Improving Coconut Using Modern Breeding Technologies: Challenges and Opportunities
Abstract
1. Introduction
2. Collection, Conservation, and Utilization of Coconut Genetic Resources
3. Coconut Breeding’s Goals, Accomplishments, and Prospects
3.1. High Yielding Coconut
3.2. Climate-Smart Coconut
3.3. Pest and Disease Resistant Coconut
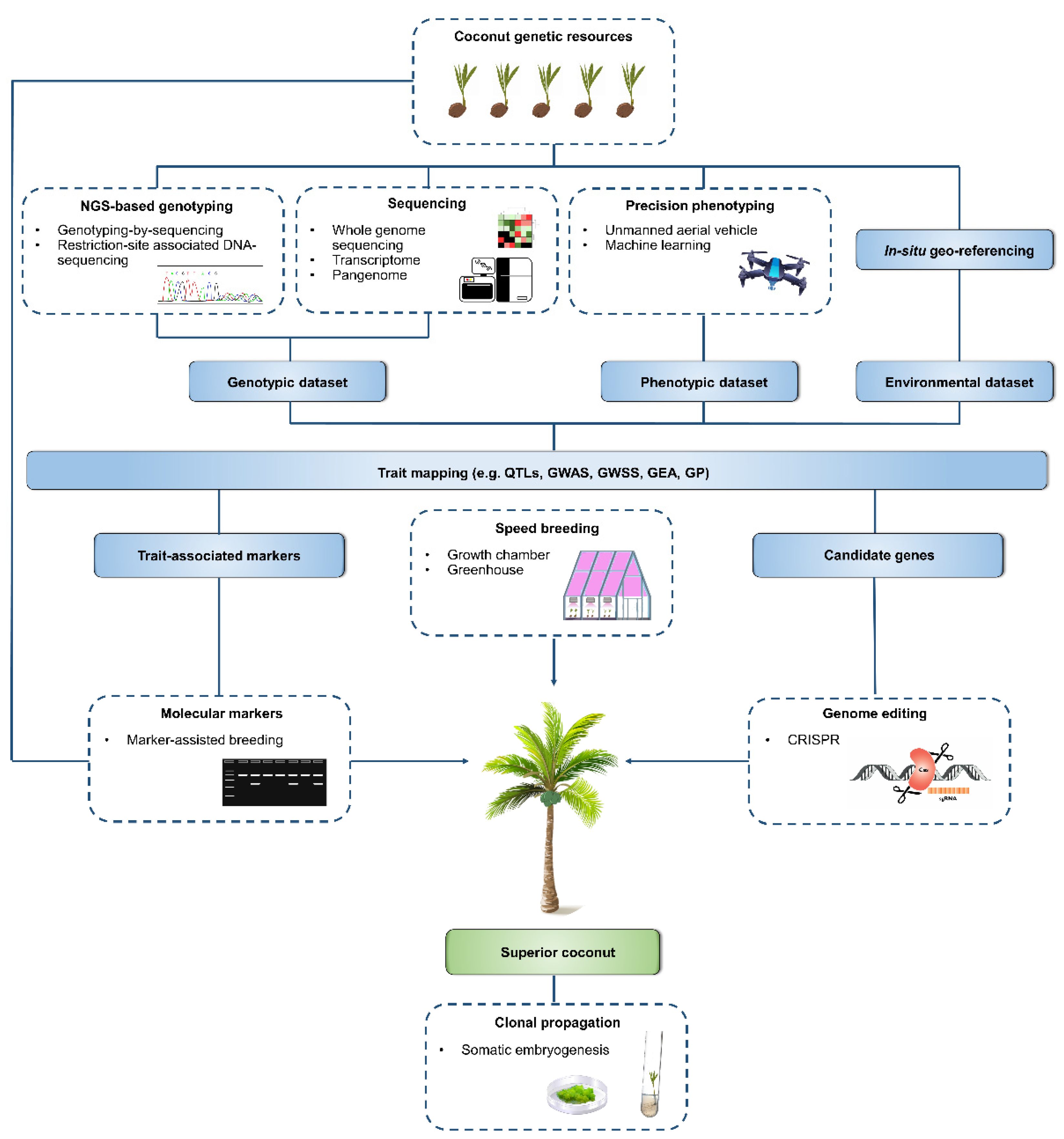
4. Progress of Current Breeding Strategies and Potentials of Biotechnological Breeding Approaches and High-Throughput Genotyping Platforms
4.1. Molecular Markers and Marker-Assisted Breeding (MAB)
4.2. Whole Genome Sequencing and Genomic-Assisted Breeding
4.3. Transcriptome Sequencing
4.4. Clonal Propagation Via Somatic Embryogenesis
4.5. Genome Editing
5. Speed Breeding to Accelerate Coconut Breeding
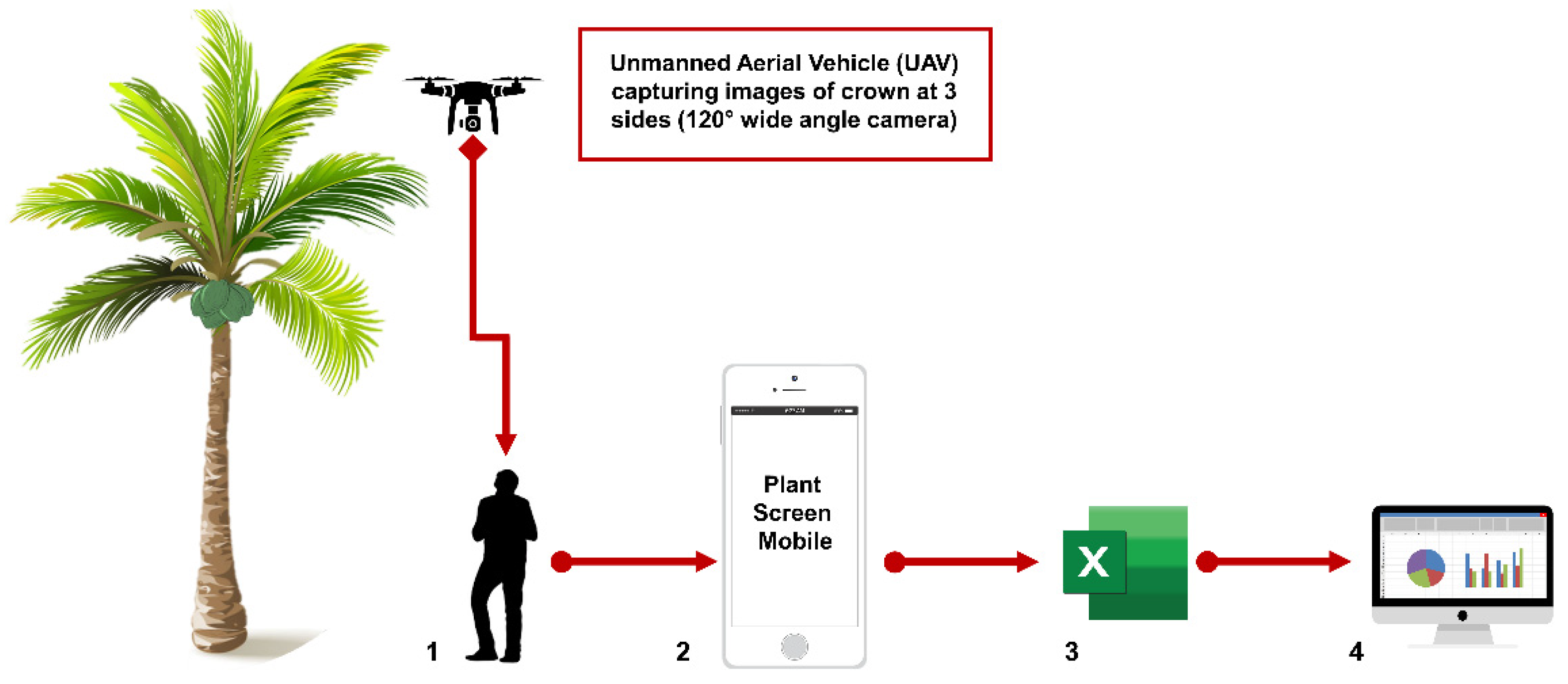
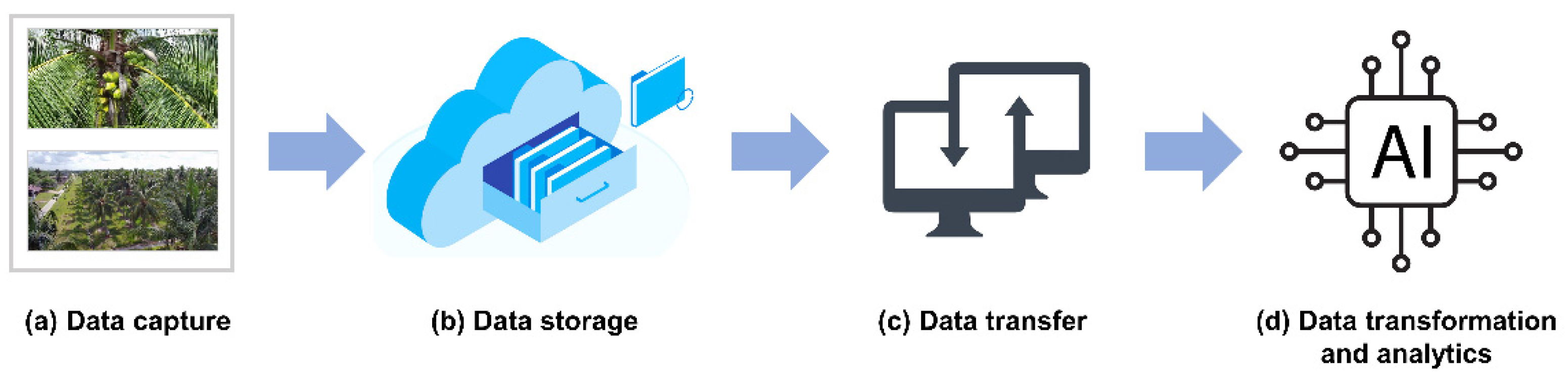
6. Next Generation Phenotyping Approaches
7. “Grower’s Experience” vs. Deep Machine Learning
8. Concluding Remarks
9. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nasir, K.H. Genetic Diversity Evaluation of MARDI’s Coconut (Cocos nucifera L.) Germplasm using Simple Sequence Repeats. Int. J. Coconut R D 2016, 32, 37–45. [Google Scholar]
- Baudouin, L.; Lebrun, P.; Konan, J.L.; Ritter, E.; Berger, A.; Billotte, N. QTL analysis of fruit components in the progeny of a Rennell Island Tall coconut (Cocos nucifera L.) individual. Theor. Appl. Genet. 2006, 112, 258–268. [Google Scholar] [CrossRef]
- Perera, L.; Perera, S.A.C.N.; Bandaranayake, C.K.; Harries, H.C. Coconut, in Oil Crops; Vollmann, J., Rajcan, I., Eds.; Springer: New York, NY, USA, 2010; pp. 369–396. [Google Scholar]
- Alrifai, O.; Marcone, M. Coconut—The Tree of Life—Improvement by Biotechnology; Elsevier: Oxford, UK, 2019; pp. 586–594. [Google Scholar]
