Identification of Rice Accessions Having Cold Tolerance at the Seedling Stage and Development of Novel Genotypic Assays for Predicting Cold Tolerance
Abstract
1. Introduction
2. Results
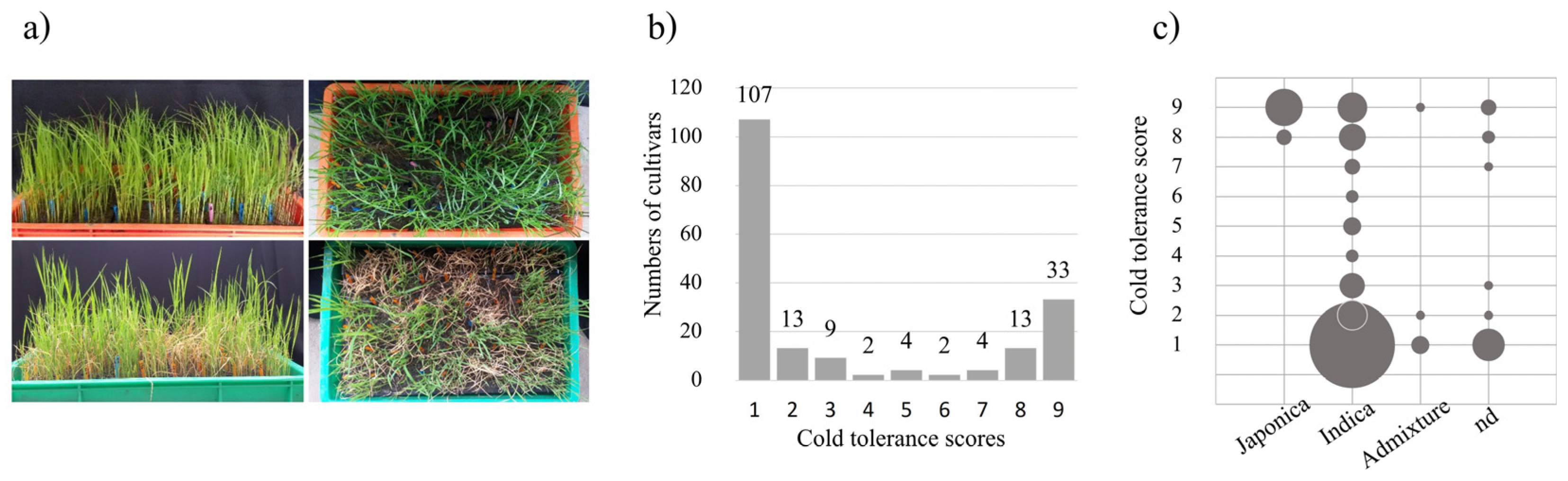
2.1. Phenotypic Variation in Response to Cold Treatment
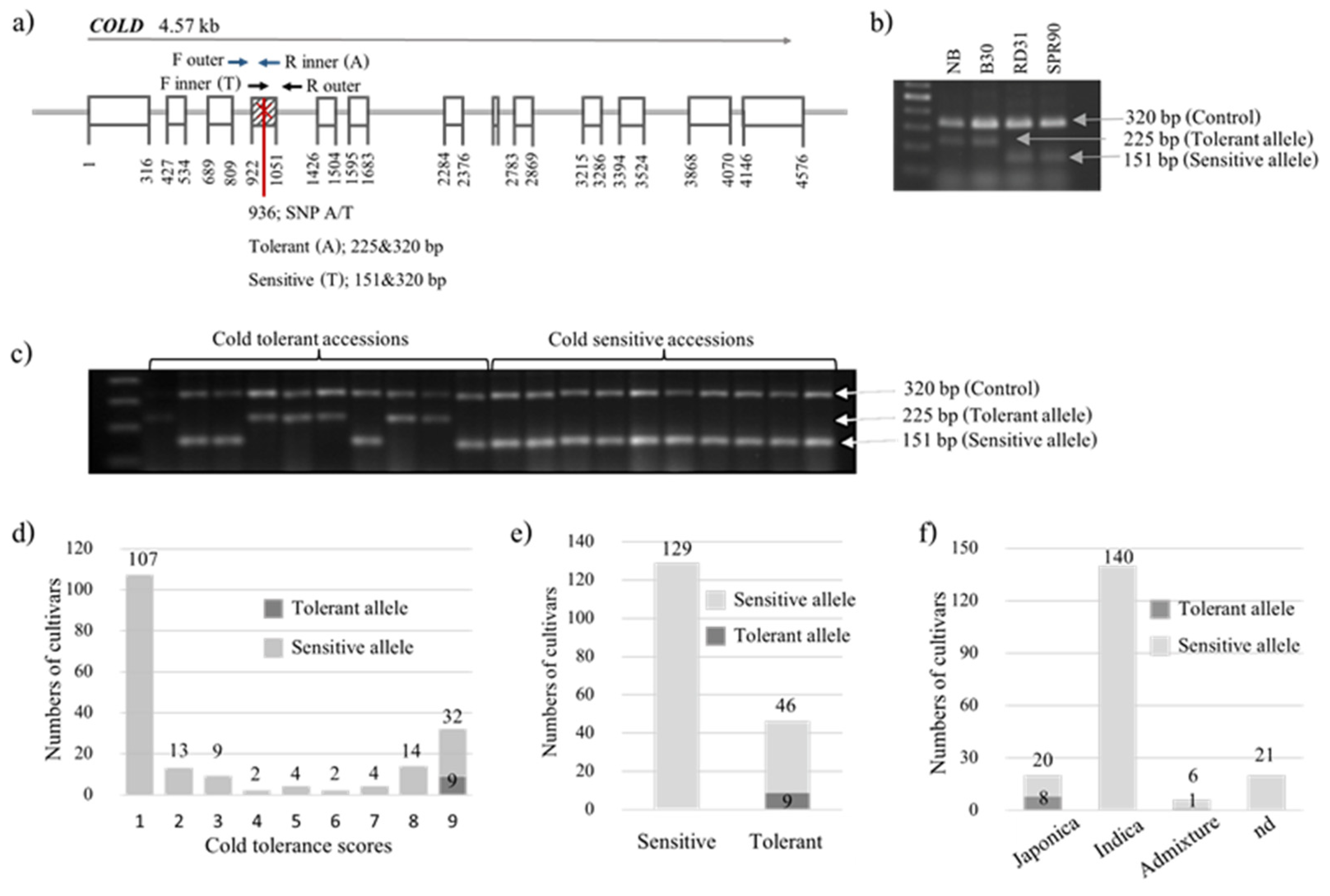
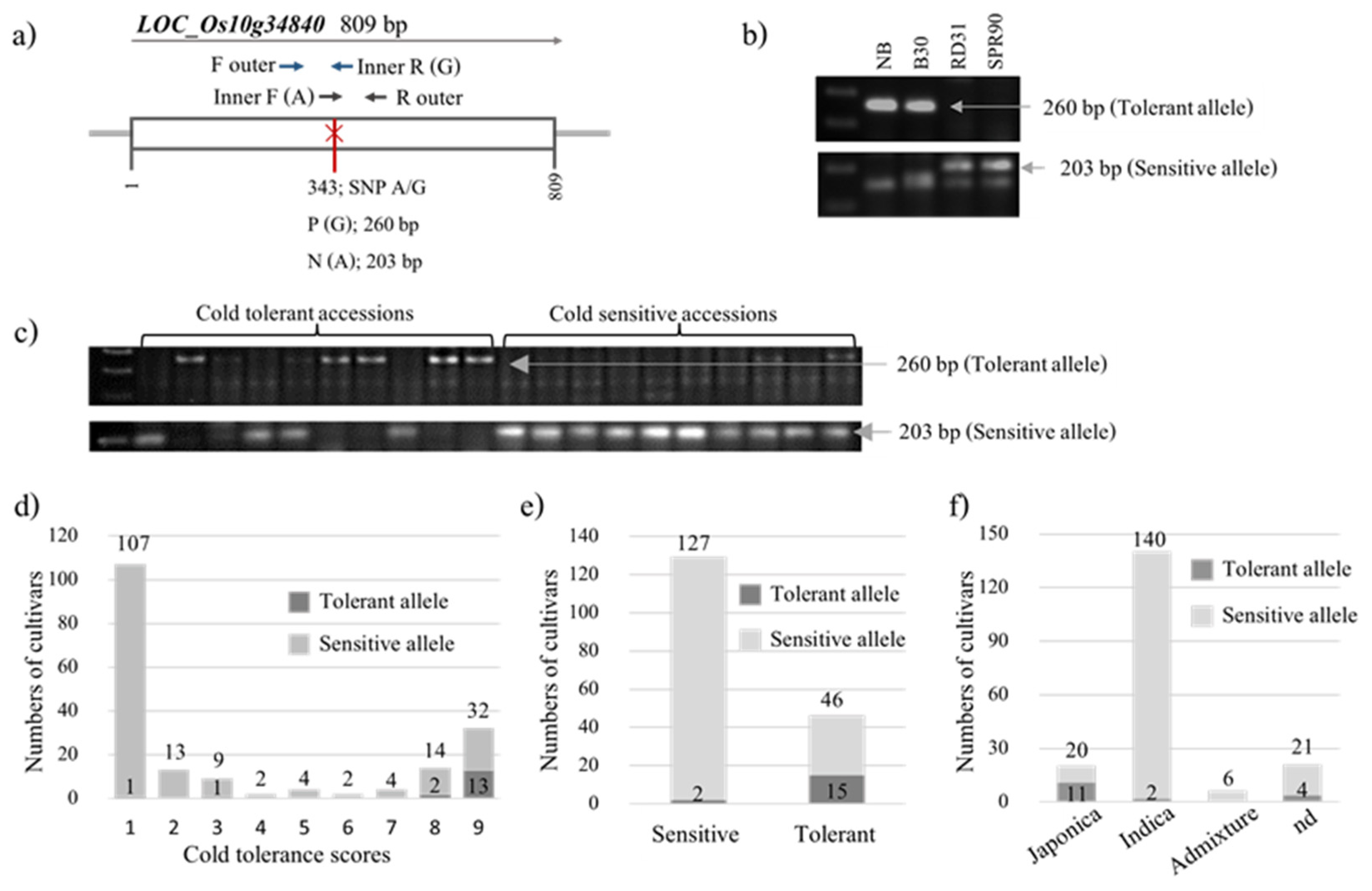
2.2. Marker Development and Genotyping
2.2.1. NAC6
2.2.2. COLD1
2.2.3. LOC_Os10g34840
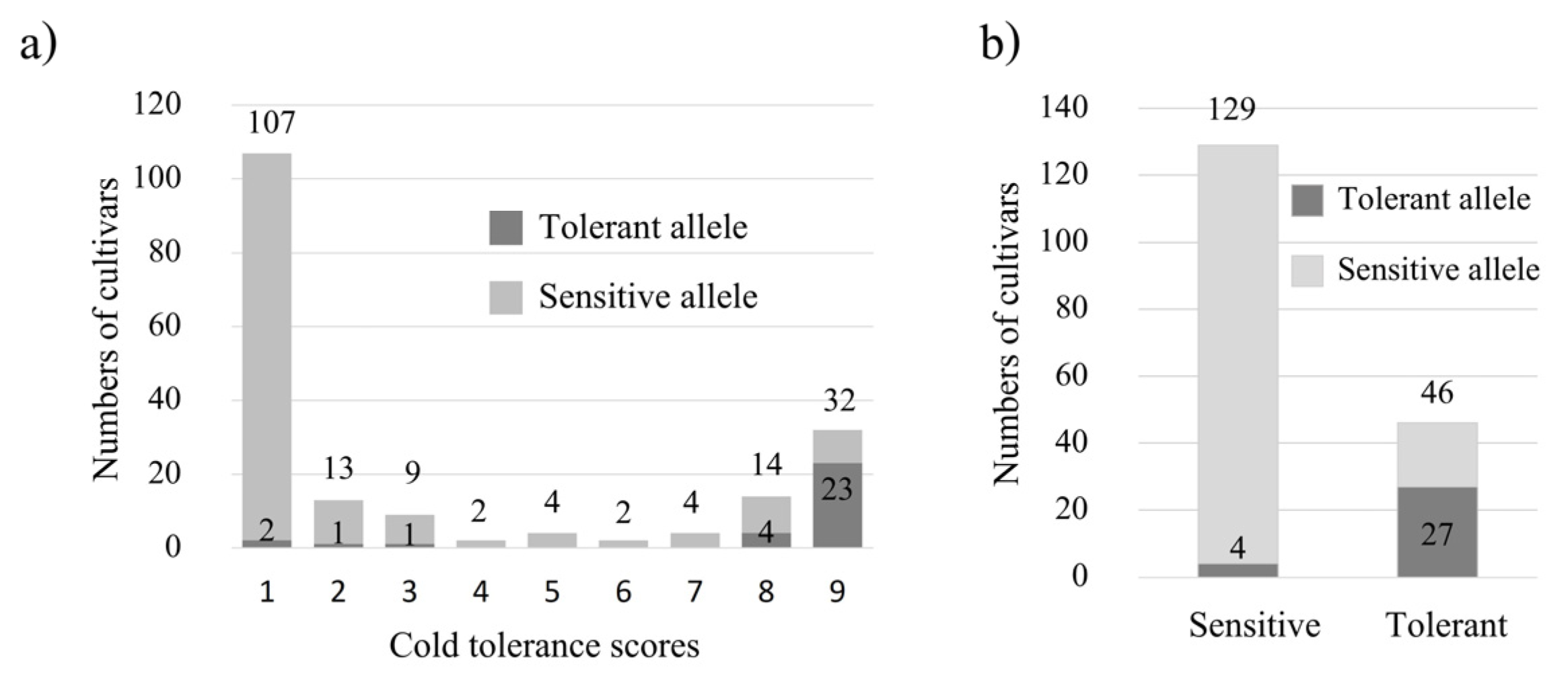
2.2.4. Marker Combination for Predicting Cold Tolerance
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Cold Treatment and Evaluation of Cold Tolerance
4.3. Marker Development and Genotyping
4.3.1. NAC6 (LOC_Os01g66120)
4.3.2. COLD1 (LOC_Os04g51180)
4.3.3. LOC_Os10g34840
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kong, W.; Zhang, C.; Qiang, Y.; Zhong, H.; Zhao, G.; Li, Y. Integrated RNA-seq analysis and Meta-QTLs mapping provide insights into cold stress response in rice seedling roots. Int. J. Mol. Sci. 2020, 21, 4615. [Google Scholar] [CrossRef]
- Guan, S.; Xu, Q.; Ma, D.; Zhang, W.; Xu, Z.; Zhao, M.; Guo, Z. Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene 2019, 685, 96–105. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 24. [Google Scholar] [CrossRef]
- Zhao, L.; Liu, F.; Xu, W.; Di, C.; Zhou, S.; Xue, Y.; Yu, J.; Su, Z. Increased expression of OsSPX1 enhances cold/subfreezing tolerance in tobacco and Arabidopsis thaliana. Plant Biotechnol. J. 2009, 7, 550–561. [Google Scholar] [CrossRef]
