Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis
Abstract
1. Introduction
2. Plants Associate with Soil Microorganisms to Access Essential Nutrients
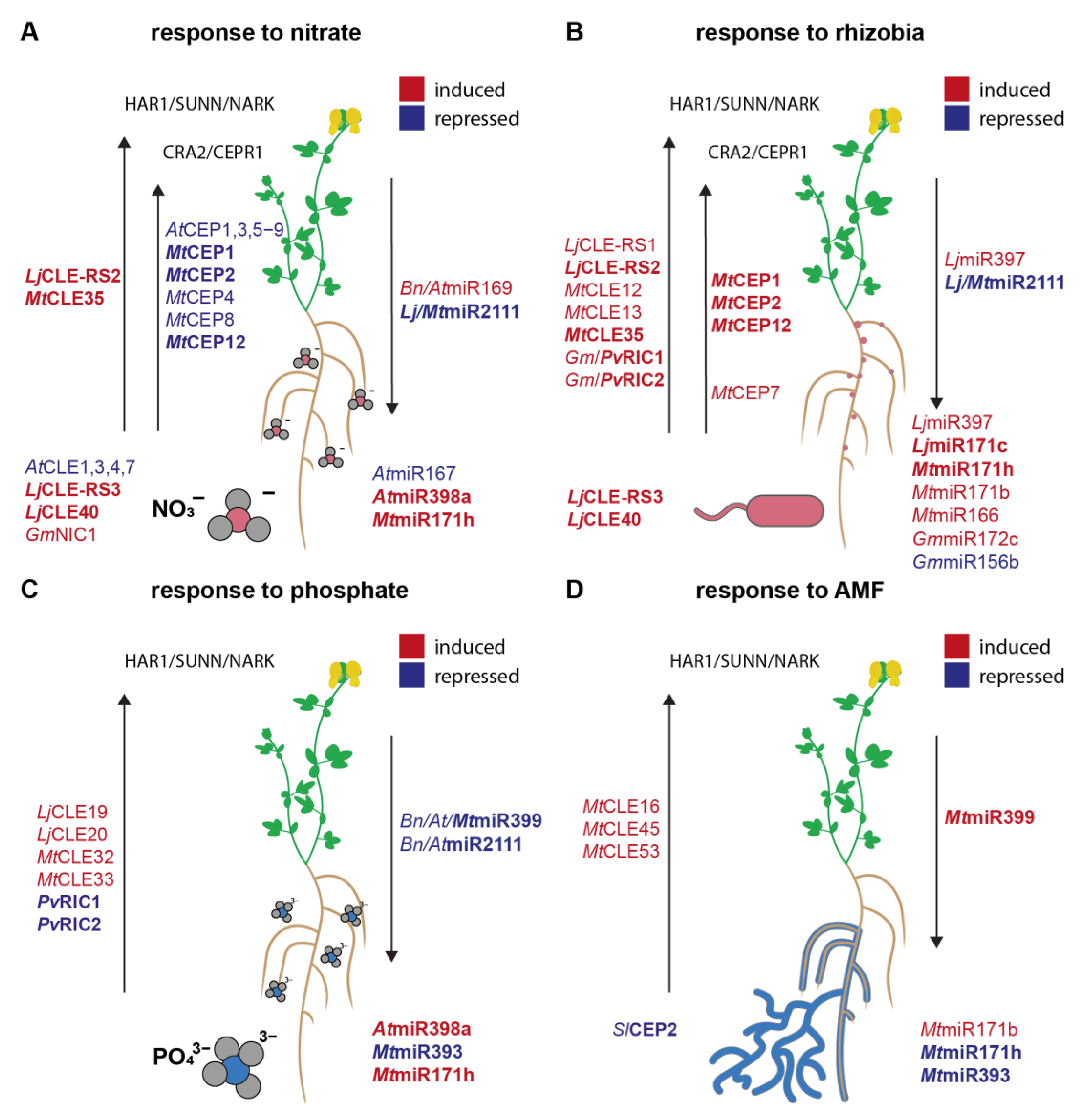
3. Plant Responses to N Availability and Rhizobial Symbiosis Involve CEP and CLE Peptide Regulation
3.1. Roles of CLE Peptides in N Homeostasis and Symbiosis Regulation
| Stimuli | Organism | Influence Range | Predominant Expression (Tissue) | Refs | |
|---|---|---|---|---|---|
| AtCLE1 AtCLE3 AtCLE4 AtCLE7 | N-deficiency induced | A. thaliana | systemic | roots | [24] |
| LjCLE-RS1 | Rhizobium-induced | L. japonicus | systemic | roots | [26] |
| LjCLE-RS2 | Rhizobium- and N-induced | L. japonicus | local and systemic | roots | [26] |
| LjCLE-RS3 LjCLE40 | Rhizobium- and N-induced | L. japonicus | roots, nodule primordia | [27] | |
| LjCLE19 LjCLE20 | P-induced | L. japonicus | roots | [44] | |
| MtCLE12 | Rhizobium-induced | M. truncatula | local and systemic | nodules | [28] |
| MtCLE13 | Rhizobium- and nod factor-induced, cytokinin-induced | M. truncatula | local and systemic | roots (symbiosis susceptible zone), inner cortical cells, nodules | [28] [45] |
| MtCLE35 | Rhizobium- and N-induced | M. truncatula | systemic | roots, nodules | [29] [30] |
| MtCLE32 | Pi-induced | M. truncatula | roots | [46] | |
| MtCLE33 | Pi-induced | M. truncatula | root vascular tissue | [46] [47] | |
| MtCLE16 MtCLE45 | AM-induced | M. truncatula | roots | [46] [47] | |
| MtCLE53 | AM-induced | M. truncatula | root vascular tissue near colonized regions | [46] [47] | |
| GmRIC1 GmRIC2 | Rhizobium-induced | G. max | systemic | roots | [31] |
| GmNIC1 | N-induced | G. max | local | roots | [31] |
| PvRIC1 PvRIC2 | Rhizobium-induced, P-deficiency increased | P. vulgaris | systemic | roots, pericycle cells of Pi-deficient roots | [32] [48] |
| AtCEP1 AtCEP3 AtCEP5 AtCEP6 AtCEP7 AtCEP8 AtCEP9 | N starvation-induced | A. thaliana | systemic | mainly roots (but also in aerial tissues) | [49] |
| MtCEP1 | Rhizobium-induced, N starvation-induced | M. truncatula | local and systemic | roots, shoots | [50,51,52] |
| MtCEP2 MtCEP12 | Rhizobium-induced, N starvation-induced | M. truncatula | mainly roots, shoots | [52] | |
| MtCEP4 MtCEP5 MtCEP6 MtCEP8 | N starvation-induced | M. truncatula | mainly roots, shoots | [52] | |
| MtCEP7 | Rhizobium- and nod factors-induced, cytokinin-induced | M. truncatula | systemic | roots, epidermal cells in colonized roots, nodule primordia, mature nodules | [45] |
| SlCEP2 | AM-reduced | S. lycopersicum | local | roots | [53] |
3.2. Roles of CEP Peptides in N Homeostasis and Symbiosis Regulation
4. CLEs and CEPs Respond to Both P and AM Fungal Infection
5. miRNAs Respond to N and P Availability and Symbiosis-Mediated Nutrient Acquisition
| Stimuli | Organism | Influence | Tissue | Target | Refs | |
|---|---|---|---|---|---|---|
| miR167 | N-repressed | A. thaliana | local | root pericycle cells | ARF8 | [65] |
| miR169 | N-limitation repressed | A. thaliana and B. napus | systemic | shoots, roots, phloem sap | NFYA5 | [67] [70] |
| miR398a | N-limitation and P-limitation repressed | A. thaliana | [70] | |||
| miR399 | P-limitation induced | A. thaliana and B. napus | systemic | vascular tissues, phloem sap | PHO2 | [70] [72] |
| miR2111 | P-limitation induced N-repressed, rhizobium-repressed | A. thaliana and B. napus L. japonicus | systemic | phloem sap leaves phloem, phloem sap | E3 ligase TML | [76] [88] |
| miR397 | nodulation-induced | L. japonicus | local and systemic | nodules, leaves | LACCASE10 | [79] |
| miR171c | nodulation-induced | L. japonicus | nodules | NSP2 | [79] | |
| miR171h | expressed in high P and N, AM-repressed, nodulation-induced | M. truncatula | roots, arbuscule-containing cells, nodules | NSP2 | [83] | |