- Burkill, I.H. A Dictionary of the Economic Products of the Malay Peninsula. In Governments of Malaysia and Singapore by the Ministry of Agriculture and Cooperatives; Ministry of Agriculture and Co-Operatives: Kuala Lumpur, Malaysia, 1966. [Google Scholar]
- Salum, U.; Foale, M.; Biddle, J.; Bazrafshan, A.; Adkins, S. Towards the Sustainability of the “Tree of Life”: An Introduction. In Coconut Biotechnology: Towards the Sustainability of the ‘Tree of Life’; Adkins, S., Foale, M., Bourdeix, R., Nguyen, Q., Biddle, J., Eds.; Springer: Cham, Switzerland, 2020; pp. 1–15. [Google Scholar]
- Rethinam, P. International Scenario of Coconut Sector. In The Coconut Palm (Cocos nucifera L.)—Research and Development Perspectives; Krishnakumar, V., Thampan, P.K., Nair, M.A., Eds.; Springer: Singapore, 2018; pp. 21–56. [Google Scholar]
- Oropeza, C.; Howard, F.W.; Ashburner, G.R. Lethal Yellowing: Research and Practical Aspects; Kluwer Academic: Norwell, MA, USA, 1995. [Google Scholar]
- Perera, L.; Perera, S.; Bandaranayake, C.K. Coconut. In Oil Crops, Handbook of Plant Breeding 4; Vollmann, J., Ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Bourdeix, R.; Engels, J. A Global Strategy for the Conservation and Use of Coconut Genetic Resources, 2018–2028; Biodiversity International: Montpellier, France, 2018. [Google Scholar]
- Engelmann, F.; Malaurie, B.; N’Nan, O.; Borges, M. Status of cryopreservation research in coconut. In Coconut Genetic Resources; Batugal, P., Rao, V.R., Oliver, J., Eds.; IPGRI-APO: Serdang, Malaysia, 2005. [Google Scholar]
- Bourdeix, R.; Batugal, P.; Oliver, J.T.; George, M.L. Catalogue of Conserved Coconut Germplasm; IPGRI: Rome, Italy, 2010. [Google Scholar]
- Cortés, A.J.; López-Hernández, F. Harnessing Crop Wild Diversity for Climate Change Adaptation. Genes 2021, 12, 783. [Google Scholar] [CrossRef]
- Sangaré, A.; le Saint, J.P.; de Nuce De Lamothe, M. The Tall coconuts at Port-Bouet (Ivory Coast). 3.Cambodian Tall, Tonga Tall, Rotuma Tall. Oleag 1984, 39, 205–213. [Google Scholar]