- Sipaseuth; Basnayake, J.; Fukai, S.; Farrell, T.C.; Senthonghae, M.; Sengkeo; Phamixay, S.; Linquist, B.; Chanphengsay, M. Opportunities to increasing dry season rice productivity in low temperature affected areas. Field Crops Res. 2007, 102, 87–97. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, H.; Zhang, D.; Li, J.; Xiong, H.; Yu, J.; Li, J.; Rashid, M.A.R.; Li, G.; Ma, X. Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS ONE 2015, 10, e0120590. [Google Scholar] [CrossRef]
- Thapa, R.; Tabien, R.E.; Thomson, M.J.; Septiningsih, E.M. Genome-wide association mapping to identify genetic loci for cold tolerance and cold recovery during germination in rice. Front. Genet. 2020, 11, 22. [Google Scholar] [CrossRef]
- Xiao, N.; Huang, W.-n.; Li, A.-h.; Gao, Y.; Li, Y.-h.; Pan, C.-h.; Ji, H.; Zhang, X.-x.; Dai, Y.; Dai, Z.-y. Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor. Appl. Genet. 2015, 128, 173–185. [Google Scholar] [CrossRef]
- Ahlawat, S.; Sharma, R.; Maitra, A.; Roy, M.; Tantia, M. Designing, optimization and validation of tetra-primer ARMS PCR protocol for genotyping mutations in caprine Fec genes. Meta Gene 2014, 2, 439–449. [Google Scholar] [CrossRef]
- Yun, K.-Y.; Park, M.R.; Mohanty, B.; Herath, V.; Xu, F.; Mauleon, R.; Wijaya, E.; Bajic, V.B.; Bruskiewich, R.; de Los Reyes, B.G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol. 2010, 10, 16. [Google Scholar] [CrossRef]
