Cytogenetic, Morphometric, and Ecological Characterization of Festuca indigesta Boiss. in the Southeast of Spain
Abstract
:1. Introduction
2. Results
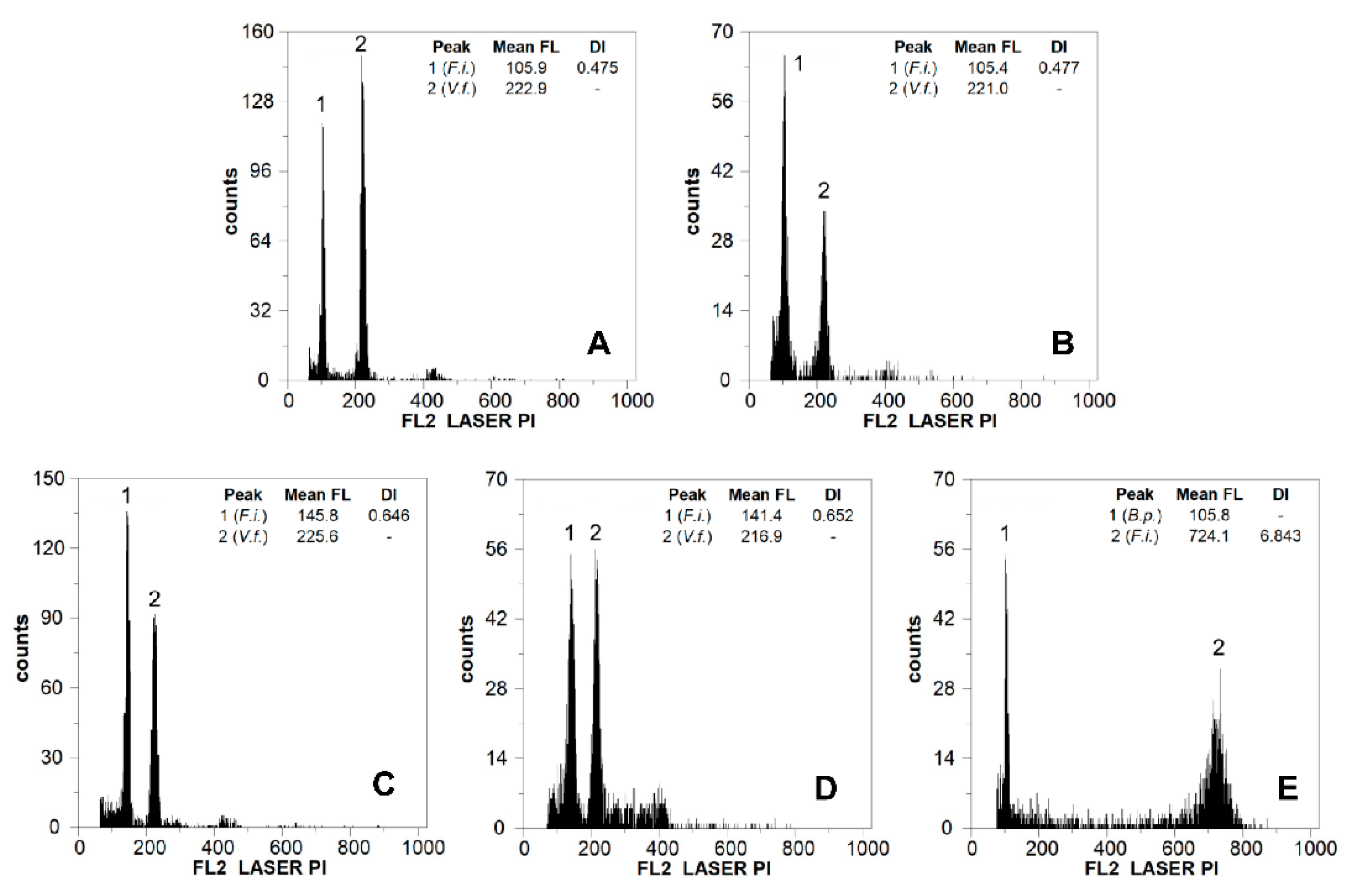
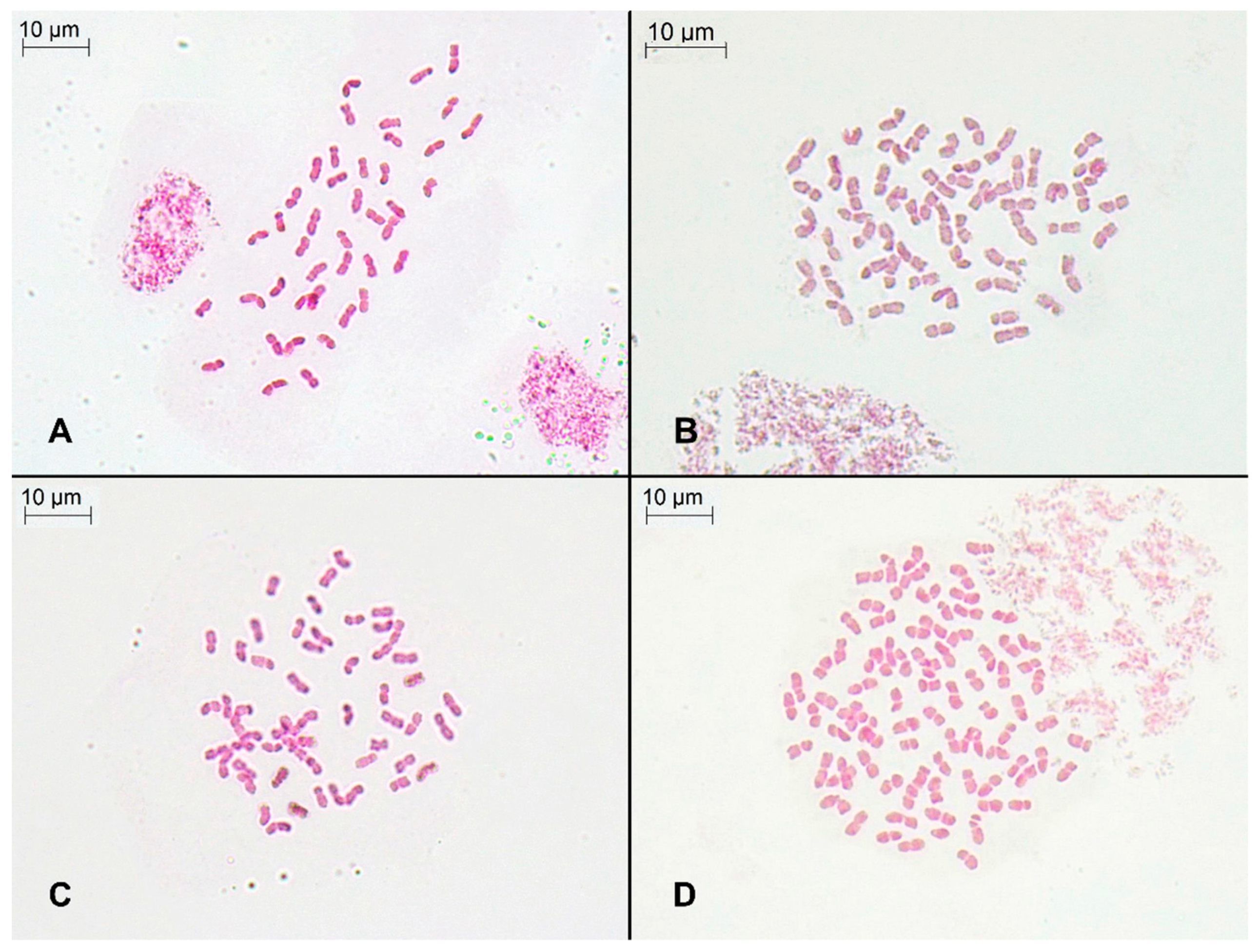
2.1. Genome Size, DNA Ploidy Level, and Chromosome Numbers
2.2. Soil Analyses and Ecological Remarks
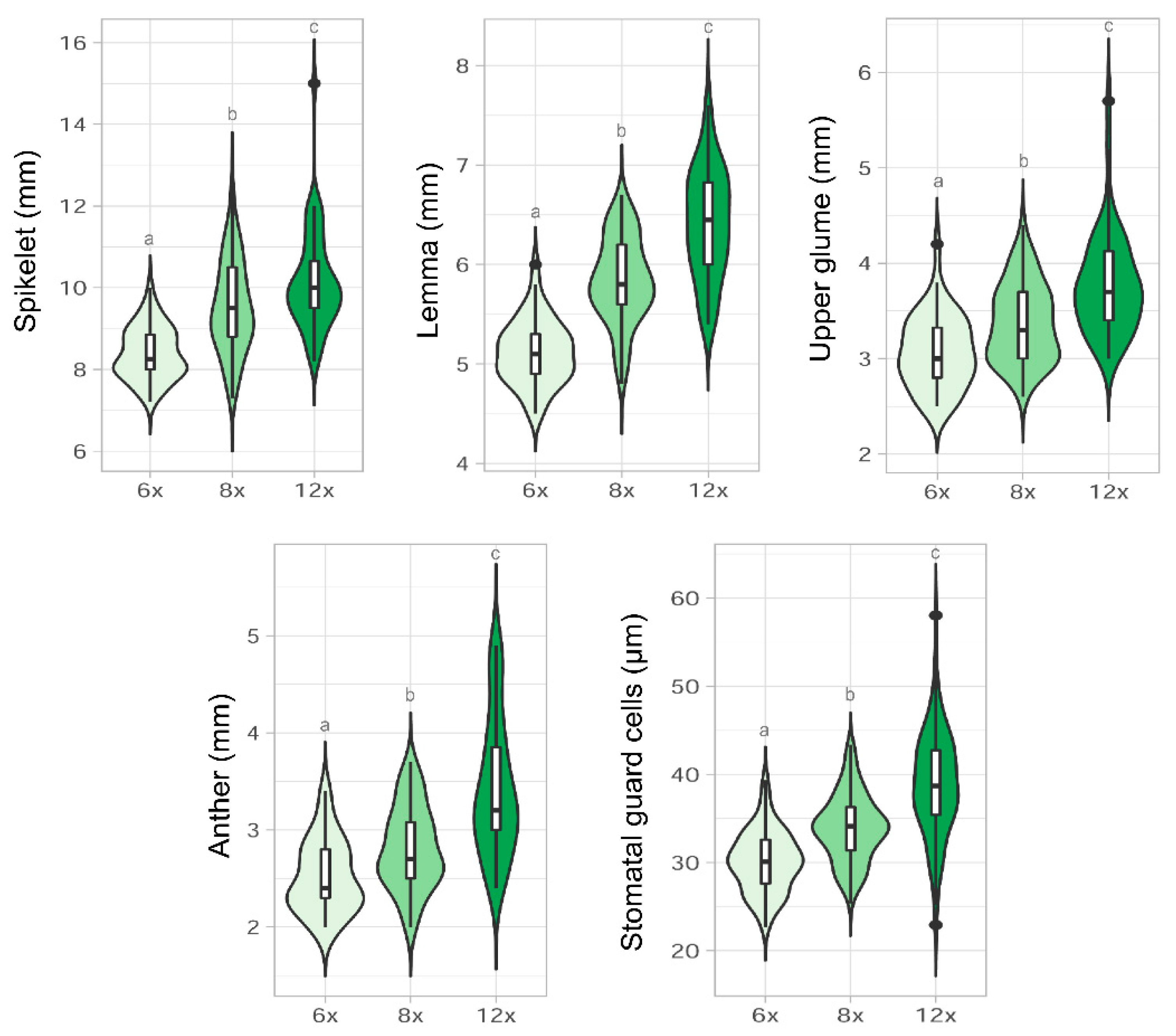
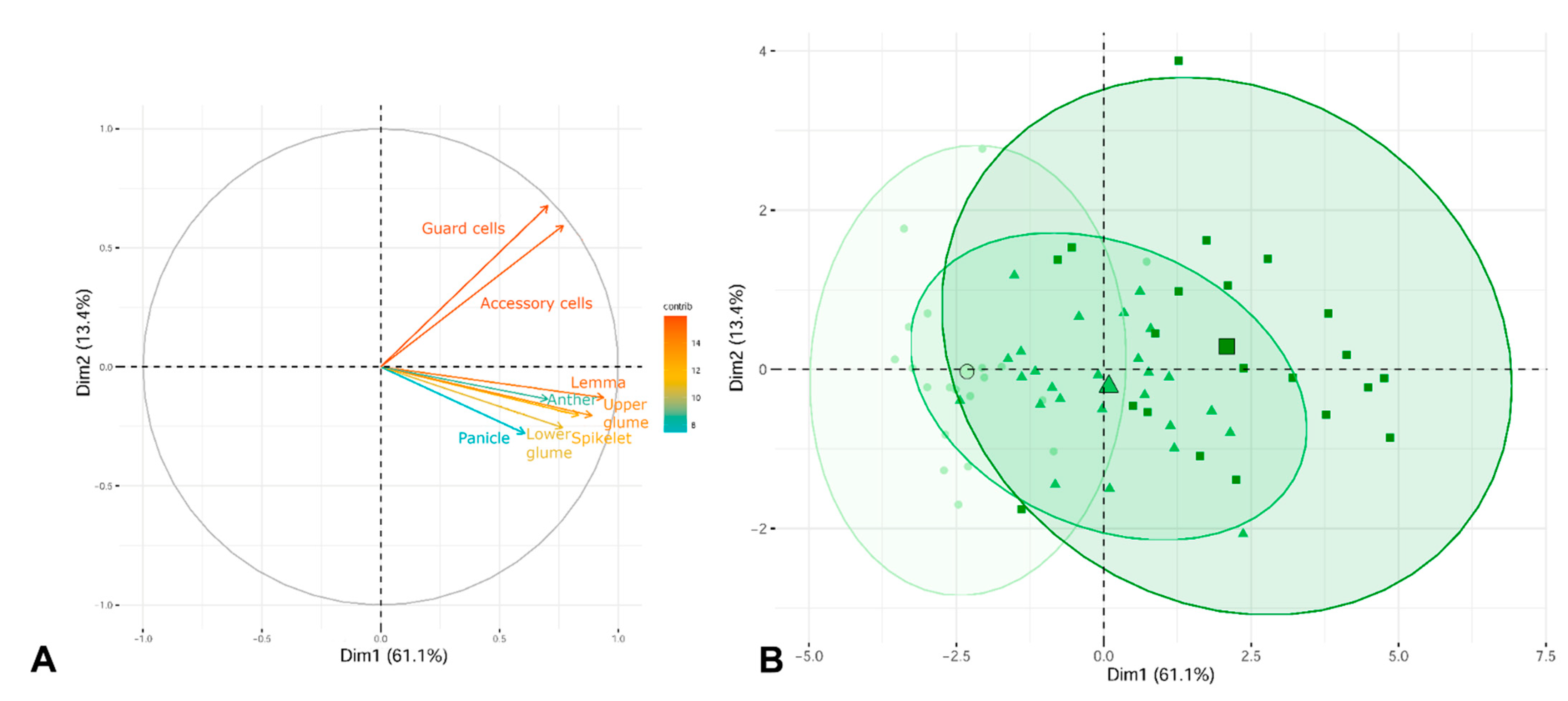
2.3. Morphological Analyses
3. Discussion
3.1. Genome Size, DNA Ploidy Level, and Chromosome Numbers
3.2. Cytotype Distribution and Ecological Parameters
3.3. Morphological Variation among Cytotypes
4. Materials and Methods
4.1. Study Area and Plant Material
4.2. Morphometric Analyses
4.3. Genome Size Estimation and DNA Ploidy Level Inference
4.4. Chromosome Counts
4.5. Soil Analyses
4.6. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catalán, P.; Torrecilla, P.; López, J.A.; Olmstead, R.G. Phylogeny of the festucoid grasses of subtribe Loliinae and allies (Poeae, Pooideae) inferred from ITS and trnL-F sequences. Mol. Phyl. Evol. 2004, 31, 517–541. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Sagarra, G.; Castro, S.; Mota, L.; Loureiro, J.; Devesa, J.A. Genome size, chromosome number and morphological data reveal unexpected infraspecific variability in Festuca (Poaceae). Genes 2021, 12, 906. [Google Scholar] [CrossRef] [PubMed]
- Marques, I.; Loureiro, J.; Draper, D.; Castro, M.; Castro, S. How much do we know about the frequency of hybridisation and polyploidy in the Mediterranean region? Plant Biol. 2018, 20, 21–37. [Google Scholar] [CrossRef]