| miR171b | AM-specific | M. truncatula | local | colonized root cells | LOM1 | [77] |
| miR393 | low-P expressed, AM-repressed | M. truncatula | local | roots | auxin receptors | [76] |
| miR399 | low P-induced, AM-induced | M. truncatula | systemic | leaves and roots | PHO2 | [75] |
| miR166 | nodulation induced | M. truncatula | local | vascular bundles, roots, nodules | HD-ZIP III | [87] |
| miR172c | rhizobium-induced, nod factors-induced | G. max | local | rhizobium-inoculated roots and nodules | NNC1 | [91] |
| miR156b | rhizobium-repressed | G. max | local | roots | GmSPL9d | [94] |
6. CEPs and CLEs and miR2111 Jointly Orchestrate Plant Responses to N and Rhizobia
7. CLE Peptide Involvement in P-Dependent Control of Nodulation
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Udvardi, M.; Poole, P.S. Transport and Metabolism in Legume-Rhizobia Symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef] [PubMed]
- Patriarca, E.J.; Tatè, R.; Iaccarino, M. Key Role of Bacterial NH4+ Metabolism in Rhizobium-Plant Symbiosis. Microbiol. Mol. Biol. Rev. 2002, 66, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, B.N.; Finnegan, P.M.; Tyerman, S.D.; Whitehead, L.F.; Bergersen, F.J.; Day, D.A.; Udvardi, M.K. Characterization of an Ammonium Transport Protein from the Peribacteroid Membrane of Soybean Nodules. Science 1998, 281, 1202–1206. [Google Scholar] [CrossRef] [PubMed]
- Salvemini, F.; Marini, A.; Riccio, A.; Patriarca, E.J.; Chiurazzi, M. Functional Characterization of an Ammonium Transporter Gene from Lotus japonicus. Gene 2001, 270, 237–243. [Google Scholar] [CrossRef]
- Maxwell, C.A.; Hartwig, U.A.; Joseph, C.M.; Phillips, D.A. A Chalcone and Two Related Flavonoids Released from Alfalfa Roots Induce Nod Genes of Rhizobium Meliloti. Plant Physiol. 1989, 91, 842–847. [Google Scholar] [CrossRef]
- Long, S.R.; Staskawicz, B.J. Prokaryotic Plant Parasites. Cell 1993, 73, 921–935. [Google Scholar] [CrossRef]
- Dénarié, J.; Debellé, F.; Promé, J.C. Rhizobium Lipo-Chitooligosaccharide Nodulation Factors: Signaling Molecules Mediating Recognition and Morphogenesis. Annu. Rev. Biochem. 1996, 65, 503–535. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The Rules of Engagement in the Legume-Rhizobial Symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef]
- Harrison, M.J.; van Buuren, M.L. A Phosphate Transporter from the Mycorrhizal Fungus Glomus Versiforme. Nature 1995, 378, 626–629. [Google Scholar] [CrossRef]
- Hijikata, N.; Murase, M.; Tani, C.; Ohtomo, R.; Osaki, M.; Ezawa, T. Polyphosphate Has a Central Role in the Rapid and Massive Accumulation of Phosphorus in Extraradical Mycelium of an Arbuscular Mycorrhizal Fungus. New Phytol. 2010, 186, 285–289. [Google Scholar] [CrossRef]
- López-Pedrosa, A.; González-Guerrero, M.; Valderas, A.; Azcón-Aguilar, C.; Ferrol, N. GintAMT1 Encodes a Functional High-Affinity Ammonium Transporter That Is Expressed in the Extraradical Mycelium of Glomus Intraradices. Fungal Genet. Biol. 2006, 43, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Cappellazzo, G.; Lanfranco, L.; Fitz, M.; Wipf, D.; Bonfante, P. Characterization of an Amino Acid Permease from the Endomycorrhizal Fungus Glomus Mosseae. Plant Physiol. 2008, 147, 429–437. [Google Scholar] [CrossRef] [PubMed]
- MacLean, A.M.; Bravo, A.; Harrison, M.J. Plant Signaling and Metabolic Pathways Enabling Arbuscular Mycorrhizal Symbiosis. Plant Cell 2017, 29, 2319–2335. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Matsuzaki, K.; Hayashi, H. Plant Sesquiterpenes Induce Hyphal Branching in Arbuscular Mycorrhizal Fungi. Nature 2005, 435, 824–827. [Google Scholar] [CrossRef]
- Maillet, F.; Poinsot, V.; André, O.; Puech-Pagès, V.; Haouy, A.; Gueunier, M.; Cromer, L.; Giraudet, D.; Formey, D.; Niebel, A.; et al. Fungal Lipochitooligosaccharide Symbiotic Signals in Arbuscular Mycorrhiza. Nature 2011, 469, 58–63. [Google Scholar] [CrossRef]