- Shen, C.; Li, D.; He, R.; Fang, Z.; Xia, Y.; Gao, J.; Shen, H.; Cao, M. Comparative transcriptome analysis of RNA-seq data for cold-tolerant and cold-sensitive rice genotypes under cold stress. J. Plant Biol. 2014, 57, 337–348. [Google Scholar] [CrossRef]
- Li, C.; Dong, S.; Beckles, D.M.; Liu, X.; Liu, D.; Miao, H.; Zhang, S.; Gu, X. Identification of QTLs controlling cold tolerance in cucumber (Cucumis sativus L.) seedlings. Sci. Hortic. 2022, 305, 111383. [Google Scholar] [CrossRef]
- Xiao, N.; Gao, Y.; Qian, H.; Gao, Q.; Wu, Y.; Zhang, D.; Zhang, X.; Yu, L.; Li, Y.; Pan, C. Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol. 2018, 177, 1108–1123. [Google Scholar] [CrossRef]
- Diao, P.; Chen, C.; Zhang, Y.; Meng, Q.; Lv, W.; Ma, N. The role of NAC transcription factor in plant cold response. Plant Signal. Behav. 2020, 15, 1785668. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Lee, D.K.; Chung, P.J.; Jeong, J.S.; Jang, G.; Bang, S.W.; Jung, H.; Kim, Y.S.; Ha, S.H.; Choi, Y.D.; Kim, J.K. The rice OsNAC6 transcription factor orchestrates multiple molecular mechanisms involving root structural adaptions and nicotianamine biosynthesis for drought tolerance. Plant Biotechnol. J. 2017, 15, 754–764. [Google Scholar] [CrossRef]
- Rabbani, M.A.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003, 133, 1755–1767. [Google Scholar] [CrossRef]
- Kumar, S.; Bink, M.C.; Volz, R.K.; Bus, V.G.; Chagné, D. Towards genomic selection in apple (Malus× domestica Borkh.) breeding programmes: Prospects, challenges and strategies. Tree Genet. Genomes 2012, 8, 1–14. [Google Scholar] [CrossRef]