- N’cho, Y.P.; le Saint, J.P.; Sangaré, A. Dwarf coconuts at Port-Bouet (Cote d’Ivoire). III. New Guinea Brown Dwarf, Thailand Green Dwarf and Polynesian Red Dwarf. Oleag 1988, 43, 55–66. [Google Scholar]
- Le Saint, J.P.; de Nuce de Lamothe, M.; Sangaré, A. The dwarf coconuts at Port-Bouet (Ivory Coast). 2. Sri Lanka Green Dwarf, and additional information on Malayan Red and Yellow Dwarfs, Equatorial Guinea Green Dwarf and Cameroon Red Dwarf [vegetative characteristics, floral biology, yield and fruit composition]. Oleag 1983, 38, 595–606. [Google Scholar]
- Teulat, B.; Aldam, C.; Trehin, R.; Lebrun, P.; Barker, J.H.A.; Arnold, G.M.; Karp, A.; Baudouin, L.; Rognon, F. An analysis of genetic diversity in coconut (Cocos nucifera) populations from across the geographic range using sequence-tagged microsatellites (SSRs) and AFLPs. Theor. Appl. Genet. 2000, 100, 764–771. [Google Scholar] [CrossRef]
- Perera, L.; Russell, J.R.; Provan, J.; Powell, W. Use of microsatellite DNA markers to investigate the level of genetic diversity and population genetic structure of coconut (Cocos nucifera L.). Genome 2000, 43, 15–21. [Google Scholar] [CrossRef]
- Perera, L.; Russell, J.; Provan, J.; Powell, W. Levels and distribution of genetic diversity of coconut (Cocos nucifera L., var. Typica form typica) from Sri Lanka assessed by microsatellite markers. Euphytica 2001, 122, 381–389. [Google Scholar] [CrossRef]
- Ashburner, G.R.; Thompson, W.K.; Halloran, G.M. RAPD Analysis of South Pacific Coconut Palm Populations. Crop Sci. 1997, 37, 992–997. [Google Scholar] [CrossRef]
- Lebrun, P.; N’Cho, Y.; Séguin, M.; Grivet, L.; Baudouin, L. Genetic diversity in coconut (Cocos nucifera L.) revealed by restriction fragment length polymorphism (RFLP) markers. Euphytica 1998, 101, 103–108. [Google Scholar] [CrossRef]
