Intraspecific Variability of Wild-Growing Common Valerian (Valeriana officinalis L.)
Abstract
1. Introduction
2. Results and Discussion
2.1. Developmental and Chemical Variabilities
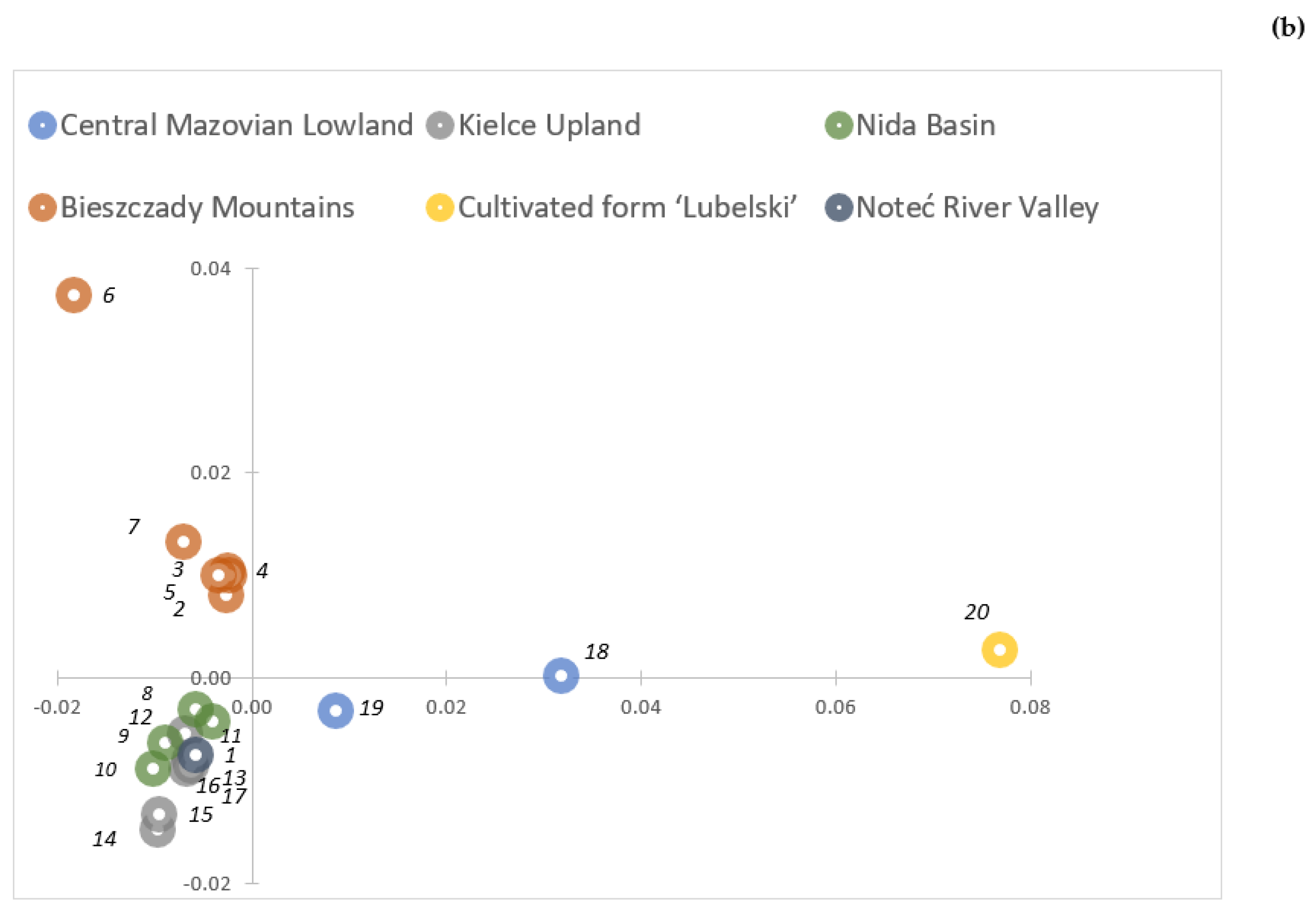
2.2. Genetic Variability
3. Materials and Methods
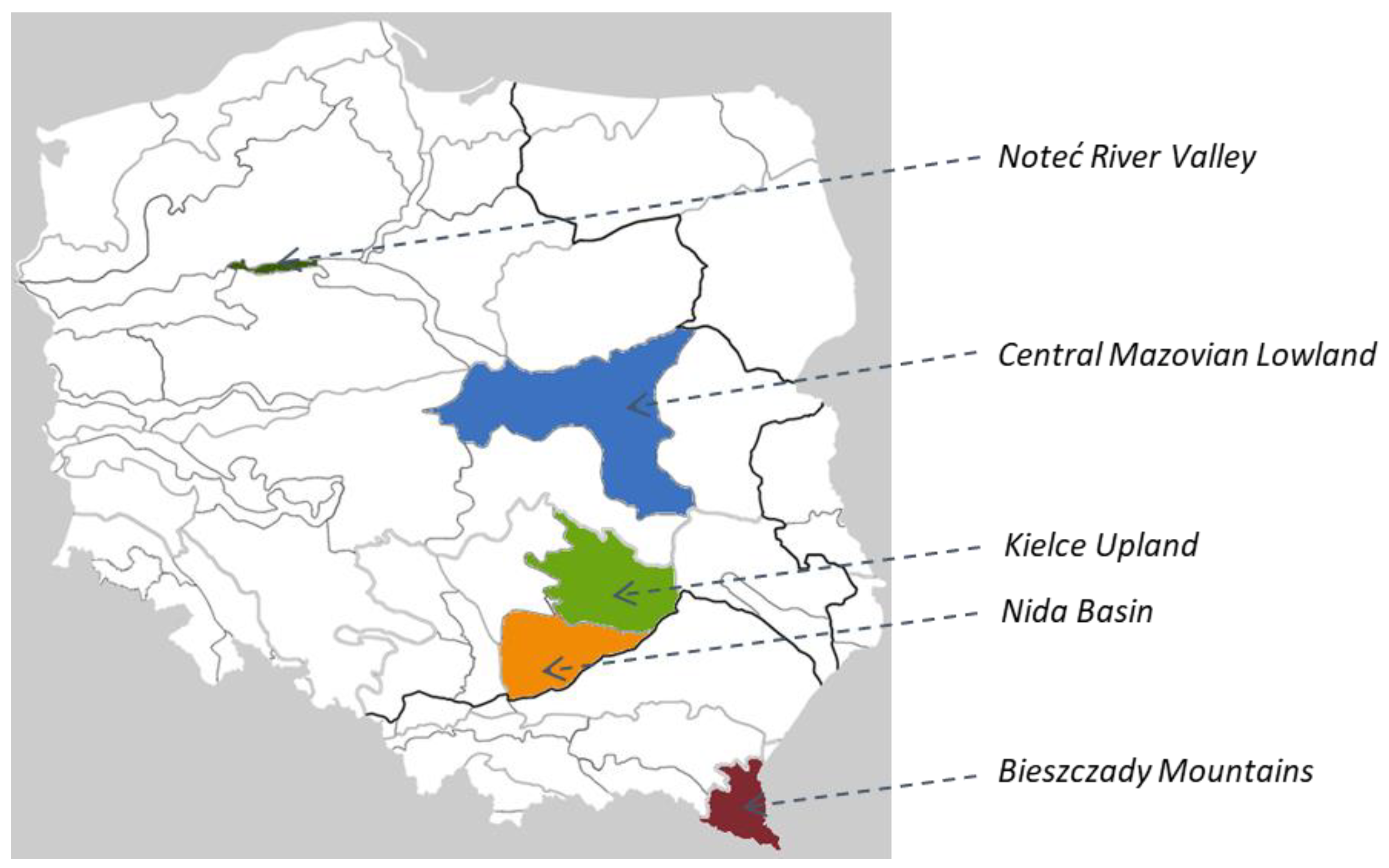
3.1. Plant Material
3.2. Developmental Characteristics
3.3. Chemical Analysis
3.4. Seed Evaluation
3.4.1. Seed Mass Test
3.4.2. Germinability and Germination Rate Tests
3.5. Molecular Analysis
3.5.1. Genotyping
3.5.2. Data Analysis
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kostrakiewicz-Gierałt, K. The variability of population and individual traits of medicinal plant Valeriana officinalis L. var. officinalis Mikan under different site conditions. Period. Biol. 2018, 120, 41–50. [Google Scholar]
- Houghton, P.J. Valerian: The Genus Valeriana; Harwood Academic Publishers: Amsterdam, The Nederlands, 1997; pp. 21–54. [Google Scholar]
- Bressler, S.; Klatte-Asselmeyer, V.; Fischer, A.; Paule, J.; Dobeš, C. Variation in genome size in the Valeriana officinalis complex resulting from multiple chromosomal evolutionary processes. Preslia 2017, 89, 41–61. [Google Scholar] [CrossRef]
- Kirschner, J.; Zeisek, V. Diploids of the Valeriana officinalis group (Valerianaceae) in Central Europe, and an attempt to unravel the nomenclatural chaos. Willdenowia 2017, 47, 189–201. [Google Scholar] [CrossRef]
- EMA (European Medicines Agency). European Union Herbal Monograph on Valerian officinalis. Available online: www.ema.europa.eu/en/documents/herbal-report/final-assessment-report-valeriana-officinalis-l-radix-valeriana-officinalis-l-aetheroleum_en.pdf (accessed on 7 November 2022).
