Metal Tolerance Protein Encoding Gene Family in Fagopyrum tartaricum: Genome-Wide Identification, Characterization and Expression under Multiple Metal Stresses
Abstract
:1. Introduction
2. Results
2.1. Identification and Physicochemical Parameters of MTPs in the Tartary Buckwheat Genome
2.2. Phylogenetic Tree of 67 MTP Proteins from Six Species
2.3. Phylogenetic Tree, Gene Structure and Motif Patterns of FtMTPs
2.4. Structure Variation of the FtMTP Proteins
2.5. Cis-Regulatory Elements of the FtMTPs
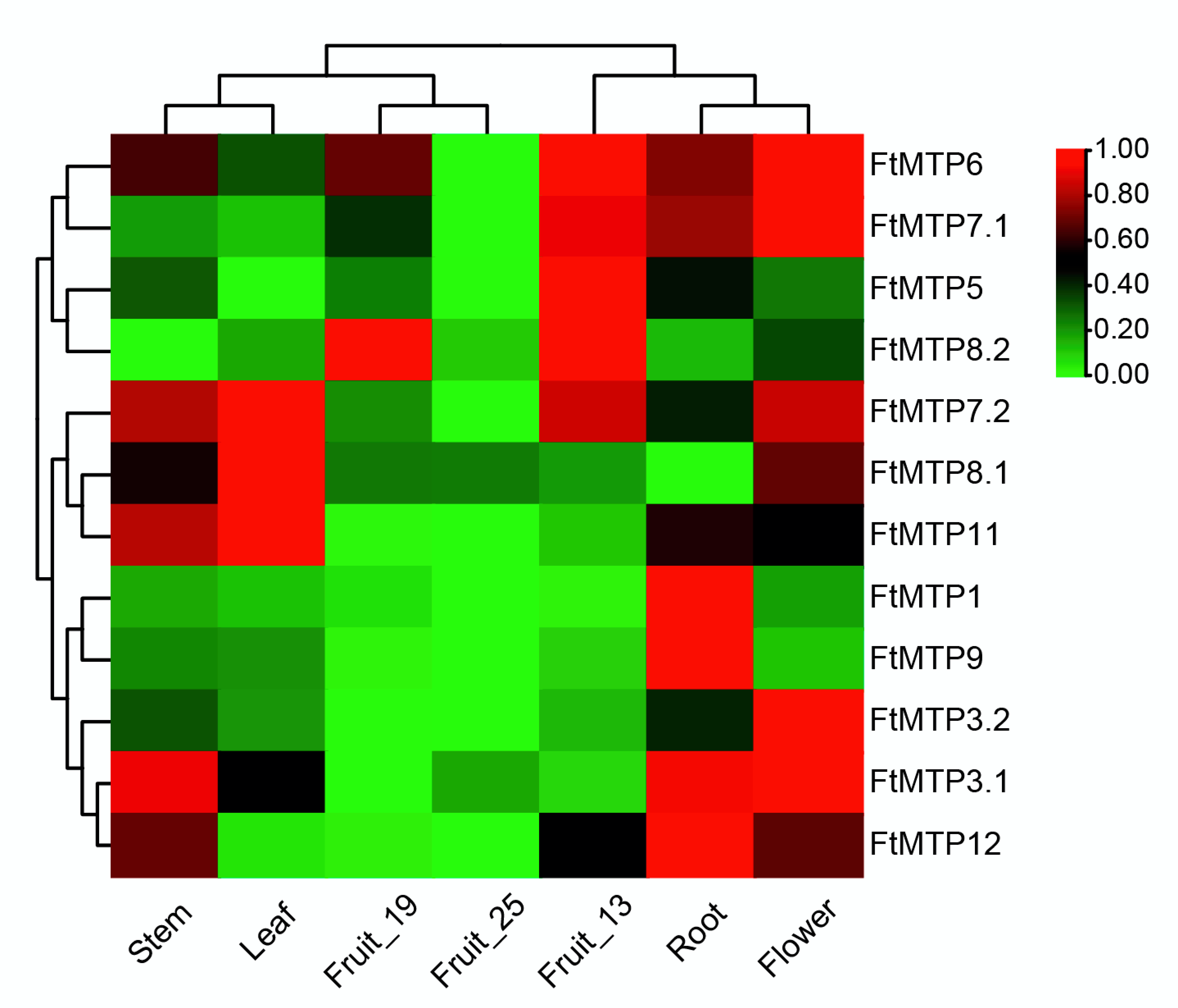
2.6. Tissues-Specific Expression Profile of FtMTPs
2.7. Expression of FtMTPs under Multiple Metal Stress
3. Discussion
4. Materials and Methods
4.1. Identification, Physicochemical Parameters of MTPs in Tartary Buckwheat
4.2. Phylogenetic Analysis
4.3. Gene Structure and Motif Analysis
4.4. Protein Modeling and Prediction
4.5. Cis-Regulatory Elements Prediction
4.6. Transcriptome Data Analysis
4.7. Plant Cultivation and Stress Treatments
4.8. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wyszkowski, M.; Brodowska, M.S. Content of Trace Elements in Soil Fertilized with Potassium and Nitrogen. Agriculture 2020, 10, 398. [Google Scholar] [CrossRef]
- Liu, J.; Gao, Y.; Tang, Y.; Wang, D.; Chen, X.; Yao, Y.; Guo, Y. Genome-Wide Identification, Comprehensive Gene Feature, Evolution, and Expression Analysis of Plant Metal Tolerance Proteins in Tobacco Under Heavy Metal Toxicity. Front. Genet. 2019, 10, 345. [Google Scholar] [CrossRef] [PubMed]
- Eid, E.M.; Alrumman, S.A.; Galal, T.M.; El-Bebany, A.F. Regression models for monitoring trace metal accumulations by Faba sativa Bernh. plants grown in soils amended with different rates of sewage sludge. Sci. Rep. 2019, 9, 5443. [Google Scholar] [CrossRef] [PubMed]
- Nishida, S.; Tanikawa, R.; Ishida, S.; Yoshida, J.; Mizuno, T.; Nakanishi, H.; Furuta, N. Elevated Expression of Vacuolar Nickel Transporter Gene IREG2 Is Associated With Reduced Root-to-Shoot Nickel Translocation in Noccaea japonica. Front. Plant Sci. 2020, 11, 610. [Google Scholar] [CrossRef] [PubMed]
- Sabra, M.; Aboulnasr, A.; Franken, P.; Perreca, E.; Wright, L.P.; Camehl, I. Beneficial Root Endophytic Fungi Increase Growth and Quality Parameters of Sweet Basil in Heavy Metal Contaminated Soil. Front. Plant Sci. 2018, 9, 1726. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.-J.; Zaidul, I.S.M.; Suzuki, T.; Mukasa, Y.; Hashimoto, N.; Takigawa, S.; Noda, T.; Matsuura-Endo, C.; Yamauchi, H. Comparison of phenolic compositions between common and tartary buckwheat (Fagopyrum) sprouts. Food Chem. 2008, 110, 814–820. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. [Google Scholar] [CrossRef] [Green Version]
