Phylogenetic and Expression Studies of Small GTP-Binding Proteins in Solanum lycopersicum Super Strain B
Abstract
1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Identification of Small GTPases from Different Species
3.2. Phylogenetic Analysis
3.3. Protein Alignments
3.4. D Modeling
3.5. Genomic Datasets
3.6. Transcriptomic Datasets
3.7. Statistical Analysis of Expression Data
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Daej, M.I. Salt Tolerance of Some Tomato (Solanum lycoversicum L.) Cultivars for Salinity under Controlled Conditions. Am. J. Plant Physiol. 2018, 13, 58–64. [Google Scholar] [CrossRef]
- Splittstoesser, W.E. Vegetable Growing Handbook, Organic and Traditional Methods, 3rd ed.; Van Nostrand Reinhold: New York, NY, USA, 1990. [Google Scholar]
- Fouda, K.F. Effect of Interaction among N Forms and Calcium Sources on Quality and Chemical Composition of Tomato (Lycopersicon esculentum). Egypt. J. Soil Sci. 2017, 57, 61–71. [Google Scholar] [CrossRef][Green Version]
- Kloepper, J.W.; Beauchamp, C.J. A review of issues related to measuring colonization of plant roots by bacteria. Can. J. Microbiol. 1992, 38, 1219–1232. [Google Scholar] [CrossRef]
- Elmerich, C.; Newton, W.E. Associative and Endophytic Nitrogen-Fixing Bacteria and Cyanobacterial Associations; Springer: Dordecht, The Netherlands, 2007. [Google Scholar]
- Dalla Via, V.D.; Traubenik, S.; Rivero, C.; Aguilar, O.M.; Zanetti, M.E.; Blanco, F.A. The monomeric GTPase RabA2 is required for progression and maintenance of membrane integrity of infection threads during root nodule symbiosis. Plant Mol. Biol. 2017, 93, 549–562. [Google Scholar] [CrossRef]
- Ke, D.; Fang, Q.; Chen, C.; Zhu, H.; Chen, T.; Chang, X.; Yuan, S.; Kang, H.; Ma, L.; Hong, Z.; et al. The Small GTPase ROP6 Interacts with NFR5 and Is Involved in Nodule Formation in Lotus japonicus. Plant Physiol. 2012, 159, 131–143. [Google Scholar] [CrossRef]
- Kiirika, L.M.; Bergmann, H.F.; Schikowsky, C.; Wimmer, D.; Korte, J.; Schmitz, U.; Niehaus, K.; Colditz, F. Silencing of the Rac1 GTPase MtROP9 in Medicago truncatula Stimulates Early Mycorrhizal and Oomycete Root Colonizations But Negatively Affects Rhizobial Infection. Plant Physiol. 2012, 159, 501–516. [Google Scholar] [CrossRef]
- Hossain, S.; Liao, J.; James, E.; Sato, S.; Tabata, S.; Jurkiewicz, A.; Madsen, L.H.; Stougaard, J.; Ross, L.; Szczyglowski, K. Lotus japonicus ARPC1 Is Required for Rhizobial Infection. Plant Physiol. 2012, 160, 917–928. [Google Scholar] [CrossRef]
- Fournier, J.; Teillet, A.; Chabaud, M.; Ivanov, S.; Genre, A.; Limpens, E.; de Carvalho-Niebel, F.; Barker, D.G. Remodeling of the Infection Chamber before Infection Thread Formation Reveals a Two-Step Mechanism for Rhizobial Entry into the Host Legume Root Hair. Plant Physiol. 2015, 167, 1233–1242. [Google Scholar] [CrossRef]
- Oldroyd, G.E.; Downie, J.A. Coordinating Nodule Morphogenesis with Rhizobial Infection in Legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Limpens, E.; Ivanov, S.; van Esse, W.; Voets, G.; Fedorova, E.; Bisseling, T. MedicagoN2-Fixing Symbiosomes Acquire the Endocytic Identity Marker Rab7 but Delay the Acquisition of Vacuolar Identity. Plant Cell 2009, 21, 2811–2828. [Google Scholar] [CrossRef]
- Memon, A.R.; Schwager, C.K.; Niehaus, K. Expression of small GTPases in the roots and nodules of Medicago truncatula cv. Jemalong. Acta Bot. Croat. 2019, 78, 1–8. [Google Scholar] [CrossRef]
- Kahn, R.A.; Der, C.J.; Bokoch, G.M. The ras superfamily of GTP-binding proteins: Guidelines on nomenclature. FASEB J. 1992, 6, 2512–2513. [Google Scholar] [CrossRef]
- Vernoud, V.; Horton, A.; Yang, Z.; Nielsen, E. Analysis of the Small GTPase Gene Superfamily of Arabidopsis. Plant Physiol. 2003, 131, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Lundquist, E.A. Small GTPases, WormBook, edn. The C. elegans Research Community, WormBook. 2006. Available online: http://www.wormbook.org/chapters/www_smallGTPases/smallGTPases.pdf (accessed on 9 December 2021).
