Short Peptides Induce Development of Root Hair Nicotiana tabacum
Abstract
:1. Introduction
2. Results
2.1. Biometric Parameters
2.2. Fluorescence and Light Microscopy
2.3. Root Hairs
2.4. Expression of Genes Controlling Root Hair Growth
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Preparation of FITC-Labeled Peptides
4.3. Fluorescence Microscopy
4.4. Light Microscopy
4.5. RNA Isolation
4.6. Real-Time PCR
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Richardson, A.; Lynch, J.; Ryan, P.; Delhaize, E.; Smith, S.; Harvey, P.; Veneklaas, E.; Lambers, H.; Oberson, A.; Culvenor, R.; et al. Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil 2011, 349, 121–156. [Google Scholar]
- Brown, L.K.; George, T.S.; Thompson, J.A.; Wright, G.; Lyon, J.; Dupuy, L.; Hubbard, S.F.; White, P.J. What are the implications of variation in root hair length on tolerance to phosphorus deficiency in combination with water stress in barley (Hordeum vulgare)? Ann. Bot. 2012, 110, 319–328. [Google Scholar] [PubMed]
- Haling, R.; Brawn, L.K.; Bengough, A.; Valentine, T.; White, P.; Young, I.; George, T. Root hair length and rhizosheath mass depend on soil porosity, strength and water content in barley genotypes. Planta 2014, 239, 643–651. [Google Scholar] [PubMed]
- Wang, Q.; Oh, J.; Lee, H.; Dhar, A.; Peng, T.; Ramos, R.; Guerrero-Juarez, C.; Wang, X.; Zhao, R. A multiscale model for hair folliclesreveals heterogeneous domains driving rapid spatiotemporal hair growth patterning. eLife 2017, 6, e22772. [Google Scholar] [PubMed]
- Meister, R.; Rajani, M.; Ruzicka, D.; Schachtman, D. Challenges of modifying root traits in crops for agriculture. Trends Plant Sci. 2014, 19, 779–788. [Google Scholar]
- Kang, Y.H.; Song, S.K.; Schiefelbein, J.; Lee, M.M. Nuclear trapping controls the position-dependent localization of CAPRICE in the root epidermis of Arabidopsis. Plant Physiol. 2013, 163, 193–204. [Google Scholar]