- European Pharmacopoeia. European Directorate for the Quality of Medicines and Health Care (EDQM), 9th ed.; Council of Europe: Strasbourg, France, 2017. [Google Scholar]
- Raal, A.; Arak, E.; Orav, A.; Kailas, T.; Müürisepp, M. Variation in the composition of the essential oil of commercial Valeriana officinalis L. roots from different countries. J. Essent. Oil Res. 2008, 20, 524–529. [Google Scholar] [CrossRef]
- Patočka, J.; Jakl, J. Biomedically relevant chemical constituents of Valeriana officinalis. J. Appl. Biomed. 2010, 8, 11–18. [Google Scholar] [CrossRef]
- Sermukhamedova, O.; Ludwiczuk, A.; Widelski, J.; Głowniak, K.; Sakipova, Z.; Ibragimova, L.; Poleszak, E.; Cordell, G.A.; Skalicka-Woźniak, K. Chemical comparison of the underground parts of Valeriana officinalis and Valeriana Turkestanica from Poland and Kazakhstan. Open Chem. 2017, 15, 75–81. [Google Scholar] [CrossRef][Green Version]
- Wichtl, M. Herbal Drugs and Phytopharmaceuticals: A Handbook of Practice on a Scientific Basis, 3rd ed.; CRC Press: Stuttgart, Germany, 2004. [Google Scholar]
- Murphy, K.; Kubin, Z.J.; Shepherd, J.N.; Ettinger, R.H. Valeriana officinalis root extracts have potent anxiolytic effects in laboratory rats. Phytomedicine 2010, 17, 674–678. [Google Scholar] [CrossRef]
- Felgentreff, F.; Becker, A.; Meier, B.; Brattström, A. Valerian extract characterized by high valerenic acid and low acetoxy valerenic acid contents demonstrates anxiolytic activity. Phytomedicine 2012, 19, 1216–1222. [Google Scholar] [CrossRef]
- Bogacz, A.; Mrozikiewicz, P.M.; Karasiewicz, M.; Bartkowiak-Wieczorek, J.; Majchrzycki, M.; Mikolajczak, P.L.; Ozarowski, M.; Grzeskowiak, E. The influence of standardized Valeriana officinalis extract on the CYP3A1 gene expression by nuclear receptors in in vivo model. BioMed Res. Int. 2014, 2014, 819093. [Google Scholar] [CrossRef]
- Chen, H.-W.; Wei, B.-J.; He, X.-H.; Liu, Y.; Wang, J. Chemical components and cardiovascular activities of Valeriana spp. Evid. Based Complement. Altern. Med. 2015, 2015, 947619. [Google Scholar] [CrossRef]
- Smith, T.; Gillespie, M.; Eckl, V.; Knepper, J.; Reynolds, C.M. Herbal supplement sales in US increase by 9.4% in 2018. J. Am. Bot. Counc. 2019, 119, 62–73. [Google Scholar]
- Nakurte, C.I.; Mežaka, I.; Taškova, I.; Primavera, A.; Vecvanags, A.; Kronberga, A. Seasonal Changes in Chemical Composition of Valeriana officinalis L. Roots in Natural Conditions and Organic Production System in Latvia. J. Plant Biol. Crop Res. 2021, 4, 1031. [Google Scholar]
- Szczepanik, M.; Wiśniewski, J.B. Porównanie plonowania oraz cech jakościowych surowca trzech odmian kozłka lekarskiego (Valeriana officinalis L.) uprawianego z rozsady. Zesz. Prob. Post. Nauk Rol. 2009, 542, 511–516. [Google Scholar]
- Nurzyńska-Wierdak, R. Biological active compounds from roots of Valeriana officinalis L. Cultivated in south-eastern region of Poland. Farmacia 2014, 62, 683–692. [Google Scholar]
- Hoppe, B. (Ed.) Handbuch des Arznei- und Gewürzpflanzenbaus. Band 1. Verein für Arznei- und Gewürzpflanzen; SALUPLANTA: Bernburg, Germany, 2009. [Google Scholar]