- Genre, A.; Chabaud, M.; Balzergue, C.; Puech-Pagès, V.; Novero, M.; Rey, T.; Fournier, J.; Rochange, S.; Bécard, G.; Bonfante, P.; et al. Short-Chain Chitin Oligomers from Arbuscular Mycorrhizal Fungi Trigger Nuclear Ca2+ Spiking in Medicago truncatula Roots and Their Production Is Enhanced by Strigolactone. New Phytol. 2013, 198, 190–202. [Google Scholar] [CrossRef]
- Nagahashi, G.; Douds, D.D., Jr. Appressorium Formation by AM Fungi on Isolated Cell Walls of Carrot Roots. New Phytol. 1997, 136, 299–304. [Google Scholar] [CrossRef]
- Bonfante, P.; Genre, A. Mechanisms Underlying Beneficial Plant–Fungus Interactions in Mycorrhizal Symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef]
- Gutjahr, C.; Parniske, M. Cell Biology: Control of Partner Lifetime in a Plant–Fungus Relationship. Curr. Biol. 2017, 27, R420–R423. [Google Scholar] [CrossRef]
- de Bang, T.C.; Lay, K.S.; Scheible, W.-R.; Takahashi, H. Small Peptide Signaling Pathways Modulating Macronutrient Utilization in Plants. Curr. Opin. Plant Biol. 2017, 39, 31–39. [Google Scholar] [CrossRef]
- de Bang, T.C.; Lundquist, P.K.; Dai, X.; Boschiero, C.; Zhuang, Z.; Pant, P.; Torres-Jerez, I.; Roy, S.; Nogales, J.; Veerappan, V.; et al. Genome-Wide Identification of Medicago Peptides Involved in Macronutrient Responses and Nodulation. Plant Physiol. 2017, 175, 1669–1689. [Google Scholar] [CrossRef] [PubMed]
- Betsuyaku, S.; Sawa, S.; Yamada, M. The Function of the CLE Peptides in Plant Development and Plant-Microbe Interactions. Arab. Book 2011, 9, e0149. [Google Scholar] [CrossRef]
- Yamaguchi, Y.L.; Ishida, T.; Sawa, S. CLE Peptides and Their Signaling Pathways in Plant Development. J. Exp. Bot. 2016, 67, 4813–4826. [Google Scholar] [CrossRef] [PubMed]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-CLAVATA1 Peptide-Receptor Signaling Module Regulates the Expansion of Plant Root Systems in a Nitrogen-Dependent Manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Chaulagain, D.; Frugoli, J. The Regulation of Nodule Number in Legumes Is a Balance of Three Signal Transduction Pathways. Int. J. Mol. Sci. 2021, 22, 1117. [Google Scholar] [CrossRef]
- Okamoto, S.; Ohnishi, E.; Sato, S.; Takahashi, H.; Nakazono, M.; Tabata, S.; Kawaguchi, M. Nod Factor/Nitrate-Induced CLE Genes That Drive HAR1-Mediated Systemic Regulation of Nodulation. Plant Cell Physiol. 2009, 50, 67–77. [Google Scholar] [CrossRef]
- Nishida, H.; Handa, Y.; Tanaka, S.; Suzaki, T.; Kawaguchi, M. Expression of the CLE-RS3 Gene Suppresses Root Nodulation in Lotus japonicus. J. Plant Res. 2016, 129, 909–919. [Google Scholar] [CrossRef]
- Mortier, V.; Den Herder, G.; Whitford, R.; Van de Velde, W.; Rombauts, S.; D’Haeseleer, K.; Holsters, M.; Goormachtig, S. CLE Peptides Control Medicago truncatula Nodulation Locally and Systemically. Plant Physiol. 2010, 153, 222–237. [Google Scholar] [CrossRef]
- Lebedeva, M.; Azarakhsh, M.; Yashenkova, Y.; Lutova, L. Nitrate-Induced CLE Peptide Systemically Inhibits Nodulation in Medicago truncatula. Plants 2020, 9, 1456. [Google Scholar] [CrossRef]
- Mens, C.; Hastwell, A.H.; Su, H.; Gresshoff, P.M.; Mathesius, U.; Ferguson, B.J. Characterisation of Medicago truncatula CLE34 and CLE35 in Nitrate and Rhizobia Regulation of Nodulation. New Phytol. 2021, 229, 2525–2534. [Google Scholar] [CrossRef]