- Gilroy, S.; Jones, D.L. Through form to function: Root hair development and nutrient uptake. Trends Plant Sci. 2000, 5, 56–60. [Google Scholar]
- Lee, R.D.W.; Cho, H.T. Auxin, the organizer of the hormonal/environmental signals for root hair growth. Front. Plant Sci. 2013, 4, 448–454. [Google Scholar]
- Tominaga-Wada, R.; Nukumizu, Y.; Sato, S.; Wada, T. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS ONE 2013, 8, e54019. [Google Scholar]
- Tominaga-Wada, R.; Wada, T. Regulation of root hair cell differentiation by R3 MYB transcription factors in tomato and Arabidopsis. Front. Plant Sci. 2014, 5, 91–95. [Google Scholar]
- Kim, D.W.; Lee, S.H.; Choi, S.B.; Won, S.K.; Heo, Y.K.; Cho, M.; Park, Y.I.; Cho, H.T. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 2006, 18, 2958–2970. [Google Scholar] [PubMed] [Green Version]
- Kim, C.M.; Dolan, L. Root hair development involves asymmetric cell division in Brachypodium distachyon and symmetric division in Oryza sativa. New Phytol. 2011, 192, 601–610. [Google Scholar] [PubMed]
- Dolan, L.; Duckett, C.M.; Grierson, C.; Linstead, P.; Schneider, K.; Lawson, E.; Dean, C.; Poethig, S.; Roberts, K. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 1994, 120, 2465–2474. [Google Scholar]
- Dolan, L.; Janmaat, K.; Willemsen, V.; Linstead, P.; Poethig, S.; Roberts, K.; Scheres, B. Cellular organisation of the Arabidopsis thaliana root. Development 1993, 119, 71–84. [Google Scholar]
- Schiefelbein, J. Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 2003, 6, 74–78. [Google Scholar] [PubMed]
- Schiefelbein, J.; Kwak, S.H.; Wieckowski, Y.; Barron, C.; Bruex, A. The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J. Exp. Bot. 2009, 60, 1515–1521. [Google Scholar]
- Baluška, F.; Salaj, J.; Mathur, J.; Braun, M.; Jasper, F.; Samaj, J.; Chua, N.H.; Barlow, P.W.; Volkmann, D. Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 2000, 227, 618–632. [Google Scholar] [PubMed] [Green Version]