- Pimpl, P.; Movafeghi, A.; Coughlan, S.; Denecke, J.; Hillmer, S.; Robinson, D.G. In Situ Localization and in Vitro Induction of Plant COPI-Coated Vesicles. Plant Cell 2000, 12, 2219–2235. [Google Scholar] [CrossRef]
- Jürgens, G. Membrane Traficking in Plants. Annu. Rev. Cell Dev. Biol. 2004, 20, 481–504. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.R. The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants. Biochim. Biophys. Acta 2004, 1664, 9–30. [Google Scholar] [CrossRef] [PubMed]
- Memon, A.R. Transcriptomics and proteomics analysis of root nodules of model legume plants. In Crop Production for Agricultural Improvement; Ashraf, M., Ozturl, M., Ahmed, S.M.A., Aksoy, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 291–315. [Google Scholar]
- Etienne-Manneville, S.; Hall, A. Rho GTPases in cell biology. Nature 2002, 420, 629–635. [Google Scholar] [CrossRef]
- Craddock, C.; Lavagi, I.; Yang, Z. New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 2012, 22, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Cavazza, T.; Vernos, I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front. Cell Dev. Biol. 2016, 3. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Ramachandran, S. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genom. 2006, 24, 235–251. [Google Scholar] [CrossRef][Green Version]
- Yuksel, B.; Memon, A.R. Legume small GTPases and their role in the establishment of symbiotic associations with Rhizobium spp. Plant Signal. Behav. 2009, 4, 257–260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rutherford, S.; Moore, I. The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 2002, 5, 518–528. [Google Scholar] [CrossRef]
- Flores, A.C.; Via, V.D.; Savy, V.; Villagra, U.M.; Zanetti, M.E.; Blanco, F. Comparative phylogenetic and expression analysis of small GTPases families in legume and non-legume plants. Plant Signal. Behav. 2018, 13, e1432956. [Google Scholar] [CrossRef]