- Perera, L.; Russell, J.R.; Provan, J.; McNicol, J.W.; Powell, W. Evaluating genetic relationships between indigenous coconut (Cocos nucifera L.) accessions from Sri Lanka by means of AFLP profiling. Theor. Appl. Genet. 1998, 96, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Forester, B.R.; Jones, M.R.; Joost, S.; Landguth, E.L.; Lasky, J.R. Detecting spatial genetic signatures of local adaptation in heterogeneous landscapes. Mol. Ecol. 2016, 25, 104–120. [Google Scholar] [CrossRef] [PubMed]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A practical guide to environmental association analysis in landscape genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef] [PubMed]
- Harries, H.C. The ‘Niu’ Indies: Long lost ‘home’ of the coconut palm. Palms 2002, 46, 97–100. [Google Scholar]
- Perera, L.; Peries, R.R.S. Report of the Genetics and Plant Breeding Division; Annual Report of the Coconut Research Institute of Sri Lanka: Lunuwila, Sri Lanka, 2005. [Google Scholar]
- Novarianto, H.; Santosa, B.; Tulalo, M.; Mawardi, S.; Maskromo, I. Varietal Improvement in Coconut in Indonesia. In Proceedings of the XLVII APCC Cocotech Conference & Exhibition, Bali, Indonesia, 26–30 September 2016. [Google Scholar]
- Arulandoo, X. Performance of Coconut Hybrids in United Plantations Berhad and Commercial Production of Seed Nuts. CORD 2014, 30, 10. [Google Scholar] [CrossRef]
- Sharma, M.; Padman, C.R.; Singh, G.; San, T.T. MAWA and MATAG: Two High Yielding Hybrids for the Revitalisation of the Malaysian Coconut Industry; United Plantations Berhad: Teluk Intan, Malaysia, 2000. [Google Scholar]
- Bourdeix, R.; Konan, J.L.; N’Cho, V. Coconut. A Guide to Traditional and Improved Varieties; Diversiflora: Montpellier, France, 2014. [Google Scholar]
- Abu Dardak, R.; Mohd Yon, R. Strengthening the Coconut Industry in Malaysia. 2021. Available online: https://ap.fftc.org.tw/article/2938 (accessed on 14 March 2022).