- Reid, D.E.; Ferguson, B.J.; Gresshoff, P.M. Inoculation- and Nitrate-Induced CLE Peptides of Soybean Control NARK-Dependent Nodule Formation. Mol. Plant-Microbe Interact. 2011, 24, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Li, D.; Hastwell, A.H.; Reid, D.E.; Li, Y.; Jackson, S.A.; Gresshoff, P.M. The Soybean (Glycine max) Nodulation-Suppressive CLE Peptide, GmRIC1, Functions Interspecifically in Common White Bean (Phaseolus vulgaris), but Not in a Supernodulating Line Mutated in the Receptor PvNARK. Plant Biotechnol. J. 2014, 12, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, S.; Shinohara, H.; Mori, T.; Matsubayashi, Y.; Kawaguchi, M. Root-Derived CLE Glycopeptides Control Nodulation by Direct Binding to HAR1 Receptor Kinase. Nat. Commun. 2013, 4, 2191. [Google Scholar] [CrossRef] [PubMed]
- Krusell, L.; Madsen, L.H.; Sato, S.; Aubert, G.; Genua, A.; Szczyglowski, K.; Duc, G.; Kaneko, T.; Tabata, S.; de Bruijn, F.; et al. Shoot Control of Root Development and Nodulation Is Mediated by a Receptor-like Kinase. Nature 2002, 420, 422–426. [Google Scholar] [CrossRef]
- Nishimura, R.; Hayashi, M.; Wu, G.-J.; Kouchi, H.; Imaizumi-Anraku, H.; Murakami, Y.; Kawasaki, S.; Akao, S.; Ohmori, M.; Nagasawa, M.; et al. HAR1 Mediates Systemic Regulation of Symbiotic Organ Development. Nature 2002, 420, 426–429. [Google Scholar] [CrossRef]
- Searle, I.R.; Men, A.E.; Laniya, T.S.; Buzas, D.M.; Iturbe-Ormaetxe, I.; Carroll, B.J.; Gresshoff, P.M. Long-Distance Signaling in Nodulation Directed by a CLAVATA1-like Receptor Kinase. Science 2003, 299, 109–112. [Google Scholar] [CrossRef]
- Moreau, C.; Gautrat, P.; Frugier, F. Nitrate-Induced CLE35 Signaling Peptides Inhibit Nodulation through the SUNN Receptor and MiR2111 Repression. Plant Physiol. 2021, 185, 1216–1228. [Google Scholar] [CrossRef]
- Ohyama, K.; Shinohara, H.; Ogawa-Ohnishi, M.; Matsubayashi, Y. A Glycopeptide Regulating Stem Cell Fate in Arabidopsis Thaliana. Nat. Chem. Biol. 2009, 5, 578–580. [Google Scholar] [CrossRef]
- Ogawa-Ohnishi, M.; Matsushita, W.; Matsubayashi, Y. Identification of Three Hydroxyproline O-Arabinosyltransferases in Arabidopsis Thaliana. Nat. Chem. Biol. 2013, 9, 726–730. [Google Scholar] [CrossRef]
- Kassaw, T.; Nowak, S.; Schnabel, E.; Frugoli, J. ROOT DETERMINED NODULATION1 Is Required for M. Truncatula CLE12, But Not CLE13, Peptide Signaling through the SUNN Receptor Kinase. Plant Physiol. 2017, 174, 2445–2456. [Google Scholar] [CrossRef]
- Imin, N.; Patel, N.; Corcilius, L.; Payne, R.J.; Djordjevic, M.A. CLE Peptide Tri-Arabinosylation and Peptide Domain Sequence Composition Are Essential for SUNN-Dependent Autoregulation of Nodulation in Medicago truncatula. New Phytol. 2018, 218, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Hastwell, A.H.; Corcilius, L.; Williams, J.T.; Gresshoff, P.M.; Payne, R.J.; Ferguson, B.J. Triarabinosylation Is Required for Nodulation-Suppressive CLE Peptides to Systemically Inhibit Nodulation in Pisum Sativum. Plant Cell Environ. 2019, 42, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Yoro, E.; Nishida, H.; Ogawa-Ohnishi, M.; Yoshida, C.; Suzaki, T.; Matsubayashi, Y.; Kawaguchi, M. PLENTY, a Hydroxyproline O-Arabinosyltransferase, Negatively Regulates Root Nodule Symbiosis in Lotus japonicus. J. Exp. Bot. 2019, 70, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Funayama-Noguchi, S.; Noguchi, K.; Yoshida, C.; Kawaguchi, M. Two CLE Genes Are Induced by Phosphate in Roots of Lotus japonicus. J. Plant Res. 2011, 124, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Laffont, C.; Ivanovici, A.; Gautrat, P.; Brault, M.; Djordjevic, M.A.; Frugier, F. The NIN Transcription Factor Coordinates CEP and CLE Signaling Peptides That Regulate Nodulation Antagonistically. Nat. Commun. 2020, 11, 3167. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.M.; Flokova, K.; Schnabel, E.; Sun, X.; Fei, Z.; Frugoli, J.; Bouwmeester, H.J.; Harrison, M.J. A CLE-SUNN Module Regulates Strigolactone Content and Fungal Colonization in Arbuscular Mycorrhiza. Nat. Plants 2019, 5, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Karlo, M.; Boschiero, C.; Landerslev, K.G.; Blanco, G.S.; Wen, J.; Mysore, K.S.; Dai, X.; Zhao, P.X.; de Bang, T.C. The CLE53-SUNN Genetic Pathway Negatively Regulates Arbuscular Mycorrhiza Root Colonization in Medicago truncatula. J. Exp. Bot. 2020, 71, 4972–4984. [Google Scholar] [CrossRef] [PubMed]