- Singh, R.J.; Hymowitz, T. The genomic relationship between Glycine max (L.) Merr. and G. soja Sieb. and Zucc. as revealed by pachytene chromosome analysis. Theor. Appl. Genet. 1988, 76, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Cheon, C.I.; Lee, N.G.; Siddique, A.B.M.; Bal, A.K.; Verma, D.P.S. Roles of plant homologs of Rab1p and Rab7p in the biogenesis of the peribacteroid membrane, a subcellular compartment formed de novo during root-nodule symbiosis. EMBO J. 1993, 12, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Borg, S.; Brandstrup, B.; Jensen, T.J.; Poulsen, C. Identification of new protein species among 33 different small GTP-binding proteins encoded by cDNAs from Lotus japonicus, and expression of corresponding mRNAs in developing root nodules. Plant J. 1997, 11, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Son, O.; Yang, H.S.; Lee, H.J.; Lee, M.Y.; Cheon, C.I. Expression of SRAB7 and SCaM genes required for endocytosis of Rhizobium in root nodules. Plant Sci. 2003, 165, 1239–1244. [Google Scholar] [CrossRef]
- Schiene, K.; Donath, S.; Brecht, M.; Pühler, A.; Niehaus, K. A Rab-related small GTP binding protein is predominantly expressed in root nodules of Medicago sativa. Mol. Genet. Genom. 2004, 272, 57–66. [Google Scholar] [CrossRef]
- MOhandas, S. Nitrogen fixation in tomato (Lycopersicon esculentum Mill ‘Pusa Ruby’). Plant Soil 1988, 107, 219–225. [Google Scholar] [CrossRef]
- Dent, D.; Cocking, E.C. Establishing symbiotic nitrogen fixation in cereals and other non-legume crops: The Greener Nitrogen Revolution. Agric. Food Secur. 2017, 6, 7. [Google Scholar] [CrossRef]