- Alvarez, N.; Thiel-Egenter, C.; Tribsch, A.; Holderegger, R.; Manel, S.; Schönswetter, P.; Taberlet, P.; Brodbeck, S.; Gaudeul, M.; Gielly, L.; et al. IntraBioDiv Consortium. History or ecology? Substrate type as a major driver of patial genetic structure in Alpine plants. Ecol. Lett. 2009, 12, 632–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, A.J.; Kadereit, J.W. The evolution of substrate differentiation in Minuartia series Laricifoliae (Caryophyllaceae) in the European Alps: In situ origin or repeated colonization? Am. J. Bot. 2013, 100, 2412–2425. [Google Scholar] [CrossRef] [Green Version]
- Buira, A.; Fernández-Mazuecos, M.; Aedo, C.; Molina-Venegas, R. The contribution of the edaphic factor as a driver of recent plant diversification in a Mediterranean biodiversity hotspot. J. Ecol. 2021, 109, 987–999. [Google Scholar] [CrossRef]
- Devesa, J.A.; Martínez-Sagarra, G.; López Nieto, E.; Muñoz Rodríguez, A.; Cebolla, C.; Ortúñez, E. Festuca L. In Flora Iberica; Devesa, J.A., Zarco, C.R., Buira, A., Quintanar, A., Aedo, C., Eds.; Real Jardín Botánico CSIC: Madrid, Spain, 2020; Volume 19, pp. 200–373. [Google Scholar]
- Mota, J.F.; Pérez-García, F.J.; Jiménez, M.L.; Amate, J.J.; Peñas, J. Phytogeographical relationships among high mountain areas in the Baetic Ranges (South Spain). Glob. Ecol. Biogeogr. 2002, 11, 497–504. [Google Scholar] [CrossRef] [Green Version]
- Médail, F.; Quézel, P. Hot-Spots Analysis for conservation of Plant Biodiversity in the Mediterranean Basin. Ann. Missouri Bot. Gard. 1997, 84, 112–127. [Google Scholar] [CrossRef]
- Boissier, E. Elenchus Plantarum Novarum Minusque Cognitarum Quas in Itinere Hispanico; Typographia Lador et Ramboz: Genevae, Switzerland, 1838; pp. 5–94. [Google Scholar]