- Nomura, T.; Itouga, M.; Kojima, M.; Kato, Y.; Sakakibara, H.; Hasezawa, S. Copper mediates auxin signalling to control cell differentiation in the copper moss Scopelophila cataractae. J. Exp. Bot. 2015, 66, 1205–1213. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Zhou, X.; Ma, Y.; Xu, E.; Yu, Y.; Liu, Y.; Chen, X.; Zhang, W. Expression of a Brassica napus metal transport protein (BnMTP3) in Arabidopsis thaliana confers tolerance to Zn and Mn. Plant Sci. 2021, 304, 110754. [Google Scholar] [CrossRef]
- Barabasz, A.; Klimecka, M.; Kendziorek, M.; Weremczuk, A.; Ruszczynska, A.; Bulska, E.; Antosiewicz, D.M. The ratio of Zn to Cd supply as a determinant of metal-homeostasis gene expression in tobacco and its modulation by overexpressing the metal exporter AtHMA4. J. Exp. Bot. 2016, 67, 6201–6214. [Google Scholar] [CrossRef] [Green Version]
- Tsunemitsu, Y.; Genga, M.; Okada, T.; Yamaji, N.; Ma, J.F.; Miyazaki, A.; Kato, S.I.; Iwasaki, K.; Ueno, D. A member of cation diffusion facilitator family, MTP11, is required for manganese tolerance and high fertility in rice. Planta 2018, 248, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Eroglu, S.; Giehl, R.F.H.; Meier, B.; Takahashi, M.; Terada, Y.; Ignatyev, K.; Andresen, E.; Kupper, H.; Peiter, E.; von Wiren, N. Metal Tolerance Protein 8 Mediates Manganese Homeostasis and Iron Reallocation during Seed Development and Germination. Plant Physiol. 2017, 174, 1633–1647. [Google Scholar] [CrossRef] [Green Version]
- Ueno, D.; Sasaki, A.; Yamaji, N.; Miyaji, T.; Fujii, Y.; Takemoto, Y.; Moriyama, S.; Che, J.; Moriyama, Y.; Iwasaki, K.; et al. A polarly localized transporter for efficient manganese uptake in rice. Nat. Plants 2015, 1, 15170. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Gu, D.; Li, Y.; Li, J.; Li, J.; Chen, X.; Zhang, W. Phylogenetic and Expression Analysis of Mn-CDF Transporters in Pear (Pyrus bretschneideri Rehd.). Plant Mol. Biol. Rep. 2019, 37, 98–110. [Google Scholar] [CrossRef]
- Li, J.; Zheng, L.; Fan, Y.; Wang, Y.; Ma, Y.; Gu, D.; Lu, Y.; Zhang, S.; Chen, X.; Zhang, W. Pear metal transport protein PbMTP8.1 confers manganese tolerance when expressed in yeast and Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2021, 208, 111687. [Google Scholar] [CrossRef] [PubMed]
- Das, U.; Haque, A.F.M.M.; Bari, M.A.; Mandal, A.; Kabir, A.H. Computational characterization and expression profile of MTP1 gene associated with zinc homeostasis across dicot plant species. Gene Rep. 2021, 23, 101073. [Google Scholar] [CrossRef]
- Arrivault, S.; Senger, T.; Krämer, U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006, 46, 861–879. [Google Scholar] [CrossRef]
- Kawachi, M.; Kobae, Y.; Mimura, T.; Maeshima, M. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J. Biol. Chem. 2008, 283, 8374–8383. [Google Scholar] [CrossRef] [Green Version]