- Isidra-Arellano, M.C.; Pozas-Rodríguez, E.A.; Del Rocío Reyero-Saavedra, M.; Arroyo-Canales, J.; Ferrer-Orgaz, S.; Del Socorro Sánchez-Correa, M.; Cardenas, L.; Covarrubias, A.A.; Valdés-López, O. Inhibition of Legume Nodulation by Pi Deficiency Is Dependent on the Autoregulation of Nodulation (AON) Pathway. Plant J. 2020, 103, 1125–1139. [Google Scholar] [CrossRef] [PubMed]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of Root-Derived Peptides by Shoot LRR-RKs Mediates Systemic N-Demand Signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef]
- Imin, N.; Mohd-Radzman, N.A.; Ogilvie, H.A.; Djordjevic, M.A. The Peptide-Encoding CEP1 Gene Modulates Lateral Root and Nodule Numbers in Medicago truncatula. J. Exp. Bot. 2013, 64, 5395–5409. [Google Scholar] [CrossRef]
- Laffont, C.; Huault, E.; Gautrat, P.; Endre, G.; Kalo, P.; Bourion, V.; Duc, G.; Frugier, F. Independent Regulation of Symbiotic Nodulation by the SUNN Negative and CRA2 Positive Systemic Pathways. Plant Physiol. 2019, 180, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Ye, Q.; Chen, H.; Dong, J.; Wang, T. Multigene Editing Reveals That MtCEP1/2/12 Redundantly Control Lateral Root and Nodule Number in Medicago truncatula. J. Exp. Bot. 2021, 72, 3661–3676. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-H.; Wei, Y.-H.; Lo, J.-C.; Pan, H.-Y.; Yang, S.-Y. Arbuscular Mycorrhizal Symbiosis Enhances Tomato Lateral Root Formation by Modulating CEP2 Peptide Expression. New Phytol. 2022, 235, 292–305. [Google Scholar] [CrossRef] [PubMed]
- Ohyama, K.; Ogawa, M.; Matsubayashi, Y. Identification of a Biologically Active, Small, Secreted Peptide in Arabidopsis by in Silico Gene Screening, Followed by LC-MS-Based Structure Analysis. Plant J. 2008, 55, 152–160. [Google Scholar] [CrossRef]
- Huault, E.; Laffont, C.; Wen, J.; Mysore, K.S.; Ratet, P.; Duc, G.; Frugier, F. Local and Systemic Regulation of Plant Root System Architecture and Symbiotic Nodulation by a Receptor-like Kinase. PLoS Genet. 2014, 10, e1004891. [Google Scholar] [CrossRef]
- Gautrat, P.; Laffont, C.; Frugier, F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the MiR2111 Systemic Effector. Curr. Biol. 2020, 30, 1339–1345. [Google Scholar] [CrossRef]
- Roy, S.; Müller, L.M. A Rulebook for Peptide Control of Legume-Microbe Endosymbioses. Trends Plant Sci 2022, 27, 870–889. [Google Scholar] [CrossRef]
- Breuillin, F.; Schramm, J.; Hajirezaei, M.; Ahkami, A.; Favre, P.; Druege, U.; Hause, B.; Bucher, M.; Kretzschmar, T.; Bossolini, E.; et al. Phosphate Systemically Inhibits Development of Arbuscular Mycorrhiza in Petunia hybrida and Represses Genes Involved in Mycorrhizal Functioning. Plant J. 2010, 64, 1002–1017. [Google Scholar] [CrossRef]
- Müller, L.M.; Harrison, M.J. Phytohormones, MiRNAs, and Peptide Signals Integrate Plant Phosphorus Status with Arbuscular Mycorrhizal Symbiosis. Curr. Opin. Plant Biol. 2019, 50, 132–139. [Google Scholar] [CrossRef]
- Meixner, C.; Ludwig-Müller, J.; Miersch, O.; Gresshoff, P.; Staehelin, C.; Vierheilig, H. Lack of Mycorrhizal Autoregulation and Phytohormonal Changes in the Supernodulating Soybean Mutant Nts1007. Planta 2005, 222, 709–715. [Google Scholar] [CrossRef]
- Wang, C.; Reid, J.B.; Foo, E. The Art of Self-Control—Autoregulation of Plant–Microbe Symbioses. Front. Plant Sci. 2018, 9, 988. [Google Scholar] [CrossRef] [PubMed]