- Schiefelbein, J.W. Constructing a plant cell. The genetic control of root hair development. Plant Physiol. 2000, 124, 1525–1531. [Google Scholar]
- Foreman, J.; Dolan, L. Root hairs as a model system for studying plant cell growth. Ann. Bot. 2001, 88, 1–7. [Google Scholar]
- Grierson, C.S.; Parker, J.S.; Kemp, A.C. Arabidopsis genes with roles in root hair development. J. Plant Nutr. Soil Sci. 2011, 164, 131–140. [Google Scholar]
- Salazar-Henao, J.E.; Schmidt, W. An inventory of nutrient-responsive genes in Arabidopsis root hairs. Front. Plant Sci. 2016, 7, 237. [Google Scholar]
- Menand, B.; Yi, K.; Jouannic, S.; Hoffmann, L.; Ryan, E.; Linstead, P.; Schaefer, D.G.; Dolan, L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science 2007, 316, 1477–1480. [Google Scholar] [PubMed] [Green Version]
- Masucci, J.D.; Schiefelbein, J.W. The rhd6 Mutation of Arabidopsis thaliana Alters Root-Hair Initiation through an Auxin- and Ethylene-Associated Process. Plant Physiol. 1994, 106, 1335–1346. [Google Scholar]
- Skinner, M.K.; Rawls, A.; Wilson-Rawls, J.; Roalson, E.H. Basic helix-loop-helix transcription factor gene family phylogenetics and nomenclature. Differentiation 2010, 80, 1–8. [Google Scholar]
- Bernhardt, C.; Zhao, M.; Gonzalez, A.; Lloyd, A.; Schiefelbein, J. The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 2005, 132, 291–298. [Google Scholar] [PubMed] [Green Version]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar]
- Keke, Y.; Menand, B.; Bell, E.; Dolan, L. A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet. 2010, 42, 264–267. [Google Scholar]
- Song, S.K.; Ryu, K.H.; Kang, Y.H.; Song, J.H.; Cho, Y.H.; Yoo, S.D.; Schiefelbein, J.; Lee, M.M. Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol. 2011, 157, 1196–1208. [Google Scholar]