- Collavino, M.M.; Cabrera, E.V.R.; Bruno, C.; Aguilar, O.M. Effect of soil chemical fertilization on the diversity and composition of the tomato endophytic diazotrophic community at different stages of growth. Braz. J. Microbiol. 2020, 51, 1965–1975. [Google Scholar] [CrossRef] [PubMed]
- Yuksel, B.; Memon, A.R. Comparative phylogenetic analysis of small GTP-binding genes of model legume plants and assessment of their roles in root nodules. J. Exp. Bot. 2008, 59, 3831–3844. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics Genet. Res. 2001, 77, 117–120. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Secco, D.; Jabnoune, M.; Walker, H.; Shou, H.; Wu, P.; Poirier, Y.; Whelan, J. Spatio-Temporal Transcript Profiling of Rice Roots and Shoots in Response to Phosphate Starvation and Recovery. Plant Cell 2013, 25, 4285–4304. [Google Scholar] [CrossRef]
- Sekhon, R.S.; Briskine, R.; Hirsch, C.N.; Myers, C.L.; Springer, N.M.; Buell, C.R.; De Leon, N.; Kaeppler, S. Maize Gene Atlas Developed by RNA Sequencing and Comparative Evaluation of Transcriptomes Based on RNA Sequencing and Microarrays. PLoS ONE 2013, 8, e61005. [Google Scholar] [CrossRef] [PubMed]
- Benedito, V.A.; Torres-Jerez, I.; Murray, J.; Andriankaja, A.; Allen, S.; Kakar, K.; Wandrey, M.; Verdier, J.; Zuber, H.; Ott, T.; et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008, 55, 504–513. [Google Scholar] [CrossRef] [PubMed]
- Høgslund, N.; Radutoiu, S.; Krusell, L.; Voroshilova, V.; Hannah, M.A.; Goffard, N.; Sanchez, D.H.; Lippold, F.; Ott, T.; Sato, S.; et al. Dissection of Symbiosis and Organ Development by Integrated Transcriptome Analysis of Lotus japonicus Mutant and Wild-Type Plants. PLoS ONE 2009, 4, e6556. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Iniguez, L.P.; Fu, F.; Bucciarelli, B.; Miller, S.S.; A Jackson, S.; E McClean, P.; Li, J.; Dai, X.; Zhao, P.X.; et al. An RNA-Seq based gene expression atlas of the common bean. BMC Genom. 2014, 15, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Severin, A.J.; Woody, J.L.; Bolon, Y.