- Dräger, D.B.; Desbrosses-Fonrouge, A.G.; Krach, C.; Chardonnens, A.N.; Meyer, R.C.; Saumitou-Laprade, P.; Krämer, U. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 2004, 39, 425–439. [Google Scholar] [CrossRef]
- Kobae, Y.; Uemura, T.; Sato, M.H.; Ohnishi, M.; Mimura, T.; Nakagawa, T.; Maeshima, M. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004, 45, 1749–1758. [Google Scholar] [CrossRef] [Green Version]
- Fujiwara, T.; Kawachi, M.; Sato, Y.; Mori, H.; Kutsuna, N.; Hasezawa, S.; Maeshima, M. A high molecular mass zinc transporter MTP 12 forms a functional heteromeric complex with MTP 5 in the Golgi in Arabidopsis thaliana. FEBS J. 2015, 282, 1965–1979. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fujii, Y.; Yamaji, N.; Masuda, S.; Takemoto, Y.; Kamiya, T.; Yusuyin, Y.; Iwasaki, K.; Kato, S.-i.; Maeshima, M. Mn tolerance in rice is mediated by MTP8. 1, a member of the cation diffusion facilitator family. J. Exp. Bot. 2013, 64, 4375–4387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takemoto, Y.; Tsunemitsu, Y.; Fujii-Kashino, M.; Mitani-Ueno, N.; Yamaji, N.; Ma, J.F.; Kato, S.-I.; Iwasaki, K.; Ueno, D. The tonoplast-localized transporter MTP8. 2 contributes to manganese detoxification in the shoots and roots of Oryza sativa L. Plant Cell Physiol. 2017, 58, 1573–1582. [Google Scholar] [CrossRef]
- Eroglu, S.; Meier, B.; von Wirén, N.; Peiter, E. The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 2016, 170, 1030–1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricachenevsky, F.K.; Menguer, P.K.; Sperotto, R.A.; Williams, L.E.; Fett, J.P. Roles of plant metal tolerance proteins (MTP) in metal storage and potential use in biofortification strategies. Front. Plant Sci. 2013, 4, 144. [Google Scholar] [CrossRef] [Green Version]
- Shirazi, Z.; Abedi, A.; Kordrostami, M.; Burritt, D.J.; Hossain, M.A. Genome-wide identification and characterization of the metal tolerance protein (MTP) family in grape (Vitis vinifera L.). 3 Biotech 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Q.; Xu, W.; Zhao, H.; Guo, F.; Wang, P.; Wang, Y.; Ni, D.; Wang, M.; Wei, C. Identification of MTP gene family in tea plant (Camellia sinensis L.) and characterization of CsMTP8. 2 in manganese toxicity. Ecotoxicol. Environ. Saf. 2020, 202, 110904. [Google Scholar] [CrossRef]
- Bonafaccia, G.; Marocchini, M.; Kreft, I. Composition and technological properties of the flour and bran from common and tartary buckwheat. Food Chem. 2003, 80, 9–15. [Google Scholar] [CrossRef]
- Peng, L.X.; Zou, L.; Wang, J.B.; Zhao, J.L.; Xiang, D.B.; Zhao, G. Flavonoids, Antioxidant Activity and Aroma Compounds Analysis from Different Kinds of Tartary Buckwheat Tea. Indian J. Pharm. Sci. 2015, 77, 661–667. [Google Scholar]
