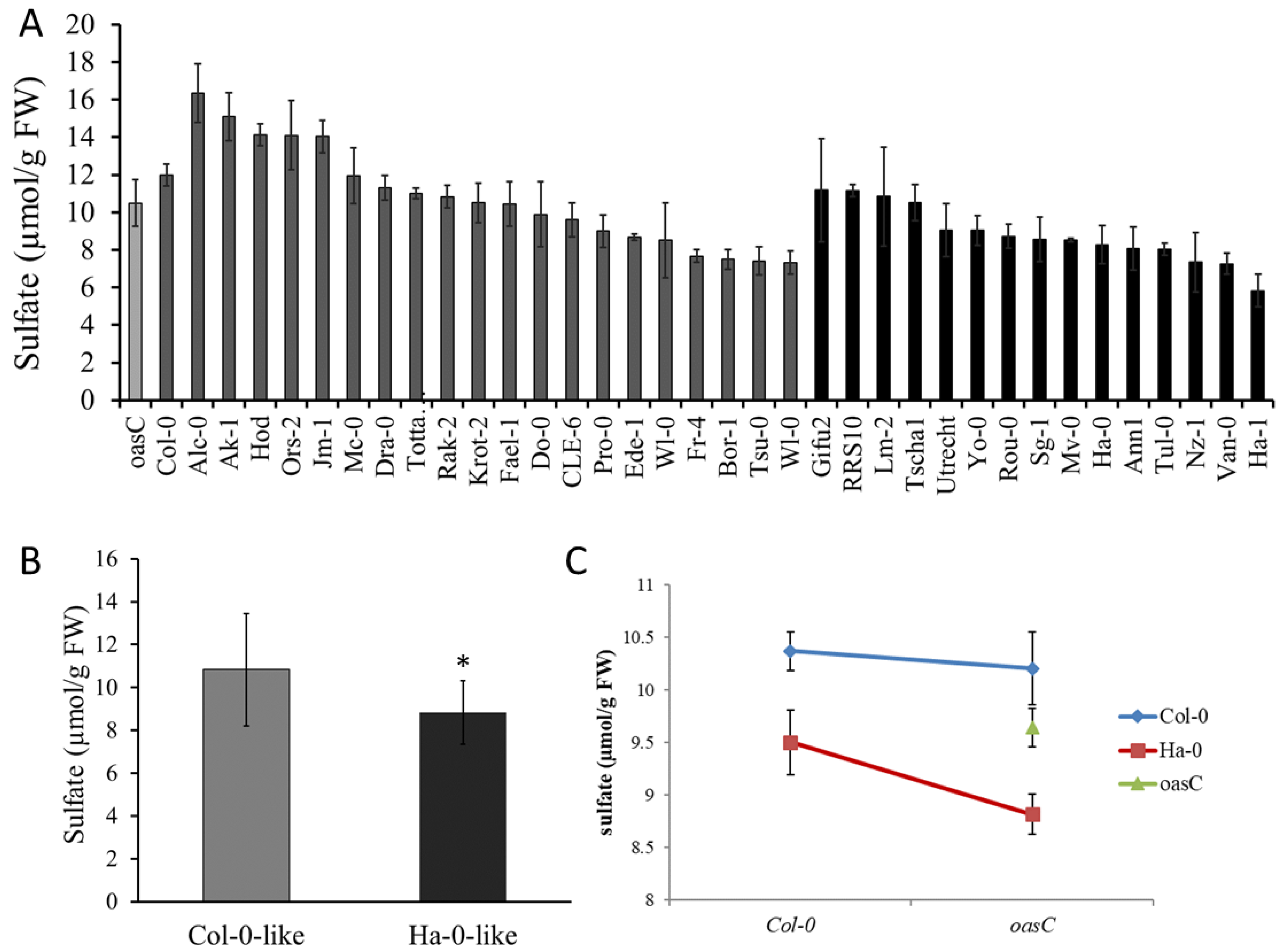
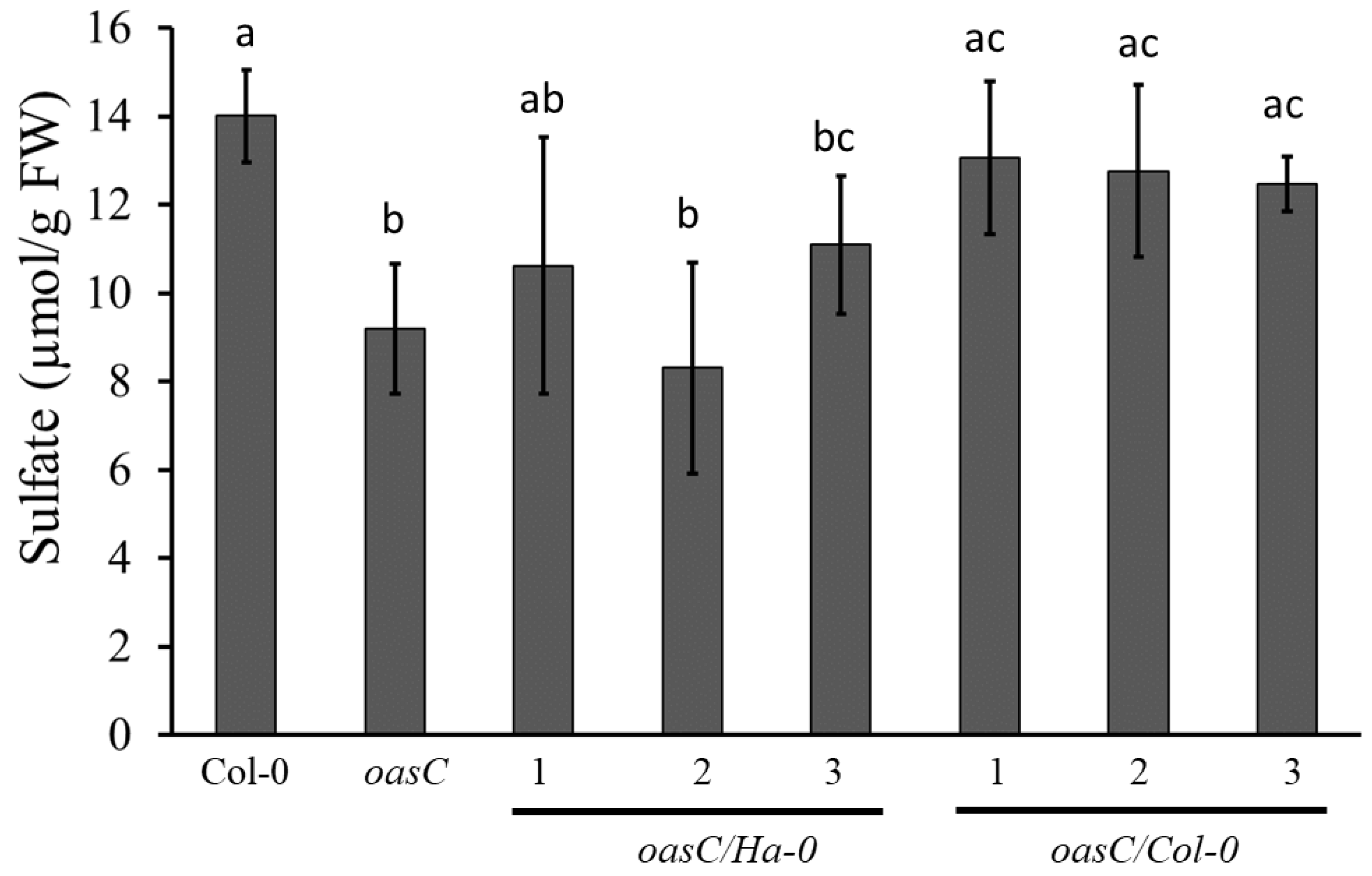
2.2. Amino Acid Variation in OASC Responsible for Variation in Sulfate Level
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Measurements of Sulfur-Containing Metabolites
4.3. Analysis of Sulfate Uptake and Flux
4.4. RNA Extraction and Quantitative PCR Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Q.; Gao, Y.; Yang, A. Sulfur Homeostasis in Plants. Int. J. Mol. Sci. 2020, 21, 8926. [Google Scholar] [CrossRef]
- Scheerer, U.; Haensch, R.; Mendel, R.R.; Kopriva, S.; Rennenberg, H.; Herschbach, C. Sulphur Flux through the Sulphate Assimilation Pathway Is Differently Controlled by Adenosine 5’-Phosphosulphate Reductase under Stress and in Transgenic Poplar Plants Overexpressing Gamma-ECS, SO, or APR. J. Exp. Bot. 2010, 61, 609–622. [Google Scholar] [CrossRef]
- Jobe, T.O.; Zenzen, I.; Karvansara, P.R.; Kopriva, S. Integration of Sulfate Assimilation with Carbon and Nitrogen Metabolism in Transition from C3 to C4 Photosynthesis. J. Exp. Bot. 2019, 70, 4211–4221. [Google Scholar] [CrossRef]
- Aarabi, F.; Kusajima, M.; Tohge, T.; Konishi, T.; Gigolashvili, T.; Takamune, M.; Sasazaki, Y.; Watanabe, M.; Nakashita, H.; Fernie, A.R.; et al. Sulfur Deficiency-Induced Repressor Proteins Optimize Glucosinolate Biosynthesis in Plants. Sci. Adv. 2016, 2, e1601087. [Google Scholar] [CrossRef]
- Krueger, S.; Donath, A.; Lopez-Martin, M.C.; Hoefgen, R.; Gotor, C.; Hesse, H. Impact of Sulfur Starvation on Cysteine Biosynthesis in T-DNA Mutants Deficient for Compartment-Specific Serine-Acetyltransferase. Amino. Acids 2010, 39, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Birke, H.; Heeg, C.; Wirtz, M.; Hell, R. Successful Fertilization Requires the Presence of at Least One Major O-Acetylserine(Thiol)Lyase for Cysteine Synthesis in Pollen of Arabidopsis. Plant Physiol. 2013, 163, 959–972. [Google Scholar] [CrossRef]
- Koprivova, A.; Giovannetti, M.; Baraniecka, P.; Lee, B.R.; Grondin, C.; Loudet, O.; Kopriva, S. Natural Variation in the ATPS1 Isoform of ATP Sulfurylase Contributes to the Control of Sulfate Levels in Arabidopsis. Plant Physiol. 2013, 163, 1133–1141. [Google Scholar] [CrossRef]