- Penzkofer, M. Breeding Support for Valeriana officinalis L. s.l.: Root Structure, Localization of Value-Determining Secondary Compounds and Mating Behavior at Open Pollination. Ph.D. Thesis, Leibniz Universität Hannover, Hannover, Germany, 7 June 2019. [Google Scholar]
- Evstatieva, L.N.; Handjieva, N.V.; Popov, S.S.; Pashankov, P.I. A biosystematic study of Valeriana officinalis (Valerianaceae) distributed in Bulgaria. Plant Syst. Evol. 1993, 185, 167–179. [Google Scholar] [CrossRef]
- Hidalgo, O.; Mathez, J.; Garcia, S.; Garnatje, T.; Pellicer, J.; Vallès, J. Genome size study in the Valerianaceae: First results and new hypotheses. J. Bot. 2010, 2010, 797246. [Google Scholar] [CrossRef]
- Hidalgo, O.; Vallès, J. First record of a natural hexaploid population for Valeriana officinalis: Genome size is confirmed to be a suitable indicator of ploidy level in the species. Caryologia 2012, 65, 243–245. [Google Scholar] [CrossRef]
- Azizi, A.; Ardalani, H.; Honermeier, B. Statistical analysis of the associations between phenolic monoterpenes and molecular markers, AFLPs and SAMPLs in the spice plant oregano. Herba Pol. 2016, 62, 42–56. [Google Scholar] [CrossRef]
- Ince, A.G.; Karaca, M.; Elmasulu, S.Y. New microsatellite and caps-microsatellite markers for clarifying taxonomic and phylogenetic relationships within Origanum L. Mol. Breed. 2014, 34, 643–654. [Google Scholar] [CrossRef]
- Jedrzejczyk, I. Study on genetic diversity between Origanum L. species based on genome size and ISSR markers. Ind. Crops Prod. 2018, 126, 201–207. [Google Scholar] [CrossRef]
- Al-Beyroutiová, M.; Sabo, M.; Sleziak, P.; Dušinský, R.; Birčák, E.; Hauptvogel, P.; Kilian, A.; Švec, M. Evolutionary relationships in the genus Secale revealed by dartseq DNA polymorphism. Plant Syst. Evol. 2016, 302, 1083–1091. [Google Scholar] [CrossRef]
- Boczkowska, M.; Bączek, K.; Kosakowska, O.; Rucińska, A.; Podyma, W.; Węglarz, Z. Genome-wide diversity analysis of Valeriana officinalis L. using DArT-seq derived SNP markers. Agronomy 2020, 10, 1346. [Google Scholar] [CrossRef]
- Seidler-Lozykowska, K.; Mielcarek, S.; Baraniak, M. Content of essential oil and valerenic acids in valerian (Valeriana offcinalis L.) roots at the selected developmental phases. J. Essent. Oil Res. 2009, 21, 413–416. [Google Scholar] [CrossRef]
- Kwiatkowski, C. Evaluation of yield quality and weed infestation of common Valerian (Valeriana officinalis L.) in dependence on weed control method and forecrop. Acta Agrobot. 2012, 63, 179–188. [Google Scholar] [CrossRef]
- Morteza, E.; Akbari, G.A.; Sanavi, S.A.M.M.; Hossein, A.F. Determination of the vegetative and reproductive characteristics of valerian (Valeriana officinalis L.) under sowing dates and planting densities at Iran. J. Med. Plants Res. 2010, 4, 857–861. [Google Scholar]
- Rostański, K. Valeriana officinalis L. Kozłek lekarski (Common valerian). In Flora Polski. Rośliny Naczyniowe Polski i Terenów Ościennych (Flora of Poland. Vascular Plants of Poland and Adjacent Territories); Pawłowski, B., Ed.; Dwuliścienne. Zrosłopłatkowe (Dicotyledones. Sympetalous), PWN: Warsaw/Krakow, Poland, 1967; Volume IX, pp. 348–349. (In Polish) [Google Scholar]