- Andersen, J.R.; Lübberstedt, T. Functional markers in plants. Trends Plant Sci. 2003, 8, 554–560. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Ndjiondjop, M.N.; Semagn, K.; Sow, M.; Manneh, B.; Gouda, A.C.; Kpeki, S.B.; Pegalepo, E.; Wambugu, P.; Sié, M.; Warburton, M.L. Assessment of genetic variation and population structure of diverse rice genotypes adapted to lowland and upland ecologies in Africa using SNPs. Front. Plant Sci. 2018, 9, 446. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, J.; Li, C.; Kang, H.; Wang, Y.; Tan, X.; Liu, M.; Deng, Y.; Wang, Z.; Liu, Y. Genome-wide association mapping of cold tolerance genes at the seedling stage in rice. Rice 2016, 9, 61. [Google Scholar] [CrossRef]
- Shakiba, E.; Edwards, J.D.; Jodari, F.; Duke, S.E.; Baldo, A.M.; Korniliev, P.; McCouch, S.R.; Eizenga, G.C. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 2017, 12, e0172133. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Perdomo, E.; Vidal, A.; Kreplak, J.; Duborjal, H.; Leveugle, M.; Duarte, J.; Desmetz, C.; Deulvot, C.; Raffiot, B.; Marget, P. Development of new genetic resources for faba bean (Vicia faba L.) breeding through the discovery of gene-based SNP markers and the construction of a high-density consensus map. Sci. Rep. 2020, 10, 6790. [Google Scholar] [CrossRef]