- Handa, Y.; Nishide, H.; Takeda, N.; Suzuki, Y.; Kawaguchi, M.; Saito, K. RNA-Seq Transcriptional Profiling of an Arbuscular Mycorrhiza Provides Insights into Regulated and Coordinated Gene Expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol. 2015, 56, 1490–1511. [Google Scholar] [CrossRef] [PubMed]
- van Zeijl, A.; Liu, W.; Xiao, T.T.; Kohlen, W.; Yang, W.-C.; Bisseling, T.; Geurts, R. The Strigolactone Biosynthesis Gene DWARF27 Is Co-Opted in Rhizobium Symbiosis. BMC Plant Biol. 2015, 15, 260. [Google Scholar] [CrossRef]
- Liang, G.; He, H.; Yu, D. Identification of Nitrogen Starvation-Responsive MicroRNAs in Arabidopsis Thaliana. PLoS ONE 2012, 7, e48951. [Google Scholar] [CrossRef] [PubMed]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-Specific Nitrogen Responses Mediate Developmental Plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutiérrez, R.A. Nitrate-Responsive MiR393/AFB3 Regulatory Module Controls Root System Architecture in Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef]
- Zhao, M.; Ding, H.; Zhu, J.-K.; Zhang, F.; Li, W.-X. Involvement of MiR169 in the Nitrogen-Starvation Responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar] [CrossRef]
- Fukuda, M.; Fujiwara, T.; Nishida, S. Roles of Non-Coding RNAs in Response to Nitrogen Availability in Plants. Int. J. Mol. Sci. 2020, 21, 8508. [Google Scholar] [CrossRef]
- Pant, B.D.; Buhtz, A.; Kehr, J.; Scheible, W.-R. MicroRNA399 Is a Long-Distance Signal for the Regulation of Plant Phosphate Homeostasis. Plant J. 2008, 53, 731–738. [Google Scholar] [CrossRef]
- Pant, B.D.; Musialak-Lange, M.; Nuc, P.; May, P.; Buhtz, A.; Kehr, J.; Walther, D.; Scheible, W.-R. Identification of Nutrient-Responsive Arabidopsis and Rapeseed MicroRNAs by Comprehensive Real-Time Polymerase Chain Reaction Profiling and Small RNA Sequencing. Plant Physiol. 2009, 150, 1541–1555. [Google Scholar] [CrossRef]
- Huen, A.; Bally, J.; Smith, P. Identification and Characterisation of MicroRNAs and Their Target Genes in Phosphate-Starved Nicotiana Benthamiana by Small RNA Deep Sequencing and 5′RACE Analysis. BMC Genom. 2018, 19, 940. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-I.; Chiang, S.-F.; Lin, W.-Y.; Chen, J.-W.; Tseng, C.-Y.; Wu, P.-C.; Chiou, T.-J. Regulatory Network of MicroRNA399 and PHO2 by Systemic Signaling. Plant Physiol. 2008, 147, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.; Lin, S.-I.; Wu, C.-C.; Huang, Y.-T.; Su, C.-L.; Chiou, T.-J. Pho2, a Phosphate Overaccumulator, Is Caused by a Nonsense Mutation in a MicroRNA399 Target Gene. Plant Physiol. 2006, 141, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-K.; Han, C.-L.; Lin, S.-I.; Chen, Y.-J.; Tsai, Y.-C.; Chen, Y.-R.; Chen, J.-W.; Lin, W.-Y.; Chen, P.-M.; Liu, T.-Y.; et al. Identification of Downstream Components of Ubiquitin-Conjugating Enzyme PHOSPHATE2 by Quantitative Membrane Proteomics in Arabidopsis Roots. Plant Cell 2013, 25, 4044–4060. [Google Scholar] [CrossRef]
- Branscheid, A.; Sieh, D.; Pant, B.D.; May, P.; Devers, E.A.; Elkrog, A.; Schauser, L.; Scheible, W.-R.; Krajinski, F. Expression Pattern Suggests a Role of MiR399 in the Regulation of the Cellular Response to Local Pi Increase during Arbuscular Mycorrhizal Symbiosis. Mol. Plant-Microbe Interact. 2010, 23, 915–926. [Google Scholar] [CrossRef]
- Etemadi, M.; Gutjahr, C.; Couzigou, J.-M.; Zouine, M.; Lauressergues, D.; Timmers, A.; Audran, C.; Bouzayen, M.; Bécard, G.; Combier, J.-P. Auxin Perception Is Required for Arbuscule Development in Arbuscular Mycorrhizal Symbiosis. Plant Physiol. 2014, 166, 281–292. [Google Scholar] [CrossRef]
- Couzigou, J.-M.; Lauressergues, D.; André, O.; Gutjahr, C.; Guillotin, B.; Bécard, G.; Combier, J.-P. Positive Gene Regulation by a Natural Protective MiRNA Enables Arbuscular Mycorrhizal Symbiosis. Cell Host Microbe 2017, 21, 106–112. [Google Scholar] [CrossRef]