- Takahashi, H.; Kopriva, S.; Giordano, M.; Saito, K.; Hell, R. Sulfur Assimilation in Photosynthetic Organisms: Molecular Functions and Regulations of Transporters and Assimilatory Enzymes. Annu. Rev. Plant Biol. 2011, 62, 157–184. [Google Scholar] [CrossRef] [PubMed]
- Riemenschneider, A.; Riedel, K.; Hoefgen, R.; Papenbrock, J.; Hesse, H. Impact of Reduced O-Acetylserine(Thiol)Lyase Isoform Contents on Potato Plant Metabolism. Plant Physiol. 2005, 137, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, M.; Hell, R. Functional Analysis of the Cysteine Synthase Protein Complex from Plants: Structural, Biochemical and Regulatory Properties. J. Plant Physiol. 2006, 163, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kusano, M.; Oikawa, A.; Fukushima, A.; Noji, M.; Saito, K. Physiological Roles of the Beta-Substituted Alanine Synthase Gene Family in Arabidopsis. Plant Physiol. 2008, 146, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Brunold, C.; Suter, M. Intracellular Localization of Serine Acetyltransferase in Spinach Leaves. Planta 1982, 155, 321–327. [Google Scholar] [CrossRef]
- Álvarez, C.; Calo, L.; Romero, L.C.; García, I.; Gotor, C. An O-Acetylserine(Thiol)Lyase Homolog with l-Cysteine Desulfhydrase Activity Regulates Cysteine Homeostasis in Arabidopsis. Plant Physiol. 2010, 152, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Hesse, H.; Lipke, J.; Altmann, T.; Höfgen, R. Molecular Cloning and Expression Analyses of Mitochondrial and Plastidic Isoforms of Cysteine Synthase (O-Acetylserine(Thiol)Lyase) FromArabidopsis Thaliana. Amino. Acids 1999, 16, 113–131. [Google Scholar] [CrossRef] [PubMed]
- Loudet, O.; Saliba-Colombani, V.; Camilleri, C.; Calenge, F.; Gaudon, V.; Koprivova, A.; North, K.A.; Kopriva, S.; Daniel-Vedele, F. Natural Variation for Sulfate Content in Arabidopsis Thaliana Is Highly Controlled by APR2. Nat. Genet. 2007, 39, 896–900. [Google Scholar] [CrossRef]