- Bomme, U. Kulturanleitung für Baldrian, 4th ed.; Bayerische Landesanstalt für Landwirtschaft: Freising, Germany, 2001. [Google Scholar]
- Heuberger, H.; Lohwasser, U.; Schmatz, R.; Tegtmeier, M. Baldrian (Valeriana officinalis L.). In Handbuch des Arznei- und Gewürzpflanzenanbaus Band 4 Arznei- und Gewürzpflanzen A-K. Verein für Arznei- und Gewürzpflanzen; Hoppe, B., Ed.; SALUPLANTA: Bernburg, Germany, 2012; pp. 164–183. [Google Scholar]
- Czarnecka, B. Strategie adaptacyjne roślin a skład gatunkowy fitocenoz. Wiadomości Botaniczne 1997, 41, 33–42. [Google Scholar]
- Matuszkiewicz, W. Przewodnik do Oznaczania Zbiorowisk Roślinnych Polski; Wydawnictwo Naukowe PWN: Warsaw, Poland, 2008. [Google Scholar]
- Wysocki, C.; Sikorski, P. Fitosocjologia Stosowana w Ochronie i Kształtowaniu Krajobrazu; Wydawnictwo SGGW: Warsaw, Poland, 2009; p. 498. [Google Scholar]
- Skalińska, M. Polyploidy in Valeriana officinalis Linn. In relation to its ecology and distribution. Bot. J. Linn. Soc. 1947, 53, 159–186. [Google Scholar] [CrossRef]
- Skalińska, M. Studies in cytoecology, geographic distribution and evolution of Valeriana L. Bull. Pol. Acad. Sci. 1951, 1, 149–175. [Google Scholar]
- Titz, W. Beitrag zur Kenntnis der österreichischen sippen desvaleriana officinalis-aggregats und ihrer chromosomenzahlen. Osterr. Bot. Z. 1969, 116, 172–180. [Google Scholar] [CrossRef]
- Kokate, C.K.; Purohit, A.P.; Gokhale, S.B. Pharmacognosy, 2nd ed.; Nirali Prakashan: Bangalore, India, 2008; pp. 5.2–5.3. [Google Scholar]
- Pierre, P.M.O.; Sousa, S.M.; Davide, L.C.; Machado, M.A.; Viccini, L.F. Karyotype analysis, DNA content and molecular screening in Lippia Alba (Verbenaceae). An. Acad. Bras. Ciências 2011, 83, 993–1006. [Google Scholar] [CrossRef]
- Sanderson, S.C. The ploidy races of Atriplex confertifolia (Chenopodiaceae). West. N. Am. Nat. 2011, 71, 67–77. [Google Scholar] [CrossRef]
- Noller, P. Untersuchung der Variabilität von Valerensäuren, Valerenal und Valeranon in Wildpopulationen und Zuchtmaterial von Valeriana officinalis L.s.l. Inaugural Dissertation, Justus-Liebig-Universität Gießen, Gießen, Germany, 1989. [Google Scholar]
- Kohlmunzer, S. Kozłek lekarski—Surowiec i lek uspokajający. Wiadomości Zielar. 1996, 11, 1–2. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Titz, W.; Titz, E. Analyse der Formenmannigfaltigkeit der Valeriana officinalis-Gruppe im zentralen und südlichen Europa. Ber. Dtsch. Bot. Gesellschaf 1982, 95, 155–164. [Google Scholar]
- Titz, W.; Titz, E. Valeriana versifolia und andere oktoploide Arznei-Baldriane in den Schweizer Alpen und angrenzenden Regionen. Bull. De La Soc. Bot. Suisse 1980, 89, 251–277. [Google Scholar]
- ISTA. International Rules for Seed Testing; International Seed Testing Association: Bassersdorf, Switzerland, 2019. [Google Scholar]
- Cruz, V.M.; Kilian, A.; Dierig, D.A. Development of Dart Marker platforms and genetic diversity assessment of the U.S. collection of the new oilseed crop lesquerella and related species. PLoS ONE 2013, 8, e64062. [Google Scholar] [CrossRef]
- Diversity Arrays Technology. Available online: https://www.diversityarrays.com/ (accessed on 2 December 2022).