- Pokhrel, S.; Meyers, B.C. Heat-responsive microRNAs and phased small interfering RNAs in reproductive development of flax. Plant Direct 2022, 6, e385. [Google Scholar] [CrossRef] [PubMed]
- Naresh Kumar, S.; Hebbar, K.B.; Kasturi Bai, K.V.; Rajagopal, V. Physiology and Biochemistry. In The Coconut Palm (Cocos nucifera L.)—Research and Development Perspectives; Krishnakumar, V., Thampan, P.K., Nair, M.A., Eds.; Springer: Singapore, 2018; pp. 443–488. [Google Scholar]
- Kole, C. Genomic Designing of Climate-Smart Fruit Crops; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Bai, K.; Rajagopal, V. Osmotic adjustment as a mechanism for drought tolerance in coconut (Cocos nucifera L.). Indian J. Plant Physiol. 2000, 5, 320–323. [Google Scholar]
- Rajagopal, V.; Shivashankar, S.; Mathew, J. Impact of dry spells on the ontogeny of coconut fruits and its relation to yield. Plant. Rech. Dev. 1996, 3, 251–255. [Google Scholar]
- Kumar, S.N.; Rajeev, M.S.; Vinayan, A.; Nagvekar, D.D.; Venkitaswamy, R.; Rao, D.V.R.; Boraiah, B.S. Gawankar, M.; Dhanapal, R.; Patil, D.V.; et al. Trends in weather and yield changes in past in coconut growing areas in India. J. Agrometeorol. 2009, 11, 15–18. [Google Scholar] [CrossRef]
- Naresh Kumar, S.; Aggarwal, P.K. Climate change and coconut plantations in India: Impacts and potential adaptation gains. Agric. Syst. 2013, 117, 45–54. [Google Scholar] [CrossRef]
- Lasky, J.R.; Marais, D.L.D.; McKAY, J.K.; Richards, J.H.; Juenger, T.E.; Keitt, T.H. Characterizing genomic variation of Arabidopsis thaliana: The roles of geography and climate. Mol. Ecol. 2012, 21, 5512–5529. [Google Scholar] [CrossRef] [PubMed]
- Stölting, K.N.; Paris, M.; Meier, C.; Heinze, B.; Castiglione, S.; Bartha, D.; Lexer, C. Genome-wide patterns of differentiation and spatially varying selection between postglacial recolonization lineages of Populus alba (Salicaceae), a widespread forest tree. New Phytol. 2015, 207, 723–734. [Google Scholar] [CrossRef] [PubMed]
- López-Hernández, F.; Cortés, A.J. Last-Generation Genome-Environment Associations Reveal the Genetic Basis of Heat Tolerance in Common Bean (Phaseolus vulgaris L.). Front. Genet. 2019, 10, 954. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; Blair, M.W. Genotyping by Sequencing and Genome-Environment Associations in Wild Common Bean Predict Widespread Divergent Adaptation to Drought. Front. Plant Sci. 2018, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Vavilov, N.I. The law of homologous series in variation. J. Genet. 1922, 12, 47–89. [Google Scholar] [CrossRef]
- Yang, Y.; Iqbal, A.; Qadri, R. Breeding of Coconut (Cocos Nucifera L.): The Tree of Life; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Navia, D.; Gondim, M.G.C.; Aratchige, N.S.; De Moraes, G.J. A review of the status of the coconut mite, Aceria guerreronis (Acari: Eriophyidae), a major tropical mite pest. Exp. Appl. Acarol. 2013, 59, 67–94. [Google Scholar] [CrossRef]
- Fernando, L.C.P.; Aratchige, N.S.; Peiris, T.S.G. Distribution patterns of coconut mite, Aceria guerreronis, and its predator Neoseiulus aff. paspalivorus in coconut palms. Exp. Appl. Acarol. 2003, 31, 71–78. [Google Scholar] [CrossRef]
- Murphy, S.T.; Briscoe, B.R. The red palm weevil as an alien invasive: Biology and the prospects for biological control as a component of IPM. Biocontrol News Inf. 1999, 20, 35–46. [Google Scholar]
- Azmi, W. A New Invasive Coconut Pest in Malaysia: The Red Palm Weevil (Curculionidae: Rhynchophorus ferrugineus). Planter 2013, 89, 97–110. [Google Scholar]