- Simon, M.; Lee, M.M.; Lin, Y.; Gish, L.; Schiefelbein, J. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 2007, 311, 566–578. [Google Scholar]
- Tominaga, R.; Iwata, M.; Sano, R.; Inoue, K.; Okada, K.; Wada, T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 2008, 135, 1335–1345. [Google Scholar]
- Lee, M.M.; Schiefelbein, J. Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 2002, 14, 611–618. [Google Scholar] [PubMed]
- Cho, H.T.; Cosgrove, D.J. Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 2002, 14, 3237–3353. [Google Scholar] [PubMed] [Green Version]
- Franciosini, A.; Rymen, B.; Shibata, M.; Favero, D.S.; Sugimoto, K. Molecular networks orchestrating plant cell growth. Curr. Opin. Plant Biol. 2017, 35, 98–104. [Google Scholar] [PubMed]
- Shibata, M.; Breuer, C.; Kawamura, A.; Clark, N.M.; Rymen, B.; Braidwood, L.; Morohashi, K.; Busch, W.; Benfey, P.N.; Sozzani, R.; et al. GTL1 and DF1 regulate root hair growth through transcriptional repression of ROOT HAIR DEFECTIVE 6-LIKE 4 in Arabidopsis. Development 2018, 145, dev159707. [Google Scholar]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar]
- Knox, K.; Grierson, C.S.; Leyser, O. AXR3 and SHY2 interact to regulate root hair development. Development 2003, 130, 5769–5777. [Google Scholar]
- Eunkyoo, O.H.; Pil, J.S.; Jungmook, K. Signaling Peptides and Receptors Coordinating Plant Root Development. Trends Plant Sci. 2018, 23, 337–351. [Google Scholar]
- Tavormina, P.; De Coninck, B.; Nikonorova, N.; De Smet, I.; Cammue, B. The plant peptidome: An expanding repertoire of structural features and biological functions. Plant Cell 2015, 27, 2095–2118. [Google Scholar]
- Motomitsu, A.; Sawa, S.; Ishida, T. Plant peptide hormone signaling. Essays Biochem. 2015, 58, 115–131. [Google Scholar]
- Vanyushin, B.F.; Ashapkin, V.V.; Akeksandryshkina, N.I. Regulatory peptides in plants. Biochemistry 2017, 82, 189–195. [Google Scholar]