- Gómez-Pugnaire, M.T.; Galindo-Zaldívar, J.; Rubatto, D.; González-Lodeiro, F.; López, V.; Jabaloy, A. A reinterpretation of the Nevado-Filábride and Alpujárride complexes (Baetic Cordillera): Field, petrography and U-Pb ages from orthogneisses (western Sierra Nevada, S Spain). Schweiz. Miner. Petrogr. Mitt. 2004, 84, 303–322. [Google Scholar]
- Martínez-Sagarra, G.; Devesa, J.A. Festuca indigesta subsp. indigesta en la Sierra de Gádor (Almería, SE de España). Acta Bot. Malac. 2015, 40, 253–254. [Google Scholar] [CrossRef]
- Gavilán, R.G.; Vilches, B.; Font, X.; Jiménez-Alfaro, B. A review of high-mountain acidophilous vegetation in the Iberian Peninsula. Appl. Veg. Sci. 2017, 20, 513–526. [Google Scholar] [CrossRef]
- San Miguel, A. 6160 Pastos orófilos mediterráneos de Festuca indigesta. In Bases Ecológicas Preliminares para la Conservación de los Tipos de Hábitat de Interés Comunitario en España; AAVV, Ed.; Ministerio de Medio Ambiente, y Medio Rural y Marino: Madrid, Spain, 2009; pp. 7–57. [Google Scholar]
- Lozano, C.C.; Rivas Ponce, M.A. Festuca indigesta Boiss. subsp. lagascae Cebolla et Rivas Ponce subsp. nova et ses relations avec le complexe Festuca indigesta Boiss. Criteres utilises lors de son etude taxonomique. Flora Mediterr. 1999, 9, 131–146. [Google Scholar]
- Sliwinska, E.; Loureiro, J.; Leitch, I.J.; Šmarda, P.; Bainard, J.; Bureš, P.; Chumová, Z.; Horová, L.; Koutecký, P.; Lučanová, M.; et al. Application-based guidelines for best practices in plant flow cytometry. Cytom. A 2021, 1–33. [Google Scholar] [CrossRef]