- Faleiro, J.R.; Rangnekar, P.A. Location specific seasonal activity of red palm weevil, Rhynchophorus ferrugineus Oliv. coconut plantations of Goa. Indian J. Appl. Entomol. 2001, 15, 7–15. [Google Scholar]
- Catley, A. The Coconut Rhinoceros Beetle Oryctes rhinoceros (L) [Coleoptera: Scarabaeidae: Dynastinae]. PANS Pest Artic. News Summ. 1969, 15, 18–30. [Google Scholar] [CrossRef]
- Marshall, S.D.; Moore, A.; Vaqalo, M.; Noble, A.; Jackson, T.A. A new haplotype of the coconut rhinoceros beetle, Oryctes rhinoceros, has escaped biological control by Oryctes rhinoceros nudivirus and is invading Pacific Islands. J. Invertebr. Pathol. 2017, 149, 127–134. [Google Scholar] [CrossRef]
- Etebari, K.; Hereward, J.; Sailo, A.; Ahoafi, E.M.; Tautua, R.; Tsatsia, H.; Jackson, G.V.; Furlong, M.J. Genetic structure of the Coconut Rhinoceros Beetle (Oryctes rhinoceros) population and the incidence of its biocontrol agent (Oryctes rhinoceros nudivirus) in the South Pacific Islands. bioRxiv 2020. [Google Scholar] [CrossRef]
- Huger, A.M. The Oryctes virus: Its detection, identification, and implementation in biological control of the coconut palm rhinoceros beetle, Oryctes rhinoceros (Coleoptera: Scarabaeidae). J. Invertebr. Pathol. 2005, 89, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Huger, A.M. A virus disease of the Indian rhinoceros beetle, Oryctes rhinoceros (linnaeus), caused by a new type of insect virus, Rhabdionvirus oryctes gen. n., sp. n. J. Invertebr. Pathol. 1966, 8, 38–51. [Google Scholar] [CrossRef]
- Bedford, G.O. Biology, Ecology, and Control of Palm Rhinoceros Beetles. Annu. Rev. Entomol. 1980, 25, 309–339. [Google Scholar] [CrossRef]
- McCoy, R.E.; Howard, F.W.; Tsai, J.H.; Donselman, H.M.; Thomas, D.L. Lethal Yellowing of Palms. In Agricultural Experiment Station Bulletin; Florida, U.O., Ed.; University of Florida, Agricultural Experiment Stations: Gainesville, FL, USA, 1983; p. 834. [Google Scholar]
- Lebrun, P.; Baudouin, L.; Myrie, W.; Berger, A.; Dollet, M. Recent lethal yellowing outbreak: Why is the Malayan Yellow Dwarf Coconut no longer resistant in Jamaica? Tree Genet. Genomes 2008, 4, 125–131. [Google Scholar] [CrossRef]
- Ramjegathesh, R.; Karthikeyan, G.; Rajendran, L.; Johnson, I.; Raguchander, T.; Samiyappan, R. Root (wilt) disease of coconut palms in South Asia—An overview. Arch. Phytopathol. Plant Prot. 2012, 45, 2485–2493. [Google Scholar] [CrossRef]
- Nair, R.V.; Jacob, P.M.; Kumar, R.A. Screening of coconut cultivars against root (wilt) disease. J. Plant. Crops 2004, 32, 59–60. [Google Scholar]
- Fan, H.; Xiao, Y.; Yang, Y.; Xia, W.; Mason, A.S.; Xia, Z.; Qiao, F.; Zhao, S.; Tang, H. RNA-Seq analysis of Cocos nucifera: Transcriptome sequencing and de novo assembly for subsequent functional genomics approaches. PLoS ONE 2013, 8, e59997. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; López-Hernández, F.; Blair, M.W. Genome-Environment Associations, an Innovative Tool for Studying Heritable Evolutionary Adaptation in Orphan Crops and Wild Relatives. Front. Genet. 2022, 13, 910386. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, M.; Jerard, B.; Preethi, P.; Thomas, R.J.; Fayas, T.; Rachana, K.; Karun, A. Development of a RAPD-derived SCAR marker associated with tall-type palm trait in coconut. Sci. Hortic. 2012, 150, 312–316. [Google Scholar] [CrossRef]
- Perera, L. Hybrid Testing and Variety Identification of Coconut (Cocos nucifera L.) in Sri Lanka Using Microsatellite Markers. Cord 2010, 26, 39–43. [Google Scholar]