- Kilian, A.; Wenzl, P.; Huttner, E.; Carling, J.; Xia, L.; Blois, H.; Caig, V.; Heller-Uszynska, K.; Jaccoud, D.; Hopper, C.; et al. Diversity arrays technology: A generic genome profiling technology on open platforms. Methods Mol. Biol. 2012, 888, 67–89. [Google Scholar]
- Melville, J.; Haines, M.L.; Boysen, K.; Hodkinson, L.; Kilian, A.; Smith Date, K.L.; Parris, K.M. Identifying hybridization and admixture using SNPs: Application of the DArTseq platform in phylogeographic research on vertebrates. R. Soc. Open Sci. 2017, 7, 161061. [Google Scholar] [CrossRef]
- Jaccard, P. Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull. Soc. Vaudoise Sci. Nat. 1901, 37, 547–579. [Google Scholar]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in excel. population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]


| Developmental Traits | The Content of the Biologically Active Compounds | |||
|---|---|---|---|---|
| Rhizome Diameter (cm) | Mass of Underground Organs (Roots + Rhizomes) (g FW × plant−1) | Sesquiterpenic Acids (%) | Essential Oil (mL/kg) | |
| Wild-growing populations | ||||
| 1 | 3.4 ± 0.78 c | 107.4 ± 36.9 d | 0.007 ± 0.001 de | 9.0 ± 1.2 a |
| 2 | 4.5 ± 0.99 ab | 164.8 ± 32.7 c | 0.010 ± 0.001 c | 9.8 ± 1.2 a |
| 3 | 4.1 ± 1.56 b | 166.3 ± 50.3 c | 0.006 ± 0.001 e | 6.7 ± 0.9 bc |
| 4 | 5.5 ± 1.27 a | 161.1 ± 45.8 c | 0.006 ± 0.001 e | 7.2 ± 1.0 b |
| 5 | 6.0 ± 1.62 a | 403.6 ± 119.2 a | 0.031 ± 0.003 a | 6.9 ± 1.0 bc |
| 6 | 4.6 ± 0.99 ab | 196.1 ± 31.3 bc | 0.008 ± 0.001 d | 7.8 ± 1.0 b |
| 7 | 3.3 ± 0.36 c | 131.1 ± 31.9 d | 0.010 ± 0.001 c | 8.0 ± 1.0 b |
| 8 | 3.5 ± 0.80 c | 186.6 ± 66.2 bc | 0.004 ± 0.000 e | 5.4 ± 0.3 c |
| 9 | 5.1 ± 1.42 ab | 220.7 ± 41.3 b | 0.017 ± 0.002 b | 5.4 ± 0.9 c |
| 10 | 3.7 ± 0.74 c | 252.5 ± 56.2 b | 0.010 ± 0.001 c | 10.4 ± 1.4 a |
| 11 | 3.9 ± 0.74 b | 160.8 ± 51.2 c | 0.011 ± 0.001 c | 7.7 ± 1.0 b |
| 12 | 1.7 ± 0.59 d | 112.8 ± 30.3 d | 0.012 ± 0.001 c | 6.0 ± 0.8 bc |
| 13 | 3.9 ± 1.13 b | 170.6 ± 61.8 c | 0.005 ± 0.001 e | 4.3 ± 0.6 cd |
| 14 | 3.1 ± 0.61 c | 247.7 ± 86.6 b | 0.008 ± 0.001 d | 8.1 ± 1.1 b |
| 15 | 4.6 ± 0.92 ab | 245.8 ± 76.5 b | 0.009 ± 0.001 d | 3.9 ± 0.5 d |
| 16 | 5.5 ± 0.43 a | 265.1 ± 55.9 b | 0.009 ± 0.001 d | 7.3 ± 1.0 b |
| 17 | 5.1 ± 0.97 ab | 187.8 ± 48.5 bc | 0.008 ± 0.001 d | 4.3 ± 0.4 cd |
| 18 | 4.8 ± 1.10 ab | 331.2 ± 88.5 ab | 0.094 ± 0.009 a | 5.1 ± 0.7 c |
| 19 | 3.9 ± 0.60 b | 190.8 ± 68.4 bc | 0.010 ± 0.001 c | 9.4 ± 1.2 a |
| mean | 4.22 | 205.4 | 0.014 | 7.0 |
| ‘Lubelski’ landrace | ||||
| CV | 0.24 | 0.35 | 1.35 | 0.27 |
| 5.4 ± 1.24 | 375.0 ± 64.5 | 0.175 ± 0.018 | 6.3 ± 0.9 | |
| No | Plant Height (cm) | Number of Flowering Shoots per Plant | Plant Lushness a | Anthocyanin Coloring of the Shoots b | Color of the Flower Buds c | Color of the Petals d | Teething of the Leaf Margin e | Width of the Leaf Sections f |