- Chao, D.Y.; Baraniecka, P.; Danku, J.; Koprivova, A.; Lahner, B.; Luo, H.; Yakubova, E.; Dilkes, B.; Kopriva, S.; Salt, D.E. Variation in Sulfur and Selenium Accumulation Is Controlled by Naturally Occurring Isoforms of the Key Sulfur Assimilation Enzyme ADENOSINE 5′-PHOSPHOSULFATE REDUCTASE2 across the Arabidopsis Species Range. Plant Physiol. 2014, 166, 1593–1608. [Google Scholar] [CrossRef]
- Koprivova, A.; Harper, A.L.; Trick, M.; Bancroft, I.; Kopriva, S. Dissection of the Control of Anion Homeostasis by Associative Transcriptomics in Brassica napus. Plant Physiol. 2014, 166, 442–450. [Google Scholar] [CrossRef]
- Ito, T.; Kitaiwa, T.; Nishizono, K.; Umahashi, M.; Miyaji, S.; Agake, S.-i.; Kuwahara, K.; Yokoyama, T.; Fushinobu, S.; Maruyama-Nakashita, A.; et al. Glutathione Degradation Activity of γ-Glutamyl Peptidase 1 Manifests Its Dual Roles in Primary and Secondary Sulfur Metabolism in Arabidopsis. Plant J. 2022, 111, 1626–1642. [Google Scholar] [CrossRef]
- 1001 Genomes Consortium. 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef]
- Chorianopoulou, S.N.; Bouranis, D.L. The Role of Sulfur in Agronomic Biofortification with Essential Micronutrients. Plants 2022, 11, 1979. [Google Scholar] [CrossRef]
- Koprivova, A.; Kopriva, S. Sulfur metabolism and its manipulation in crops. J. Genet. Genom. 2016, 43, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Jobe, T.O.; Karvansara, P.R.; Zenzen, I.; Kopriva, S. Ensuring Nutritious Food Under Elevated CO2 Conditions: A Case for Improved C4 Crops. Front. Plant Sci. 2020, 11, 1267. [Google Scholar] [CrossRef] [PubMed]
- Raboanatahiry, N.; Li, H.; Yu, L.; Li, M. Rapeseed (Brassica Napus): Processing, Utilization, and Genetic Improvement. Agronomy 2021, 11, 1776. [Google Scholar] [CrossRef]
- Weese, A.; Pallmann, P.; Papenbrock, J.; Riemenschneider, A. Brassica Napus L. Cultivars Show a Broad Variability in Their Morphology, Physiology and Metabolite Levels in Response to Sulfur Limitations and to Pathogen Attack. Front. Plant Sci. 2015, 6, 9. [Google Scholar] [CrossRef]
- Feller, U.; Kopriva, S.; Vassileva, V. Plant Nutrient Dynamics in Stressful Environments: Needs Interfere with Burdens. Agriculture 2018, 8, 97. [Google Scholar] [CrossRef]