- Qiu, Y.; Hamernick, S.; Ortiz, J.B.; Watkins, E. DNA content and ploidy estimation of Festuca ovina accessions by flow cytometry. Crop Sci. 2020, 60, 2757–2767. [Google Scholar] [CrossRef]
- Rewicz, A.; Tomczyk, P.P.; Kiedrzyński, M.; Zielińska, K.M.; Jędrzejczyk, I.; Rewers, M.; Kiedrzyńska, E.; Rewicz, T. Morphometric traits in the fine-leaved fescues depend on ploidy level: The case of Festuca amethystina L. PeerJ 2018, 6, e5576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šmarda, P.; Bures, P.; Horová, L.; Rotreklová, O. Intrapopulation genome size dynamics in Festuca pallens. Ann. Bot. 2008, 102, 599–607. [Google Scholar] [CrossRef] [Green Version]
- Fuente, V.; Ferrero, L.M.; Ortúñez, E. Chromosome counts in the genus Festuca L. section Festuca (Poaceae) in the Iberian Peninsula. Bot. J. Linn. Soc. 2001, 137, 385–398. [Google Scholar] [CrossRef]
- Loureiro, J.; Kopecký, D.; Castro, S.; Santos, C.; Silveira, P. Flow cytometric and cytogenetic analyses of Iberian Peninsula Festuca spp. Plant Syst. Evol. 2007, 269, 89–105. [Google Scholar] [CrossRef]
- Šmarda, P. DNA ploidy levels and intraspecific DNA content variability in Romanian fescues (Festuca, Poaceae) measured in fresh and herbarium material. Folia Geobot. 2006, 41, 417–432. [Google Scholar] [CrossRef]
- Šmarda, P.; Bureš, P.; Horová, L.; Foggi, B.; Rossi, G. Genome size and GC content evolution of Festuca: Ancestral expansion and subsequent reduction. Ann. Bot. 2008, 101, 421–433. [Google Scholar] [CrossRef] [Green Version]
- Küpfer, P. Nouvelles prospections caryologiques dans la flore orophile des Pyrénées et de la Sierra Nevada. Bull. Soc. Neuchâteloise Sci. Nat. 1968, 91, 87–104. [Google Scholar] [CrossRef]