- Pirzadah, T.B.; Malik, B.; Tahir, I.; Irfan, Q.M.; Rehman, R.U. Characterization of mercury-induced stress biomarkers in Fagopyrum tataricum plants. Int. J. Phytoremediation 2018, 20, 225–236. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, Q.-f.; Li, J.; Xiong, J.; Zhou, L.-n.; He, S.-l.; Zhang, J.-q.; Chen, Z.-a.; He, S.-g.; Liu, H. Effects of exogenous sulfur on alleviating cadmium stress in tartary buckwheat. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirzadah, T.B.; Malik, B.; Tahir, I.; Hakeem, K.R.; Alharby, H.F.; Rehman, R.U. Lead toxicity alters the antioxidant defense machinery and modulate the biomarkers in Tartary buckwheat plants. Int. Biodeterior. Biodegrad. 2020, 151, 104992. [Google Scholar] [CrossRef]
- Wang, L.; Zheng, B.; Yuan, Y.; Xu, Q.; Chen, P. Transcriptome profiling of Fagopyrum tataricum leaves in response to lead stress. BMC Plant Biol. 2020, 20, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, Z.; Huang, Y.; Jiang, Y.; Liu, Y.; Wen, W.; Li, H.; Shao, J.; Wang, C.; Zhu, X. Antioxidant capacity, metal contents, and their health risk assessment of tartary buckwheat teas. ACS Omega 2020, 5, 9724–9732. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, F.; Liu, J.; Xie, W.; Zhang, L.; Chen, Z.; Peng, Z.; Ou, Y.; Yao, Y. Genome-Wide Identification of Metal Tolerance Protein Genes in Populus trichocarpa and Their Roles in Response to Various Heavy Metal Stresses. Int. J. Mol. Sci. 2020, 21, 1680. [Google Scholar] [CrossRef] [Green Version]
- Shingu, Y.; Kudo, T.; Ohsato, S.; Kimura, M.; Ono, Y.; Yamaguchi, I.; Hamamoto, H. Characterization of genes encoding metal tolerance proteins isolated from Nicotiana glauca and Nicotiana tabacum. Biochem. Biophys. Res. Commun. 2005, 331, 675–680. [Google Scholar] [CrossRef]
- Fu, X.-Z.; Tong, Y.-H.; Zhou, X.; Ling, L.-L.; Chun, C.-P.; Cao, L.; Zeng, M.; Peng, L.-Z. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene 2017, 629, 1–8. [Google Scholar] [CrossRef]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Sun, W.; Ma, Z.; Zheng, T.; Huang, L.; Wu, Q.; Zhao, G.; Tang, Z.; Bu, T.; Li, C. Genome-wide investigation of the AP2/ERF gene family in tartary buckwheat (Fagopyum Tataricum). BMC Plant Biol. 2019, 19, 84. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Huang, L.; Ma, Z.; Sun, W.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 342. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Ma, Z.; Wang, A.; Zheng, T.; Huang, L.; Sun, W.; Zhang, Y.; Jin, W.; Zhan, J.; Cai, Y. Genome-wide investigation of the auxin response factor gene family in Tartary buckwheat (Fagopyrum tataricum). Int. J. Mol. Sci. 2018, 19, 3526. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Wen, Y.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, phylogeny, evolutionary expansion and expression analyses of bZIP transcription factor family in tartary buckwheat. BMC Genom. 2019, 20, 483. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Jin, X.; Ma, Z.; Chen, H.; Liu, M. Basic helix–loop–helix (bHLH) gene family in Tartary buckwheat (Fagopyrum tataricum): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses. Int. J. Biol. Macromol. 2020, 155, 1478–1490. [Google Scholar] [CrossRef]