- Saensuk, C.; Wanchana, S.; Choowongkomon, K.; Wongpornchai, S.; Kraithong, T.; Imsabai, W.; Chaichoompu, E.; Ruanjaichon, V.; Toojinda, T.; Vanavichit, A.; et al. De novo transcriptome assembly and identification of the gene conferring a "pandan-like" aroma in coconut (Cocos nucifera L.). Plant Sci. 2016, 252, 324–334. [Google Scholar] [CrossRef]
- Vongvanrungruang, A.; Mongkolsiriwatana, C.; Boonkaew, T.; Sawatdichaikul, O.; Srikulnath, K.; Peyachoknagul, S. Single base substitution causing the fragrant phenotype and development of a type-specific marker in aromatic coconut (Cocos nucifera). Genet. Mol. Res. 2016, 15, gmr.15038748. [Google Scholar] [CrossRef]
- Preethi, P.P.; Rajesh, M.K.; Rahul, C.U.; Jerard, B.A.; Samsudeen, K.; Thomas, R.J.; Karun, A. Identification and utilization of informative EST-SSR markers for genetic purity testing of coconut hybrids. J. Plant. Crops 2016, 44, 77–84. [Google Scholar]
- Azevedo, A.O.N.; Azevedo, C.D.D.O.; Santos, P.H.A.D.; Ramos, H.C.C.; Boechat, M.S.B.; Arêdes, F.A.S.; Ramos, S.R.R.; Mirizola, L.; Perera, L.; Aragão, W.M.; et al. Selection of legitimate dwarf coconut hybrid seedlings using DNA fingerprinting. Crop Breed. Appl. Biotechnol. 2018, 18, 409. [Google Scholar] [CrossRef]
- Xiao, Y.; Xu, P.; Fan, H.; Baudouin, L.; Xia, W.; Bocs, S.; Xu, J.; Li, Q.; Guo, A.; Zhou, L.; et al. The genome draft of coconut (Cocos nucifera). Gigascience 2017, 6, 1–11. [Google Scholar] [CrossRef]
- Lantican, D.V.; Strickler, S.R.; Canama, A.O.; Gardoce, R.R.; Mueller, L.A.; Galvez, H.F. De Novo Genome Sequence Assembly of Dwarf Coconut (Cocos nucifera L. ’Catigan Green Dwarf’) Provides Insights into Genomic Variation Between Coconut Types and Related Palm Species. G3 (Bethesda) 2019, 9, 2377–2393. [Google Scholar] [CrossRef]
- Wang, S.; Xiao, Y.; Zhou, Z.-W.; Yuan, J.; Guo, H.; Yang, Z.; Yang, J.; Sun, P.; Sun, L.; Deng, Y.; et al. High-quality reference genome sequences of two coconut cultivars provide insights into evolution of monocot chromosomes and differentiation of fiber content and plant height. Genome Biol. 2021, 22, 304. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.K.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.H.A.D.; Venâncio, T.M.; Ramos, H.C.C.; Arêdes, F.A.S.; Azevedo, A.O.N.; Boechat, M.S.B.; Filho, G.A.D.S.; Ramos, S.R.R.; Mirisola, L.A.; Aragão, W.M.; et al. Genotyping-by-sequencing technology reveals directions for coconut (Cocos nucifera L.) breeding strategies for water production. Euphytica 2020, 216, 45. [Google Scholar] [CrossRef]
- Riangwong, K.; Wanchana, S.; Aesomnuk, W.; Saensuk, C.; Nubankoh, P.; Ruanjaichon, V.; Kraithong, T.; Toojinda, T.; Vanavichit, A.; Arikit, S. Mining and validation of novel genotyping-by-sequencing (GBS)-based simple sequence repeats (SSRs) and their application for the estimation of the genetic diversity and population structure of coconuts (Cocos nucifera L.) in Thailand. Hortic. Res. 2020, 7, 156. [Google Scholar] [CrossRef]
- Varshney, R.K.; Bohra, A.; Yu, J.; Graner, A.; Zhang, Q.; Sorrells, M.E. Designing Future Crops: Genomics-Assisted Breeding Comes of Age. Trends Plant Sci. 2021, 26, 631–649. [Google Scholar] [CrossRef]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef]
- Jain, M. Next-generation sequencing technologies for gene expression profiling in plants. Brief. Funct. Genom. 2012, 11, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Hamilton, M.; Jacobi, J.L.; Ngam, P.; Devitt, N.; Schilkey, F.; Ben-Hur, A.; Reddy, A.S.N. A survey of the sorghum transcriptome using single-molecule long reads. Nat. Commun. 2016, 7, 11706. [Google Scholar] [CrossRef]