- Kerguélen, M. Les Gramineae (Poaceae) de la flore française. Essai de mise au point taxonomique et nomenclaturale. Lejeunia Ser. 2 1975, 75, 1–343. [Google Scholar]
- Fuente, V.; Ortúñez, E.; Ferrero, L. A new species of Festuca L. (Poaceae) from the Iberian Peninsula. Parlatorea 1999, 3, 65–77. [Google Scholar]
- Kruckerberg, A.R. An essay: Geoedaphics and island biogeography for vascular plants. Aliso 1991, 13, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Mota, J.F.; Garrido-Becerra, J.A.; Merlo, M.E.; Medina-Cazorla, J.M.; Sánchez-Gómez, P. The edaphism: Gypsum, dolomite and serpentine flora and vegetation. In The Vegetation of the Iberian Peninsula; Loidi, J., Ed.; Springer: Cham, Switzerland, 2017; pp. 277–354. [Google Scholar] [CrossRef]
- Kolář, F.; Lučanová, M.; Záveská, E.; Fuxová, G.; Mandáková, T.; Španiel, S.; Senko, D.; Svitok, M.; Kolník, M.; Gudžinskas, Z.; et al. Ecological segregation does not drive the intricate parapatric distribution of diploid and tetraploid cytotypes of the Arabidopsis arenosa group (Brassicaceae). Biol. J. Linn. Soc. 2016, 119, 673–688. [Google Scholar] [CrossRef] [Green Version]
- Pauli, H.; Gottfried, M.; Dullinger, S.; Abdaladze, O.; Akhalkatsi, M.; Benito Alonso, J.L.; Coldea, G.; Dick, J.; Erschbamer, B.; Fernández Calzado, R.; et al. Recent plant diversity changes on Europe’s mountain summits. Science 2012, 336, 353–355. [Google Scholar] [CrossRef] [Green Version]
- Weiss-Schneeweiss, H.; Emadzade, K.; Jang, T.S.; Schneeweiss, G.M. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet. Genome Res. 2013, 140, 137–150. [Google Scholar] [CrossRef]