- Liu, M.; Fu, Q.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide investigation of the MADS gene family and dehulling genes in tartary buckwheat (Fagopyrum tataricum). Planta 2019, 249, 1301–1318. [Google Scholar] [CrossRef]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 2019, 19, 299. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, H.; Huang, J.; Shi, T.; Meng, Z.; Chen, Q.; Deng, J. Genome-wide analysis of BBX gene family in Tartary buckwheat (Fagopyrum tataricum). PeerJ 2021, 9, e11939. [Google Scholar] [CrossRef]
- Yan, H.; Liu, C.; Zhao, J.; Ye, X.; Wu, Q.; Yao, T.; Peng, L.; Zou, L.; Zhao, G. Genome-wide analysis of the NF-Y gene family and their roles in relation to fruit development in Tartary buckwheat (Fagopyrum tataricum). Int. J. Biol. Macromol. 2021, 190, 487–498. [Google Scholar] [CrossRef]
- Desbrosses-Fonrouge, A.-G.; Voigt, K.; Schröder, A.; Arrivault, S.; Thomine, S.; Krämer, U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005, 579, 4165–4174. [Google Scholar] [CrossRef]
- Blaudez, D.; Kohler, A.; Martin, F.; Sanders, D.; Chalot, M. Poplar metal tolerance protein 1 confers zinc tolerance and is an oligomeric vacuolar zinc transporter with an essential leucine zipper motif. Plant Cell 2003, 15, 2911–2928. [Google Scholar] [CrossRef] [Green Version]
- Peiter, E.; Montanini, B.; Gobert, A.; Pedas, P.; Husted, S.; Maathuis, F.J.; Blaudez, D.; Chalot, M.; Sanders, D. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 8532–8537. [Google Scholar] [CrossRef] [Green Version]
- Pedas, P.; Schiller Stokholm, M.; Hegelund, J.N.; Ladegård, A.H.; Schjoerring, J.K.; Husted, S. Golgi localized barley MTP8 proteins facilitate Mn transport. PLoS ONE 2014, 9, e113759. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Gruber, B.D.; Pittman, J.K.; White, R.G.; Leung, H.; Miao, Y.; Jiang, L.; Ryan, P.R.; Richardson, A.E. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007, 51, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Goodstein, D.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2011, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Vatansever, R.; Filiz, E.; Eroglu, S. Genome-wide exploration of metal tolerance protein (MTP) genes in common wheat (Triticum aestivum): Insights into metal homeostasis and biofortification. Biometals 2017, 30, 217–235. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes11Edited by F. Cohen. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [Green Version]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 2007, 100, 665–678. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, K.-C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2005, 21, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 7, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Ma, Z.; Hu, Y.; Chen, H. Tartary buckwheat database (TBD): An integrative platform for gene analysis of and biological information on Tartary buckwheat. J. Zhejiang Univ.-Sci. B 2021, 22, 954–958. [Google Scholar] [CrossRef] [PubMed]





| Gene Names | Peptide Length | pI a | MW b (KDa) | TMHs c | GRAVY d | SL e |
|---|---|---|---|---|---|---|
| FtMTP1 | 445 | 5.98 | 49.58 | 6 | −0.041 | V f |
| FtMTP3.1 | 408 | 6.32 | 44.74 | 6 | 0.050 | V |
| FtMTP3.2 | 431 | 6.19 | 46.95 | 6 | −0.016 | V |
| FtMTP5 | 230 | 6.03 | 25.41 | 4 | 0.374 | V |
| FtMTP6 | 256 | 5.9 | 28.54 | 1 | 0.009 | V |
| FtMTP7.1 | 460 | 9.23 | 51.39 | 4 | 0.002 | V |
| FtMTP7.2 | 467 | 6.85 | 51.10 | 4 | 0.066 | V |
| FtMTP8.1 | 405 | 5.11 | 45.61 | 5 | −0.065 | V |
| FtMTP8.2 | 415 | 5.57 | 46.58 | 4 | 0.016 | V |
| FtMTP9 | 384 | 6.88 | 43.78 | 6 | −0.018 | C g/V |
| FtMTP11 | 403 | 5.08 | 45.40 | 4 | 0.037 | V |
| FtMTP12 | 722 | 6.62 | 80.51 | 10 | 0.032 | V |
| Metal Ion | Concentration (mg L−1) | Source |
|---|---|---|
| Zn | 2.0 | ZnSO4·7H2O |
| Cu | 5.0 | CuSO4·5H2O |
| Mn | 5.0 | MnSO4 |
| Fe | 2.5 | FeCl3·6H2O |
| Cd | 2.0 | CdCl2·2.5H2O |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Wang, C.; Wang, K.; Zhao, J.; Shao, J.; Chen, H.; Zhou, M.; Zhu, X. Metal Tolerance Protein Encoding Gene Family in Fagopyrum tartaricum: Genome-Wide Identification, Characterization and Expression under Multiple Metal Stresses. Plants 2022, 11, 850. https://doi.org/10.3390/plants11070850
Li Z, Wang C, Wang K, Zhao J, Shao J, Chen H, Zhou M, Zhu X. Metal Tolerance Protein Encoding Gene Family in Fagopyrum tartaricum: Genome-Wide Identification, Characterization and Expression under Multiple Metal Stresses. Plants. 2022; 11(7):850. https://doi.org/10.3390/plants11070850
Chicago/Turabian StyleLi, Zhiqiang, Chenglong Wang, Kaiyi Wang, Jiayu Zhao, Jirong Shao, Hui Chen, Meiliang Zhou, and Xuemei Zhu. 2022. "Metal Tolerance Protein Encoding Gene Family in Fagopyrum tartaricum: Genome-Wide Identification, Characterization and Expression under Multiple Metal Stresses" Plants 11, no. 7: 850. https://doi.org/10.3390/plants11070850
APA StyleLi, Z., Wang, C., Wang, K., Zhao, J., Shao, J., Chen, H., Zhou, M., & Zhu, X. (2022). Metal Tolerance Protein Encoding Gene Family in Fagopyrum tartaricum: Genome-Wide Identification, Characterization and Expression under Multiple Metal Stresses. Plants, 11(7), 850. https://doi.org/10.3390/plants11070850