- Bus, A.; Körber, N.; Parkin, I.A.P.; Samans, B.; Snowdon, R.J.; Li, J.; Stich, B. Species- and Genome-Wide Dissection of the Shoot Ionome in Brassica Napus and Its Relationship to Seedling Development. Front. Plant Sci. 2014, 5, 485. [Google Scholar] [CrossRef] [PubMed]
- Maillard, A.; Etienne, P.; Diquélou, S.; Trouverie, J.; Billard, V.; Yvin, J.-C.; Ourry, A. Nutrient Deficiencies Modify the Ionomic Composition of Plant Tissues: A Focus on Cross-Talk between Molybdenum and Other Nutrients in Brassica Napus. J. Exp. Bot. 2016, 67, 5631–5641. [Google Scholar] [CrossRef]
- Courbet, G.; D’Oria, A.; Lornac, A.; Diquélou, S.; Pluchon, S.; Arkoun, M.; Koprivova, A.; Kopriva, S.; Etienne, P.; Ourry, A. Specificity and Plasticity of the Functional Ionome of Brassica Napus and Triticum Aestivum Subjected to Macronutrient Deprivation. Front. Plant Sci. 2021, 12, 641648. [Google Scholar] [CrossRef]
- Heeg, C.; Kruse, C.; Jost, R.; Gutensohn, M.; Ruppert, T.; Wirtz, M.; Hell, R. Analysis of the Arabidopsis O-Acetylserine(Thiol)Lyase Gene Family Demonstrates Compartment-Specific Differences in the Regulation of Cysteine Synthesis. Plant Cell 2008, 20, 168–185. [Google Scholar] [CrossRef]
- Lee, B.-R.; Huseby, S.; Koprivova, A.; Chételat, A.; Wirtz, M.; Mugford, S.T.; Navid, E.; Brearley, C.; Saha, S.; Mithen, R.; et al. Effects of Fou8/Fry1 Mutation on Sulfur Metabolism: Is Decreased Internal Sulfate the Trigger of Sulfate Starvation Response? PLoS ONE 2012, 7, e39425. [Google Scholar] [CrossRef]
- Haas, F.H.; Heeg, C.; Queiroz, R.; Bauer, A.; Wirtz, M.; Hell, R. Mitochondrial Serine Acetyltransferase Functions as a Pacemaker of Cysteine Synthesis in Plant Cells. Plant Physiol. 2008, 148, 1055–1067. [Google Scholar] [CrossRef]
- Harper, A.L.; Trick, M.; Higgins, J.; Fraser, F.; Clissold, L.; Wells, R.; Hattori, C.; Werner, P.; Bancroft, I. Associative Transcriptomics of Traits in the Polyploid Crop Species Brassica Napus. Nat. Biotechnol. 2012, 30, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Dietzen, C.; Koprivova, A.; Whitcomb, S.J.; Langen, G.; Jobe, T.O.; Hoefgen, R.; Kopriva, S. The Transcription Factor EIL1 Participates in the Regulation of Sulfur-Deficiency Response. Plant Physiol. 2020, 184, 2120–2136. [Google Scholar] [CrossRef] [PubMed]