- Cheon, K.-S.; Baek, J.; Cho, Y.-i.; Jeong, Y.-M.; Lee, Y.-Y.; Oh, J.; Won, Y.J.; Kang, D.-Y.; Oh, H.; Kim, S.L. Single nucleotide polymorphism (SNP) discovery and kompetitive allele-specific PCR (KASP) marker development with Korean japonica rice varieties. Plant Breed. Biotech. 2018, 6, 391–403. [Google Scholar] [CrossRef]
- Park, J.; Lee, J.; Ban, C.; Kim, W.J. An approach toward SNP detection by modulating the fluorescence of DNA-templated silver nanoclusters. Biosens. Bioelectron. 2013, 43, 419–424. [Google Scholar] [CrossRef]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar] [CrossRef]
- Tian, D.; Chen, Z.; Chen, Z.; Zhou, Y.; Wang, Z.; Wang, F.; Chen, S. Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice 2016, 9, 19. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]

| Gene | Sequence (5’–3’) | Tm °C | Allele | Size (bp) |
|---|---|---|---|---|
| NAC6 | F-GCTTTGGGCCGCAGAAATTA | 60 | Tolerant | 651 |
| R-TGTTGTAAATCCGGCACAGCA | Sensitive (deleted) | 429 | ||
| COLD1 | Inner F-TCCTGGCTTACAGGGAAATTGATGAGAT | 57 | Control | 320 |
| Inner R-AGCTGCCTTTCCAATGTTTTGATGTTCT | Tolerant (A) | 225 | ||
| Outer F-AGTGTCATGGCTGTTCTTTCTGGTTTTG | Sensitive (T) | 151 | ||
| Outer R-AGCACACCTCTTCTGATCCTTGAATCCT | ||||
| LOC_Os10g34840 | Outer F-CGGCAGCGGAGGCGGTGGCTTC | 70 | Tolerant (G) | 260 |
| Inner R-TCCCCGTGCAGGACGGCGCTCC | ||||
| Inner F-CGACGGCGGGGACGGCGTCA | 70 | Sensitive (A) | 203 | |