- Czyzewicz, N.; Yue, K.; Beeckman, T.; De Smet, I. Message in a bottle: Small signalling peptide outputs during growth and development. J. Exp. Bot. 2013, 64, 5281–5296. [Google Scholar] [PubMed] [Green Version]
- Wang, G.; Zhang, G.; Wu, M. CLE peptide signaling and crosstalk with phytohormones and environmental stimuli. Front. Plant Sci. 2016, 6, 1211. [Google Scholar] [PubMed] [Green Version]
- Fedoreyevaa, L.I.; Kononenko, N.V.; Baranova, E.N.; Dilovarova, T.A.; Smirnova, E.A.; Vanyushin, B.F. Dipeptides and Glycine Modulate Development of Seedlings and Regenerants of Tobacco Nicotiana tabacum L. Biol. Bull. 2020, 47, 351–360. [Google Scholar]
- Hsiao, Y.-C.; Yamada, M. The Roles of Peptide Hormones and Their Receptors during Plant Root Development. Genes 2021, 12, 22. [Google Scholar]
- Ryan, C.A.; Pearce, G. Systemin: A polypeptide signal for plant defensive genes. Annu. Rev. Cell Dev. Biol. 1998, 14, 1–17. [Google Scholar]
- Clark, S.E.; Running, M.P.; Meyerowit, E.M. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA. Development 1995, 121, 2057–2067. [Google Scholar]
- Vetsch, M.; Janzik, I.; Schaller, A. Characterization of prosystemin expressed in the baculovirus/insect cell system reveals biological activity of the systemin precursor. Planta 2000, 211, 91–97. [Google Scholar]
- Komori, R.; Amano, Y.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 15067–15072. [Google Scholar]
- Amano, Y.; Tsubouchi, H.; Shinohara, H.; Ogawa, Y.; Matsubayashi, M. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 18333–18338. [Google Scholar]
- Kouchi, H.; Takane, K.; So, R.B.; Ladha, J.K. Rice ENOD40: Isolation and expression analysis in rice and transgenic soybean root nodules. Plant J. 1999, 18, 121–129. [Google Scholar]
- Haruta, M.; Sabat, G.; Stecker, K.; Minkoff, B.B.; Sussman, M.R. A Peptide Hormone and Its Receptor Protein Kinase Regulate Plant Cell Expansion. Science 2014, 343, 408–411. [Google Scholar] [PubMed] [Green Version]