- Huseby, S.; Koprivova, A.; Lee, B.R.; Saha, S.; Mithen, R.; Wold, A.B.; Bengtsson, G.B.; Kopriva, S. Diurnal and Light Regulation of Sulphur Assimilation and Glucosinolate Biosynthesis in Arabidopsis. J. Exp. Bot. 2013, 64, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Mugford, S.G.; Lee, B.R.; Koprivova, A.; Matthewman, C.; Kopriva, S. Control of Sulfur Partitioning between Primary and Secondary Metabolism. Plant J. 2011, 65, 96–105. [Google Scholar] [CrossRef]
- Koprivova, A.; Schuck, S.; Jacoby, R.P.; Klinkhammer, I.; Welter, B.; Leson, L.; Martyn, A.; Nauen, J.; Grabenhorst, N.; Mandelkow, J.F.; et al. Root-Specific Camalexin Biosynthesis Controls the Plant Growth-Promoting Effects of Multiple Bacterial Strains. Proc. Natl. Acad. Sci. USA 2019, 116, 15735–15744. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koprivova, A.; Elkatmis, B.; Gerlich, S.C.; Trick, M.; Harper, A.L.; Bancroft, I.; Kopriva, S. Natural Variation in OASC Gene for Mitochondrial O-Acetylserine Thiollyase Affects Sulfate Levels in Arabidopsis. Plants 2023, 12, 35. https://doi.org/10.3390/plants12010035
Koprivova A, Elkatmis B, Gerlich SC, Trick M, Harper AL, Bancroft I, Kopriva S. Natural Variation in OASC Gene for Mitochondrial O-Acetylserine Thiollyase Affects Sulfate Levels in Arabidopsis. Plants. 2023; 12(1):35. https://doi.org/10.3390/plants12010035
Chicago/Turabian StyleKoprivova, Anna, Büsra Elkatmis, Silke C. Gerlich, Martin Trick, Andrea L. Harper, Ian Bancroft, and Stanislav Kopriva. 2023. "Natural Variation in OASC Gene for Mitochondrial O-Acetylserine Thiollyase Affects Sulfate Levels in Arabidopsis" Plants 12, no. 1: 35. https://doi.org/10.3390/plants12010035
APA StyleKoprivova, A., Elkatmis, B., Gerlich, S. C., Trick, M., Harper, A. L., Bancroft, I., & Kopriva, S. (2023). Natural Variation in OASC Gene for Mitochondrial O-Acetylserine Thiollyase Affects Sulfate Levels in Arabidopsis. Plants, 12(1), 35. https://doi.org/10.3390/plants12010035