- Hodálová, I.; Grulich, V.; Horová, L.; Valachovič, M.; Marhold, K. Occurrence of tetraploid and octoploid cytotypes in Senecio jacobaea ssp. jacobaea (Asteraceae) in Pannonia and the Carpathians. Bot. J. Linn. Soc. 2007, 153, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Španiel, S.; Marhold, K.; Hodálová, I.; Lihová, J. Diploid and tetraploid cytotypes of Centaurea stoebe (Asteraceae) in Central Europe: Morphological differentiation and cytotype distribution patterns. Folia Geobot. 2008, 43, 131–158. [Google Scholar] [CrossRef]
- Inda, L.A.; Segarra-Moragues, J.G.; Müller, J.; Peterson, P.M.; Catalán, P. Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Mol. Phylogenet. Evol. 2008, 46, 932–957. [Google Scholar] [CrossRef]
- Van Santen, E.; Casler, M.D. Evaluation of indirect ploidy indicators in Dactylis L. subspecies. Crop Sci. 1986, 26, 848–852. [Google Scholar] [CrossRef]
- Joachimiak, A.; Grabowska-Joachimiak, A. Stomatal cell length and ploidy level in four taxa belonging to the Phleum sect. Phleum. Acta Biol. Cracov. Ser. Bot. 2000, 42, 103–107. [Google Scholar]
- Beaulieu, J.M.; Leitch, I.J.; Patel, S.; Pendharkar, A.; Knight, C.A. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 2008, 179, 975–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodgson, J.G.; Sharafi, M.; Jalili, A.; Díaz, S.; Montserrat-Martí, G.; Palmer, C.; Cerabolini, B.; Pierce, S.; Hamzehee, B.; Asri, Y.; et al. Stomatal vs. genome size in angiosperms: The somatic tail wagging the genomic dog? Ann. Bot. 2010, 105, 573–584. [Google Scholar] [CrossRef]
- Thiers, B. Index Herbariorum: A Global Directory of Public Herbaria and Associated Staff. New York Botanical Garden’s Virtual Herbarium. Available online: http://sweetgum.nybg.org/science/ih/ (accessed on 18 January 2022).