|---|---|---|---|---|---|---|---|---|
| Wild-growing populations | ||||||||
| 1 | 135 ± 3 ab | 15 ± 2 a | 3 | 2, 3 | 3 | 2, 3 | 1, 2 | 1 |
| 2 | 110 ± 1 c | 5 ± 2 d | 1, 2 | 1, 2 | 2 | 2 | 2 | 1, 2 |
| 3 | 107 ± 7 c | 7 ± 2 c | 1, 2 | 1, 2 | 2 | 1, 2 | 1, 2, 3 | 1, 2 |
| 4 | 122 ± 5 b | 10 ± 1 b | 2, 3 | 1 | 2, 3 | 1, 2 | 1, 3 | 2 |
| 5 | 124 ± 6 b | 12 ± 2 ab | 2, 3 | 1 | 1, 2 | 1, 2 | 2, 3 | 1 |
| 6 | 124 ± 2 b | 8 ± 0 c | 1, 2, 3 | 1, 2 | 1, 2 | 1, 2 | 2, 3 | 1 |
| 7 | 134 ± 10 ab | 11 ± 2 b | 1, 3 | 1, 2, 3 | 1, 2 | 1, 2 | 1, 2 | 1 |
| 8 | 129 ± 6 b | 14 ± 3 a | 2, 3 | 2, 3 | 2 | 1, 2 | 1, 2 | 1 |
| 9 | 122 ± 6 b | 9 ± 1 c | 1, 2, 3 | 2, 3 | 2 | 1, 2 | 2 | 1 |
| 10 | 125 ± 9 b | 9 ± 1 c | 1, 2 | 2, 3 | 2 | 2 | 2, 3 | 1, 2 |
| 11 | 122 ± 9 b | 13 ± 1 ab | 1, 3 | 2 | 1, 2 | 1, 2 | 2, 3 | 1 |
| 12 | 120 ± 4 b | 8 ± 2 c | 1, 2, 3 | 1, 2, 3 | 1, 2 | 1 | 1, 2, 3 | 1, 2 |
| 13 | 134 ± 11 ab | 11 ± 1 b | 2, 3 | 1, 2, 3 | 1, 2 | 1 | 2, 3 | 1, 2 |
| 14 | 117 ± 2 c | 11 ± 1 b | 2, 3 | 1, 2 | 1, 2 | 1, 2 | 2 | 1 |
| 15 | 131 ± 3 ab | 10 ± 0 b | 2, 3 | 2 | 1, 2 | 1, 2 | 2 | 1, 2 |
| 16 | 133 ± 0 ab | 5 ± 0 d | 1 | 1, 2 | 2, 3 | 1, 2 | 1, 2 | 1, 2 |
| 17 | 122 ± 1 b | 13 ± 1 ab | 2, 3 | 1, 2, 3 | 1, 2 | 1, 2 | 2 | 1 |
| 18 | 143 ± 3 a | 12 ± 1 ab | 2 | 1, 2, 3 | 1, 2 | 1 | 1, 2 | 1 |
| 19 | 114 ± 6 c | 11 ± 1 b | 2 | 2, 3 | 1 | 1 | 1, 2 | 1 |
| mean | 124.63 | 10.21 | - | - | - | - | - | - |
| CV | 0.07 | 0.27 | - | - | - | - | - | - |
| ‘Lubelski’ landrace | ||||||||
| 128 ± 7 | 13 ± 1 | 2, 3 | 1, 2, 3 | 1, 2 | 1, 2 | 1, 2 | 1 | |
| 1000 Seeds Mass (g) | Germinability (%) | Germination Rate (days) | |
|---|---|---|---|
| Wild-growing populations | |||
| 1 | 0.2805 ± 0.0369 c | 44.50 ± 3.23 c | 6.00 c |
| 2 | 0.4785 ± 0.0493 a | 47.75 ± 1.10 c | 7.39 b |
| 3 | 0.4210 ± 0.0362 a | 45.50 ± 3.34 c | 7.73 b |
| 4 | 0.4393 ± 0.0442 a | 47.75 ± 1.96 c | 8.10 b |
| 5 | 0.1380 ± 0.0223 d | 42.25 ± 2.71 c | 6.11 c |
| 6 | 0.3335 ± 0.0367 b | 60.25 ± 2.59 a | 13.00 a |
| 7 | 0.3295 ± 0.0396 b | 34.25 ± 3.51 cd | 8.76 b |
| 8 | 0.2765 ± 0.0335 c | 30.25 ± 1.16 d | 7.00 bc |
| 9 | 0.3468 ± 0.0402 b | 28.75 ± 1.09 d | 7.00 bc |
| 10 | 0.2890 ± 0.0218 c | 52.00 ± 3.42 b | 5.88 c |
| 11 | 0.3375 ± 0.0260 b | 48.00 ± 1.92 c | 7.75 b |
| 12 | 0.3233 ± 0.0291 b | 56.50 ± 3.77 ab | 6.45 c |
| 13 | 0.2933 ± 0.0323 bc | 53.00 ± 2.85 b | 6.54 c |
| 14 | 0.3538 ± 0.0312 b | 25.25 ± 1.03 d | 5.43 a |
| 15 | 0.3270 ± 0.0353 b | 58.25 ± 3.63 ab | 4.64 a |
| 16 | 0.3623 ± 0.0417 b | 62.25 ± 2.55 a | 6.39 c |
| 17 | 0.3080 ± 0.0496 b | 44.25 ± 3.08 c | 8.41 b |
| 18 | 0.3348 ± 0.0330 b | 53.50 ± 2.89 b | 11.14 a |