- Lauressergues, D.; Delaux, P.-M.; Formey, D.; Lelandais-Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J.-P. The MicroRNA MiR171h Modulates Arbuscular Mycorrhizal Colonization of Medicago truncatula by Targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef]
- De Luis, A.; Markmann, K.; Cognat, V.; Holt, D.B.; Charpentier, M.; Parniske, M.; Stougaard, J.; Voinnet, O. Two MicroRNAs Linked to Nodule Infection and Nitrogen-Fixing Ability in the Legume Lotus japonicus. Plant Physiol. 2012, 160, 2137–2154. [Google Scholar] [CrossRef]
- Kaló, P.; Gleason, C.; Edwards, A.; Marsh, J.; Mitra, R.M.; Hirsch, S.; Jakab, J.; Sims, S.; Long, S.R.; Rogers, J.; et al. Nodulation Signaling in Legumes Requires NSP2, a Member of the GRAS Family of Transcriptional Regulators. Science 2005, 308, 1786–1789. [Google Scholar] [CrossRef]
- Heckmann, A.B.; Lombardo, F.; Miwa, H.; Perry, J.A.; Bunnewell, S.; Parniske, M.; Wang, T.L.; Downie, J.A. Lotus japonicus Nodulation Requires Two GRAS Domain Regulators, One of Which Is Functionally Conserved in a Non-Legume. Plant Physiol. 2006, 142, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Kohlen, W.; Lillo, A.; Op den Camp, R.; Ivanov, S.; Hartog, M.; Limpens, E.; Jamil, M.; Smaczniak, C.; Kaufmann, K.; et al. Strigolactone Biosynthesis in Medicago truncatula and Rice Requires the Symbiotic GRAS-Type Transcription Factors NSP1 and NSP2. Plant Cell 2011, 23, 3853–3865. [Google Scholar] [CrossRef] [PubMed]
- Hofferek, V.; Mendrinna, A.; Gaude, N.; Krajinski, F.; Devers, E.A. MiR171h Restricts Root Symbioses and Shows like Its Target NSP2 a Complex Transcriptional Regulation in Medicago truncatula. BMC Plant Biol. 2014, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Hoang, N.T.; Tóth, K.; Stacey, G. The Role of MicroRNAs in the Legume-Rhizobium Nitrogen-Fixing Symbiosis. J. Exp. Bot. 2020, 71, 1668–1680. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Hossain, M.S.; Arikit, S.; Valdés-López, O.; Zhai, J.; Wang, J.; Libault, M.; Ji, T.; Qiu, L.; Meyers, B.C.; et al. Identification of MicroRNAs and Their MRNA Targets during Soybean Nodule Development: Functional Analysis of the Role of MiR393j-3p in Soybean Nodulation. New Phytol. 2015, 207, 748–759. [Google Scholar] [CrossRef] [PubMed]
- Lelandais-Brière, C.; Naya, L.; Sallet, E.; Calenge, F.; Frugier, F.; Hartmann, C.; Gouzy, J.; Crespi, M. Genome-Wide Medicago truncatula Small RNA Analysis Revealed Novel MicroRNAs and Isoforms Differentially Regulated in Roots and Nodules. Plant Cell 2009, 21, 2780–2796. [Google Scholar] [CrossRef]
- Boualem, A.; Laporte, P.; Jovanovic, M.; Laffont, C.; Plet, J.; Combier, J.-P.; Niebel, A.; Crespi, M.; Frugier, F. MicroRNA166 Controls Root and Nodule Development in Medicago truncatula. Plant J. 2008, 54, 876–887. [Google Scholar] [CrossRef]
- Tsikou, D.; Yan, Z.; Holt, D.B.; Abel, N.B.; Reid, D.E.; Madsen, L.H.; Bhasin, H.; Sexauer, M.; Stougaard, J.; Markmann, K. Systemic Control of Legume Susceptibility to Rhizobial Infection by a Mobile MicroRNA. Science 2018, 362, 233–236. [Google Scholar] [CrossRef]
- Magori, S.; Oka-Kira, E.; Shibata, S.; Umehara, Y.; Kouchi, H.; Hase, Y.; Tanaka, A.; Sato, S.; Tabata, S.; Kawaguchi, M. Too Much Love, a Root Regulator Associated with the Long-Distance Control of Nodulation in Lotus japonicus. Mol. Plant-Microbe Interact. 2009, 22, 259–268. [Google Scholar] [CrossRef]