| Outer R-GTCGCTGTCGCAGCCGCGCTGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yongbin, Q.; Summat, P.; Panyawut, N.; Sikaewtung, K.; Ditthab, K.; Tongmark, K.; Chakhonkaen, S.; Sangarwut, N.; Wasinanon, T.; Kaewmungkun, K.; et al. Identification of Rice Accessions Having Cold Tolerance at the Seedling Stage and Development of Novel Genotypic Assays for Predicting Cold Tolerance. Plants 2023, 12, 215. https://doi.org/10.3390/plants12010215
Yongbin Q, Summat P, Panyawut N, Sikaewtung K, Ditthab K, Tongmark K, Chakhonkaen S, Sangarwut N, Wasinanon T, Kaewmungkun K, et al. Identification of Rice Accessions Having Cold Tolerance at the Seedling Stage and Development of Novel Genotypic Assays for Predicting Cold Tolerance. Plants. 2023; 12(1):215. https://doi.org/10.3390/plants12010215
Chicago/Turabian StyleYongbin, Qi, Patcharaporn Summat, Natjaree Panyawut, Kannika Sikaewtung, Khanittha Ditthab, Keasinee Tongmark, Sriprapai Chakhonkaen, Numphet Sangarwut, Thiwawan Wasinanon, Kanokwan Kaewmungkun, and et al. 2023. "Identification of Rice Accessions Having Cold Tolerance at the Seedling Stage and Development of Novel Genotypic Assays for Predicting Cold Tolerance" Plants 12, no. 1: 215. https://doi.org/10.3390/plants12010215
APA StyleYongbin, Q., Summat, P., Panyawut, N., Sikaewtung, K., Ditthab, K., Tongmark, K., Chakhonkaen, S., Sangarwut, N., Wasinanon, T., Kaewmungkun, K., & Muangprom, A. (2023). Identification of Rice Accessions Having Cold Tolerance at the Seedling Stage and Development of Novel Genotypic Assays for Predicting Cold Tolerance. Plants, 12(1), 215. https://doi.org/10.3390/plants12010215