- Zhu, S.; Martínez, J.; Pacheco, J.; Estevez, J.M.; Yu, F. Autocrine regulation of root hair size by the RaLF FERONIA-RSL4 signaling pathway. New Phytol. 2020, 227, 45–49. [Google Scholar]
- Khavinson, V.K.; Tendler, S.M.; Vanyushin, B.F.; Kasyanenko, N.A.; Kvetnoy, I.M.; Linkova, N.S.; Ashapkin, V.V.; Polyakova, V.O.; Basharina, V.S.; Bernadotte, A. Peptide regulation of gene expression and protein synthesis in bronchial epithelium. Lung 2014, 192, 781–791. [Google Scholar] [PubMed]
- Khavinson, V.K.; Tendler, S.M.; Kasyanenko, N.A.; Tarnovskaya, S.I.; Linkova, N.S.; Ashapkin, V.V.; Yakutseni, P.P.; Vanyushin, B.F. Tetrapeptide KEDW interacts with DNA and regulates gene expression. Am. J. Biomed. Sci. 2015, 7, 156–169. [Google Scholar]
- Fedoreyeva, L.I.; Vanyushin, B.F. Gly, GlyGly, and GlyAsp Modulate Expression of Genes of the SNF2 Family and DNA Methyltransferases in Regenerants from Calluses of Tobacco Nicotiana tabacum. Biol. Bull. Russ. Acad. Sci. 2021, 48, 450–458. [Google Scholar]
- Archacki, R.; Yatusevich, R.; Buszewicz, D.; Krzyczmonik, K.; Patryn, J.; Iwanicka-Nowicka, R.; Biecek, P.; Wilczynski, B.; Koblowska, M.; Jerzmanowski, A.; et al. Arabidopsis SWI/SNF chromatin remodeling complex binds both promoters and terminators to regulate gene expression. Nucleic Acids Res. 2017, 45, 3116–3129. [Google Scholar]
- Kwon, C.S.; Wagner, D. Unwinding chromatin for development and growth: A few genes at a time. Trends Genet. 2007, 23, 403–412. [Google Scholar]
- Flaus, A.; Owen-Hughes, T. Mechanisms for ATP-dependent chromatin remodeling: The means to the end. FEBS J. 2011, 278, 3579–3595. [Google Scholar]
- Kanno, T.; Aufsatz, W.; Jaligot, E.; Mette, M.F.; Matzke, M.; Matzke, A.J. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 2005, 6, 649–655. [Google Scholar]
- Fedoreyeva, L.I.; Vanyushin, B.F.; Baranova, E.N. Peptide AEDL alters chromatin conformation via histone binding. AIMS Biophys. 2020, 7, 1–16. [Google Scholar]