- Takahara, M.; Magori, S.; Soyano, T.; Okamoto, S.; Yoshida, C.; Yano, K.; Sato, S.; Tabata, S.; Yamaguchi, K.; Shigenobu, S.; et al. Too Much Love, a Novel Kelch Repeat-Containing F-Box Protein, Functions in the Long-Distance Regulation of the Legume–Rhizobium Symbiosis. Plant Cell Physiol. 2013, 54, 433–447. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Zou, Y.; Chen, L.; Cai, Z.; Zhang, S.; Zhao, F.; Tian, Y.; Jiang, Q.; Ferguson, B.J.; et al. Soybean MiR172c Targets the Repressive AP2 Transcription Factor NNC1 to Activate ENOD40 Expression and Regulate Nodule Initiation. Plant Cell 2014, 26, 4782–4801. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, Z.; Su, C.; Wang, Y.; Yan, Q.; Chen, J.; Ott, T.; Li, X. A GmNINa-MiR172c-NNC1 Regulatory Network Coordinates the Nodulation and Autoregulation of Nodulation Pathways in Soybean. Mol. Plant 2019, 12, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Holt, D.B.; Gupta, V.; Meyer, D.; Abel, N.B.; Andersen, S.U.; Stougaard, J.; Markmann, K. Micro RNA 172 (MiR172) Signals Epidermal Infection and Is Expressed in Cells Primed for Bacterial Invasion in Lotus japonicus Roots and Nodules. New Phytol. 2015, 208, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Sun, Z.; Jiang, Q.; Wang, Y.; Wang, C.; Luo, Y.; Zhang, F.; Li, X. The MiR156b-GmSPL9d Module Modulates Nodulation by Targeting Multiple Core Nodulation Genes in Soybean. New Phytol. 2022, 233, 1881–1899. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Amyot, L.; Tian, L.; Xu, Z.; Gruber, M.Y.; Hannoufa, A. Ectopic Expression of MiR156 Represses Nodulation and Causes Morphological and Developmental Changes in Lotus japonicus. Mol. Genet. Genom. 2015, 290, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Okuma, N.; Soyano, T.; Suzaki, T.; Kawaguchi, M. MIR2111-5 Locus and Shoot-Accumulated Mature MiR2111 Systemically Enhance Nodulation Depending on HAR1 in Lotus japonicus. Nat. Commun. 2020, 11, 5192. [Google Scholar] [CrossRef]
- Skopelitis, D.S.; Hill, K.; Klesen, S.; Marco, C.F.; von Born, P.; Chitwood, D.H.; Timmermans, M.C.P. Gating of MiRNA Movement at Defined Cell-Cell Interfaces Governs Their Impact as Positional Signals. Nat. Commun. 2018, 9, 3107. [Google Scholar] [CrossRef]
- Jakobsen, I. The Role of Phosphorus in Nitrogen Fixation by Young Pea Plants (Pisum sativum). Physiol. Plant. 1985, 64, 190–196. [Google Scholar] [CrossRef]
- Kuang, R.-B.; Liao, H.; Yan, X.-L.; Dong, Y.-S. Phosphorus and Nitrogen Interactions in Field-Grown Soybean as Related to Genetic Attributes of Root Morphological and Nodular Traits. J. Integr. Plant Biol. 2005, 47, 549–559. [Google Scholar] [CrossRef]
- Divito, G.A.; Sadras, V.O. How Do Phosphorus, Potassium and Sulphur Affect Plant Growth and Biological Nitrogen Fixation in Crop and Pasture Legumes? A Meta-Analysis. Field Crops Res. 2014, 156, 161–171. [Google Scholar] [CrossRef]
- Gentili, F.; Huss-Danell, K. Local and Systemic Effects of Phosphorus and Nitrogen on Nodulation and Nodule Function in Alnus incana. J. Exp. Bot. 2003, 54, 2757–2767. [Google Scholar] [CrossRef] [PubMed]
- Isidra-Arellano, M.C.; Reyero-Saavedra, M.D.R.; Sánchez-Correa, M.D.S.; Pingault, L.; Sen, S.; Joshi, T.; Girard, L.; Castro-Guerrero, N.A.; Mendoza-Cozatl, D.G.; Libault, M.; et al. Phosphate Deficiency Negatively Affects Early Steps of the Symbiosis between Common Bean and Rhizobia. Genes 2018, 9, 498. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valmas, M.I.; Sexauer, M.; Markmann, K.; Tsikou, D. Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis. Plants 2023, 12, 187. https://doi.org/10.3390/plants12010187
Valmas MI, Sexauer M, Markmann K, Tsikou D. Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis. Plants. 2023; 12(1):187. https://doi.org/10.3390/plants12010187
Chicago/Turabian StyleValmas, Marios I., Moritz Sexauer, Katharina Markmann, and Daniela Tsikou. 2023. "Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis" Plants 12, no. 1: 187. https://doi.org/10.3390/plants12010187
APA StyleValmas, M. I., Sexauer, M., Markmann, K., & Tsikou, D. (2023). Plants Recruit Peptides and Micro RNAs to Regulate Nutrient Acquisition from Soil and Symbiosis. Plants, 12(1), 187. https://doi.org/10.3390/plants12010187