-T.; Joseph, B.; Diers, B.W.; Farmer, A.D.; Muehlbauer, G.J.; Nelson, R.T.; Grant, D.; Specht, J.E.; et al. RNA-Seq Atlas of Glycine max: A guide to the soybean transcriptome. BMC Plant Biol. 2010, 10, 160. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
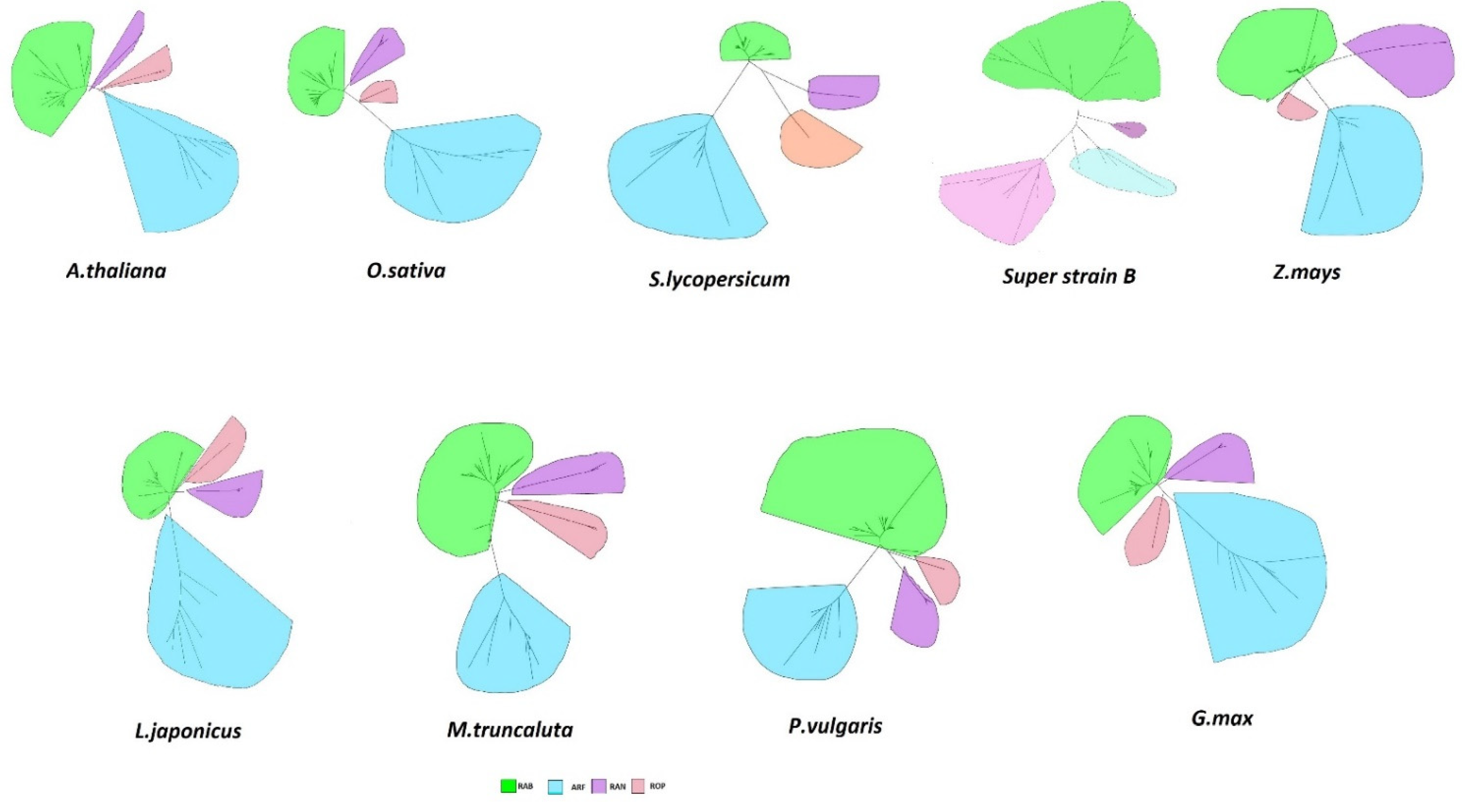


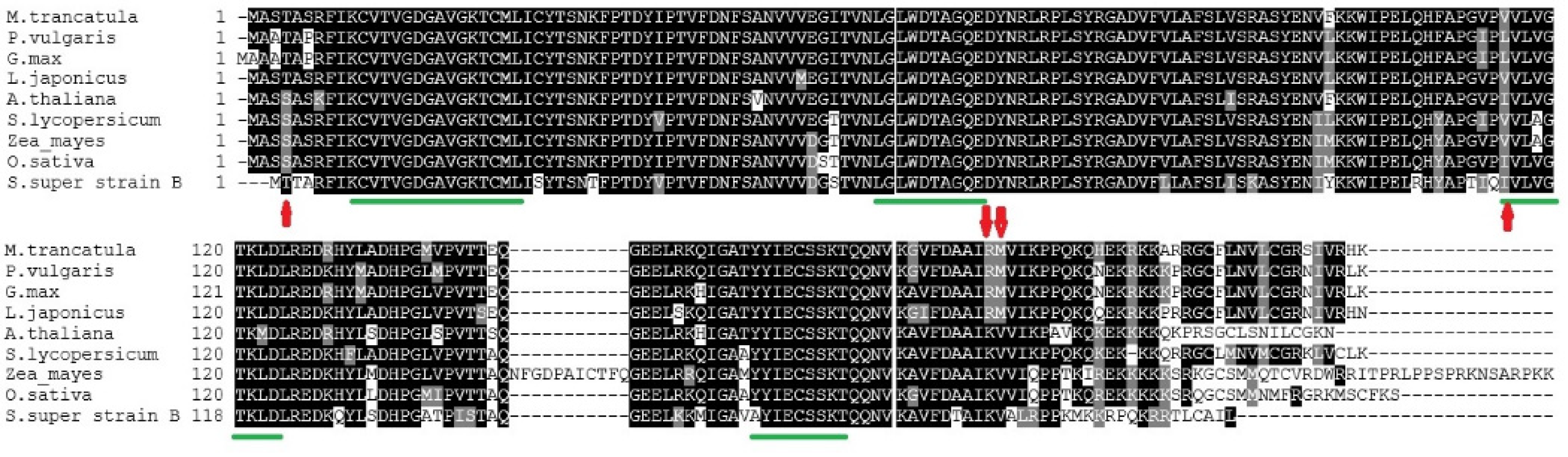

| GTPases | S. lycopersicum Super Strain “B” | Non-Legumes | Legumes | |||||
|---|---|---|---|---|---|---|---|---|
| A. thaliana | O. sativa | Z. mays | L. japonicus | M. truncatula | P. vulgaris | G. max | ||
| RAB | 46 | 57 | 37 | 53 | 30 | 64 | 50 | 94 |
| ARF | 21 | 21 | 21 | 25 | 13 | 19 | 20 | 41 |
| ROP | 9 | 11 | 8 | 9 | 8 | 7 | 11 | 20 |
| RAN | 4 | 4 | 2 | 3 | 2 | 4 | 3 | 7 |
| Total | 80 | 93 | 68 | 90 | 53 | 94 | 84 | 162 |
| Group | S. lycopersicum Super Strain “B” | Non-Legumes | Legumes | |||||
|---|---|---|---|---|---|---|---|---|
| A. thaliana | O. sativa | Z. mays | L. japonicus | M. truncatula | P. vulgaris | G. max | ||
| A | 21 | 26 | 17 | 23 | 12 | 23 | 23 | 41 |
| B | 2 | 3 | 3 | 4 | 2 | 7 | 1 | 4 |
| C | 3 | 3 | 0 | 3 | 4 | 6 | 5 | 11 |
| D | 5 | 4 | 4 | 6 | 1 | 4 | 4 | 7 |
| E | 5 | 5 | 3 | 5 | 3 | 6 | 5 | 8 |
| F | 4 | 3 | 4 | 3 | 3 | 5 | 4 | 7 |
| G | 3 | 8 | 4 | 5 | 3 | 9 | 4 | 8 |
| H | 3 | 5 | 2 | 4 | 2 | 4 | 4 | 8 |
| Total | 46 | 57 | 37 | 53 | 30 | 64 | 50 | 94 |
| Group | S. lycopersicum Super Strain “B” | Non-Legumes | Legumes | |||||
|---|---|---|---|---|---|---|---|---|
| A. thaliana | O. sativa | Z. mays | L. japonicus | M. truncatula | P. vulgaris | G. max | ||
| A | 5 | 6 | 6 | 6 | 4 | 5 | 4 | 10 |
| B + C + D | 8 | 6 | 6 | 9 | 4 | 7 | 6 | 12 |
| ARLA | 3 | 4 | 2 | 4 | 1 | 3 | 4 | 5 |
| ARLB | 1 | 1 | 2 | 2 | 1 | 0 | 1 | 2 |
| ARLC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 |