- Fedoreyeva, L.I.; Dilovarova, T.A.; Kononenko, N.V.; Baranova, E.N.; Smirnova, E.A.; Vanyushin, B.F. Influence of glycylglycine, glycine, and glycylaspartic acid on growth, development, and gene expression in a tobacco (Nicotiana tabacum) callus culture. Biol. Bull. 2018, 45, 351–358. [Google Scholar]
- Kononenko, N.; Baranova, E.; Dilovarova, T.; Akanov, E.; Fedoreyeva, L. Oxidative Damage to Various Root and Shoot Tissues of Durum and Soft Wheat Seedlings during Salinity. Agriculture 2020, 10, 55–71. [Google Scholar]
- Bell, K.S.; Mitchell, S.; Paultre, D.; Posch, M.; Oparka, K. Correlative Imaging of Fluorescent Proteins in Resin-Embedded Plant Material. Plant Physiol. 2013, 161, 1595–1603. [Google Scholar] [PubMed] [Green Version]
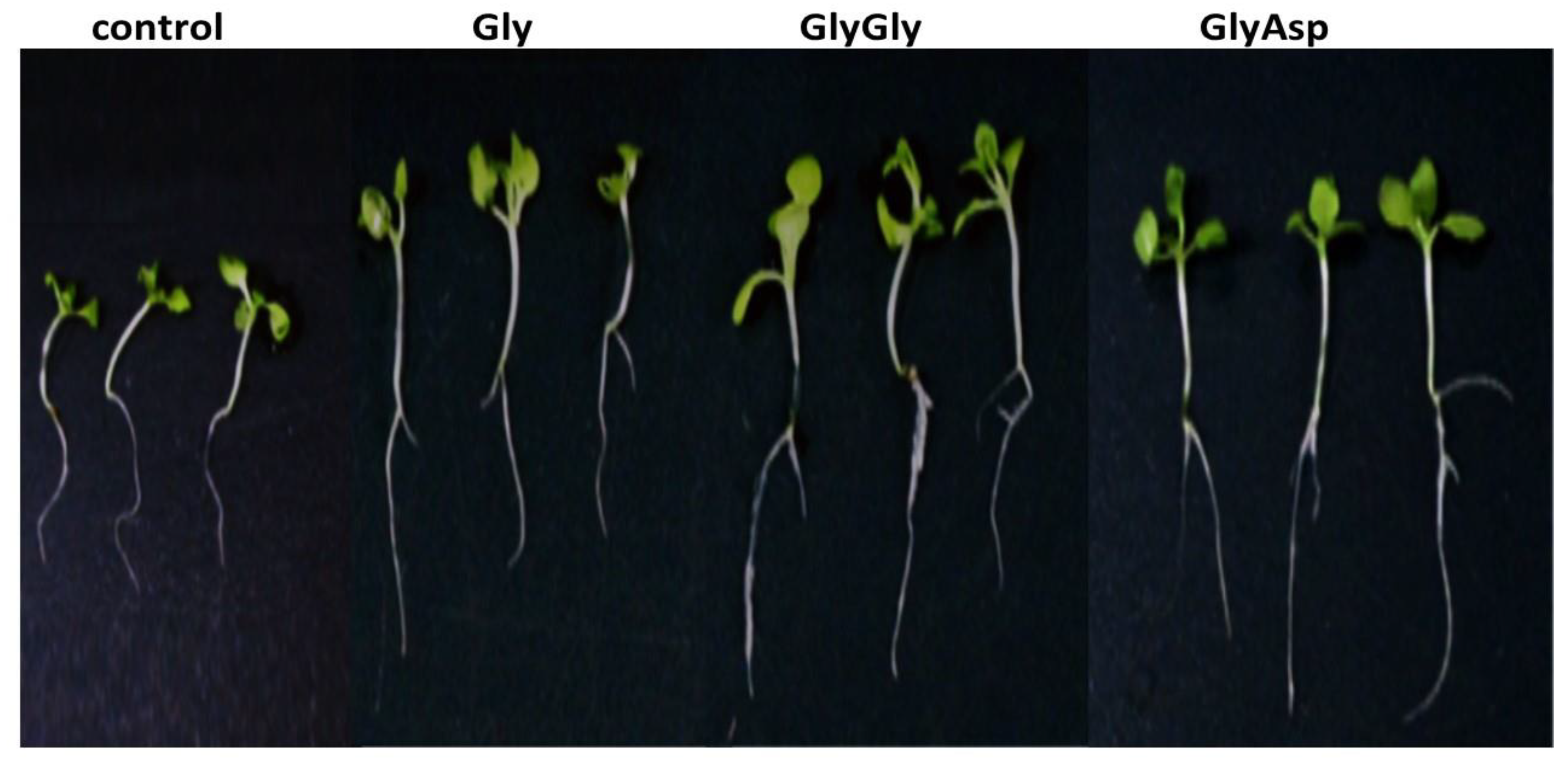
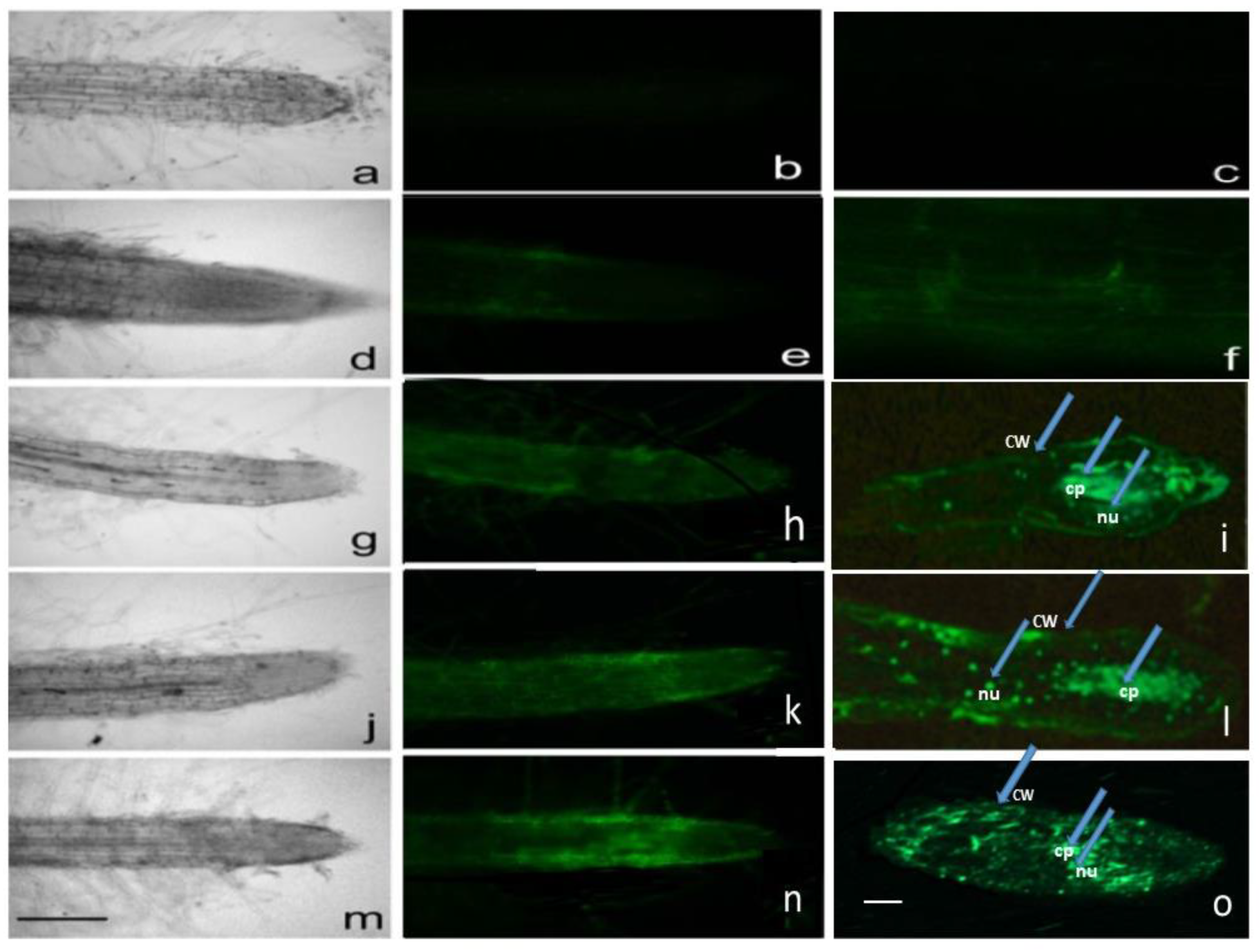

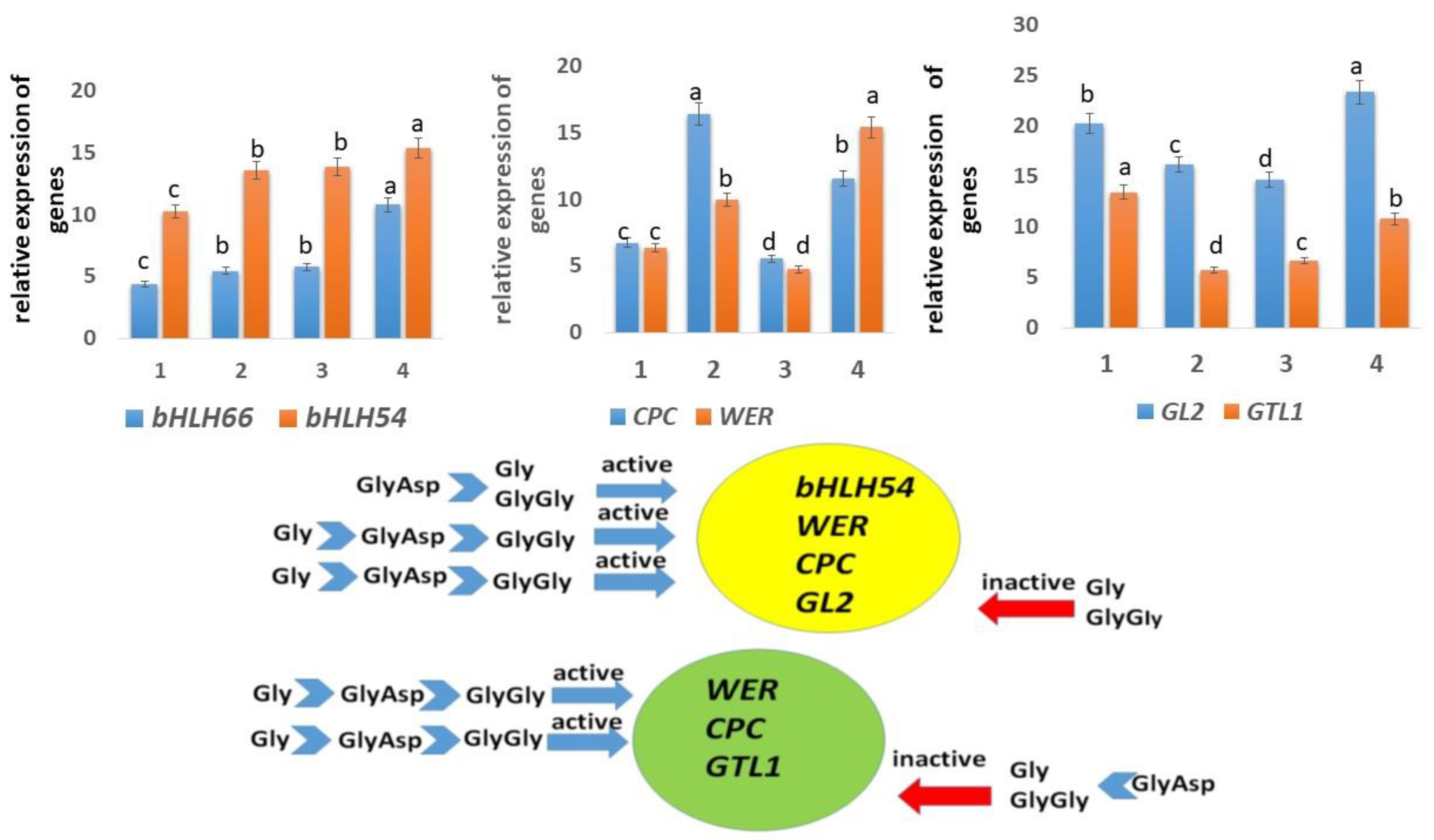
| Additives | Mean ± Standard Error (n = 20) | ||
|---|---|---|---|
| Seedling Fresh Weight (mg) | Total Seedling Length (mm) | Main Root Length (mm) | |
| Control | 0.412 ± 0.03 d | 21.6 ± 1.2 b | 12.4 ± 0.6 c |
| Gly | 0.440 ± 0.03 c | 23.7 ± 1.6 a | 14.8 ± 1.1 b |
| GlyGly | 0.587 ± 0.04 a | 24.5 ± 1.6 a | 17.9 ± 0.8 a |
| GlyAsp | 0.561 ± 0.04 b | 25.0 ± 1.5 a | 17.8 ± 1.0 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedoreyeva, L.I.; Chaban, I.A.; Kononenko, N.V. Short Peptides Induce Development of Root Hair Nicotiana tabacum. Plants 2022, 11, 852. https://doi.org/10.3390/plants11070852
Fedoreyeva LI, Chaban IA, Kononenko NV. Short Peptides Induce Development of Root Hair Nicotiana tabacum. Plants. 2022; 11(7):852. https://doi.org/10.3390/plants11070852
Chicago/Turabian StyleFedoreyeva, Larisa I., Inna A. Chaban, and Neonila V. Kononenko. 2022. "Short Peptides Induce Development of Root Hair Nicotiana tabacum" Plants 11, no. 7: 852. https://doi.org/10.3390/plants11070852