- Doležel, J.; Sgorbati, S.; Lucretii, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Loureiro, J.; Rodriguez, E.; Doležel, J.; Santos, C. Two new nuclear isolation buffers for plant DNA flow cytometry: A test with 37 species. Ann. Bot. 2007, 100, 875–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galbraith, D.W.; Harkins, K.R.; Maddox, J.M.; Ayres, N.M.; Sharma, D.P.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef]
- Greilhuber, J.; Temsch, E.; Loureiro, J. Nuclear DNA content measurement. In Flow Cytometry with Plant Cells; Doležel, J., Greilhuber, J., Suda, J., Eds.; Wiley-VCH: Weinheim, Germany, 2007; pp. 67–101. [Google Scholar]
- Snow, R. Alcoholic hydrochloric acid-carmine as stain for chromosomes in squash preparations. Stain Technol. 1963, 38, 9–13. [Google Scholar] [CrossRef]
- Pierce, F.J.; Fortin, M.-C.; Staton, M.J. Periodic Plowing Effects on Soil Properties in a No-Till Farming System. Soil Sci. Soc. Am. J. 1994, 58, 1782–1787. [Google Scholar] [CrossRef]
- Hueso-González, P.; Ruiz-Sinoga, J.D.; Martínez-Murillo, J.F.; Lavee, H. Overland flow generation mechanisms affected by topsoil treatment: Application to soil conservation. Geomorphology 2015, 228, 796–804. [Google Scholar] [CrossRef]
- FAO (Organización de las Naciones Unidas para la Agricultura y la Alimentación). Guía para la Descripción de Suelos; FAO: Roma, Italy, 2009; pp. 1–99. [Google Scholar]
- Mapa Geológico de España E 1:50.000. Segunda Serie (MAGNA), Primera Edición, IGME. Available online: info.igme.es/cartografiadigital/geologica/Magna50.aspx (accessed on 23 October 2021).