| 19 | 0.3665 ± 0.0532 b | 42.75 ± 1.24 c | 6.82 bc |
| mean | 0.3336 | 46.16 | 7.40 |
| CV | 0.21 | 0.22 | 0.26 |
| ‘Lubelski’ landrace | |||
| 0.6463 ± 0.0582 | 81.25 ± 3.49 | 6.10 | |
| Population No | Region | Accession No. | Latitude | Longitude | Altitude |
|---|---|---|---|---|---|
| 1 | Noteć River Valley | 401930 | N 53 03 102 | E 17 11 506 | 50 |
| 2 | Bieszczady Mountains | 401935 | N 49 26 752 | E 22 03 516 | 475 |
| 3 | Bieszczady Mountains | 403179 | N 49 21 325 | E 22 09 286 | 600 |
| 4 | Bieszczady Mountains | 403181 | N 49 25 056 | E 22 07 692 | 450 |
| 5 | Bieszczady Mountains | 403182 | N 49 39 298 | E 21 59 682 | 350 |
| 6 | Bieszczady Mountains | 403183 | N 49 36 334 | E 22 12 478 | 360 |
| 7 | Bieszczady Mountains | 403184 | N 49 41 278 | E 22 15 037 | 275 |
| 8 | Nida Basin | 403186 | N 50 43 330 | E 20 31 539 | 250 |
| 9 | Nida Basin | 403187 | N 50 43 059 | E 20 31 474 | 250 |
| 10 | Nida Basin | 401938 | N 50 46 849 | E 20 32 340 | 235 |
| 11 | Nida Basin | 401939 | N 50 52 805 | E 20 56 694 | 375 |
| 12 | Kielce Upland | 401941 | N 50 93 333 | E 20 56 673 | 290 |
| 13 | Kielce Upland | 401942 | N 51 01 020 | E 20 26 141 | 260 |
| 14 | Kielce Upland | 403190 | N 51 01 025 | E 20 28 164 | 260 |
| 15 | Kielce Upland | 401945 | N 51 04 365 | E 20 23 061 | 250 |
| 16 | Kielce Upland | 401946 | N 51 14 630 | E 20 22 354 | 210 |
| 17 | Kielce Upland | 403191 | N 51 06 126 | E 20 15 914 | 225 |
| 18 | Mazovian Lowland | 406738 | N 52 07 282 | E 21 05 541 | 80 |
| 19 | Mazovian Lowland | 403192 | N 52 07 254 | E 21 05 421 | 80 |
| 20 | ‘Lubelski’ landrace | 404920 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bączek, K.B.; Kosakowska, O.; Boczkowska, M.; Bolc, P.; Chmielecki, R.; Pióro-Jabrucka, E.; Raj, K.; Węglarz, Z. Intraspecific Variability of Wild-Growing Common Valerian (Valeriana officinalis L.). Plants 2022, 11, 3455. https://doi.org/10.3390/plants11243455
Bączek KB, Kosakowska O, Boczkowska M, Bolc P, Chmielecki R, Pióro-Jabrucka E, Raj K, Węglarz Z. Intraspecific Variability of Wild-Growing Common Valerian (Valeriana officinalis L.). Plants. 2022; 11(24):3455. https://doi.org/10.3390/plants11243455
Chicago/Turabian StyleBączek, Katarzyna Barbara, Olga Kosakowska, Maja Boczkowska, Paulina Bolc, Rafał Chmielecki, Ewelina Pióro-Jabrucka, Kavana Raj, and Zenon Węglarz. 2022. "Intraspecific Variability of Wild-Growing Common Valerian (Valeriana officinalis L.)" Plants 11, no. 24: 3455. https://doi.org/10.3390/plants11243455
APA StyleBączek, K. B., Kosakowska, O., Boczkowska, M., Bolc, P., Chmielecki, R., Pióro-Jabrucka, E., Raj, K., & Węglarz, Z. (2022). Intraspecific Variability of Wild-Growing Common Valerian (Valeriana officinalis L.). Plants, 11(24), 3455. https://doi.org/10.3390/plants11243455