| SARA | 3 | 3 | 4 | 3 | 2 | 3 | 4 | 10 |
| Total | 21 | 21 | 21 | 25 | 13 | 19 | 20 | 41 |
| Plant Species | Rab | Arf | Rop | Ran | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − | N | + | − | N | + | − | N | + | − | N | + | |
| S. lycopersicum super strain “B” a | 8 | 24 | 14 | 8 | 8 | 5 | 1 | 3 | 5 | 0 | 4 | 0 |
| A. thalianaa | 16 | 36 | 5 | 6 | 13 | 2 | 4 | 7 | 0 | 0 | 4 | 0 |
| O. sativab | 14 | 20 | 2 | 4 | 16 | 1 | 2 | 4 | 2 | 0 | 1 | 1 |
| Z. maysc | 17 | 34 | 0 | 5 | 18 | 1 | 0 | 8 | 1 | 0 | 3 | 0 |
| L. japonicusd | 1 | 29 | 0 | 0 | 13 | 0 | 2 | 6 | 0 | 0 | 1 | 1 |
| M. trancatulaa | 17 | 26 | 2 | 5 | 13 | 0 | 2 | 5 | 0 | 2 | 2 | 0 |
| P. vulgarisa | 14 | 28 | 3 | 5 | 13 | 0 | 1 | 6 | 4 | 1 | 2 | 0 |
| G. maxa | 50 | 42 | 2 | 19 | 20 | 2 | 5 | 9 | 6 | 4 | 3 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Zahrani, H.S.; Moussa, T.A.A.; Alsamadany, H.; Hafez, R.M.; Fuller, M.P. Phylogenetic and Expression Studies of Small GTP-Binding Proteins in Solanum lycopersicum Super Strain B. Plants 2022, 11, 641. https://doi.org/10.3390/plants11050641
Al-Zahrani HS, Moussa TAA, Alsamadany H, Hafez RM, Fuller MP. Phylogenetic and Expression Studies of Small GTP-Binding Proteins in Solanum lycopersicum Super Strain B. Plants. 2022; 11(5):641. https://doi.org/10.3390/plants11050641
Chicago/Turabian StyleAl-Zahrani, Hassan S., Tarek A. A. Moussa, Hameed Alsamadany, Rehab M. Hafez, and Michael P. Fuller. 2022. "Phylogenetic and Expression Studies of Small GTP-Binding Proteins in Solanum lycopersicum Super Strain B" Plants 11, no. 5: 641. https://doi.org/10.3390/plants11050641
APA StyleAl-Zahrani, H. S., Moussa, T. A. A., Alsamadany, H., Hafez, R. M., & Fuller, M. P. (2022). Phylogenetic and Expression Studies of Small GTP-Binding Proteins in Solanum lycopersicum Super Strain B. Plants, 11(5), 641. https://doi.org/10.3390/plants11050641