| Locality | n | 2C Mean ± SD (pg) | G.s. Range (Min–Max, pg) | Cv Sample (%) | Cv Standard (%) | Chro. No. | Ploidy Level |
|---|---|---|---|---|---|---|---|
| Granada: Sierra Nevada core, Peñones de San Francisco (p1) | 8 | 12.99 ± 0.34 a | 12.56–13.61 | 4.69 | 3.38 | 42 | 6x |
| Granada: Sierra Nevada core, Collado de Las Sabinas (p2) | 6 | 13.25 ± 0.86 a | 12.35–14.34 | 4.74 | 3.65 | 42 | 6x |
| Granada: Sierra Nevada, Puerto de la Ragua (p3) | 7 | 17.69 ± 0.40 b | 17.35–18.33 | 4.08 | 3.36 | 56 | 8x |
| Almería: Sierra de Gádor, Los Morrones (p4) | 5 | 17.06 ± 0.44 b | 16.59–17.54 | 4.55 | 3.65 | 56 | 8x |
| Málaga: Sierra de las Nieves, Los Ventisqueros (p5) | 5 | 25.33 ± 1.03 c | 23.73–26.37 | 3.83 | 4.00 | 84 | 12x |
| Mountain Range | Population | UTM Coordinates | Elevation (m.a.s.l.) | Lithology | pH | Ca | Mg | Na | CaCO3 Presence |
|---|---|---|---|---|---|---|---|---|---|
| Sierra Nevada | Peñones de San Francisco (p1) | 30S 465525 4105791 | 2543 | Micaschist | 6.30 | 3.70 | 0.70 | 2.90 | - |
| Sierra Nevada | Collado de Las Sabinas (p2) | 30S 462602 4107780 | 2195 | Marble | 8.30 | 7.90 | 1.30 | 2.90 | - |
| Sierra Nevada | Puerto de La Ragua (p3) | 30S 497515 4107549 | 2070 | Micaschist | 6.10 | 3.40 | 0.50 | 2.90 | - |
| Sierra de Gádor | Pozo Lupión | 30S 510391 4083210 | 1912 | Limestone and dolomite | 7.70 | 17.40 | 3.20 | 4.80 | + |
| Sierra de Gádor | Pozo Lupión | 30S 510336 4083172 | 1910 | Limestone and dolomite | 8.20 | 15.10 | 2.30 | 4.70 | + |
| Sierra de Gádor | Los Morrones (p4) | 30S 512984 4083216 | 1933 | Dolomite and limestone | 7.90 | 17.70 | 2.50 | 2.90 | + |
| Sierra de las Nieves | Los Ventisqueros (p5) | 30S 320761 4063517 | 1709 | Limestone | 7.90 | 27.00 | 0.50 | 4.90 | + |
| Sierra de las Nieves | Los Ventisqueros (p5) | 30S 321094 4064038 | 1744 | Limestone | 7.80 | 17.20 | 0.50 | 4.90 | + |
| Sierra de las Nieves | Los Ventisqueros (p5) | 30S 320843 4063724 | 1725 | Limestone | 7.40 | 16.30 | 0.70 | 4.80 | + |
| Sierra de las Nieves | Los Ventisqueros (p5) | 30S 322526 4064861 | 1595 | Limestone | 7.95 | 30.11 | 0.64 | 0.23 | + |
| Sierra de las Nieves | Los Ventisqueros (p5) | 30S 319202 4062159 | 1740 | Limestone | 8.01 | 21.83 | 4.82 | 0.11 | + |
| Sierra de las Nieves | Cerro del Viento | 30S 318261 4072666 | 1345 | Dolomite | 7.40 | 42.62 | 4.03 | 0.21 | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Sagarra, G.; Casimiro-Soriguer, F.; Castro, S.; Loureiro, J.; Devesa, J.A. Cytogenetic, Morphometric, and Ecological Characterization of Festuca indigesta Boiss. in the Southeast of Spain. Plants 2022, 11, 693. https://doi.org/10.3390/plants11050693
Martínez-Sagarra G, Casimiro-Soriguer F, Castro S, Loureiro J, Devesa JA. Cytogenetic, Morphometric, and Ecological Characterization of Festuca indigesta Boiss. in the Southeast of Spain. Plants. 2022; 11(5):693. https://doi.org/10.3390/plants11050693
Chicago/Turabian StyleMartínez-Sagarra, Gloria, Federico Casimiro-Soriguer, Sílvia Castro, João Loureiro, and Juan A. Devesa. 2022. "Cytogenetic, Morphometric, and Ecological Characterization of Festuca indigesta Boiss. in the Southeast of Spain" Plants 11, no. 5: 693. https://doi.org/10.3390/plants11050693
APA StyleMartínez-Sagarra, G., Casimiro-Soriguer, F., Castro, S., Loureiro, J., & Devesa, J. A. (2022). Cytogenetic, Morphometric, and Ecological Characterization of Festuca indigesta Boiss. in the Southeast of Spain. Plants, 11(5), 693. https://doi.org/10.3390/plants11050693