- Wang, B.; Tseng, E.; Regulski, M.; Clark, T.A.; Hon, T.; Jiao, Y.; Lu, Z.; Olson, A.; Stein, J.C.; Ware, D. Unveiling the complexity of the maize transcriptome by single-molecule long-read sequencing. Nat. Commun. 2016, 7, 11708. [Google Scholar] [CrossRef]
- Cloonan, N.; Grimmond, S.M. Transcriptome content and dynamics at single-nucleotide resolution. Genome Biol. 2008, 9, 234. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef]
- Rajesh, M.K.; Fayas, T.P.; Naganeeswaran, S.; Rachana, K.E.; Bhavyashree, U.; Sajini, K.K.; Karun, A. De novo assembly and characterization of global transcriptome of coconut palm (Cocos nucifera L.) embryogenic calli using Illumina paired-end sequencing. Protoplasma 2016, 253, 913–928. [Google Scholar] [CrossRef]
- Birol, I.; Jackman, S.; Nielsen, C.B.; Qian, J.Q.; Varhol, R.; Stazyk, G.; Morin, R.D.; Zhao, Y.; Hirst, M.; Schein, J.E.; et al. De novo transcriptome assembly with ABySS. Bioinformatics 2009, 25, 2872–2877. [Google Scholar] [CrossRef] [PubMed]
- Orshinsky, A.M.; Hu, J.; Opiyo, S.O.; Reddyvari-Channarayappa, V.; Mitchell, T.K.; Boehm, M. RNA-Seq analysis of the Sclerotinia homoeocarpa-creeping bentgrass pathosystem. PLoS ONE 2012, 7, e41150. [Google Scholar] [CrossRef]
- Xu, H.; Gao, Y.; Wang, J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS ONE 2012, 7, e30646. [Google Scholar] [CrossRef]
- Rajesh, M.K.; Rachana, K.E.; Kulkarni, K.; Sahu, B.B.; Thomas, R.J.; Karun, A. Comparative transcriptome profiling of healthy and diseased Chowghat Green Dwarf coconut palms from root (wilt) disease hot spots. Eur. J. Plant Pathol. 2018, 151, 173–193. [Google Scholar] [CrossRef]
- Nejat, N.; Cahill, D.M.; Vadamalai, G.; Ziemann, M.; Rookes, J.; Naderali, N. Transcriptomics-based analysis using RNA-Seq of the coconut (Cocos nucifera) leaf in response to yellow decline phytoplasma infection. Mol. Genet. Genom. 2015, 290, 1899–1910. [Google Scholar] [CrossRef]
- Huang, Y.-Y.; Lee, C.-P.; Fu, J.L.; Chang, B.C.-H.; Matzke, A.J.M.; Matzke, M. De novo transcriptome sequence assembly from coconut leaves and seeds with a focus on factors involved in RNA-directed DNA methylation. G3 (Bethesda) 2014, 4, 2147–2157. [Google Scholar] [CrossRef]
- Belmonte, M.F.; Kirkbride, R.C.; Stone, S.L.; Pelletier, J.M.; Bui, A.Q.; Yeung, E.C.; Hashimoto, M.; Fei, J.; Harada, C.M.; Munoz, M.D.; et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 2013, 110, E435–E444. [Google Scholar] [CrossRef]
- Gehring, M. Genomic imprinting: Insights from plants. Annu. Rev. Genet. 2013, 47, 187–208. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Bocs, S.; Fan, H.; Armero, A.; Baudouin, L.; Xu, P.; Xu, J.; This, D.; Hamelin, C.; Iqbal, A.; et al. Coconut genome assembly enables evolutionary analysis of palms and highlights signaling pathways involved in salt tolerance. Commun. Biol. 2021, 4, 105. [Google Scholar] [CrossRef]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Jha, S.K.; Bagri, J.; Pandey, G.K. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 2015, 10, e0125168. [Google Scholar] [CrossRef]
- Branton, R.L.; Blake, J. Development of Organized Structures in Callus Derived from Explants of Cocos nucifera L. Ann. Bot. 1983, 52, 673–678. [Google Scholar] [CrossRef]
- Eeuwens, C.J.; Blake, J. Culture of Coconut and Date Palm Tissue with a View to Vegetative Propagation; International Society for Horticultural Science (ISHS): Leuven, Belgium, 1977. [Google Scholar]
- Gupta, P.K.; Kendurkar, S.V.; Kulkarni, V.M.; Shirgurkar, M.V.; Mascarenhas, A.F. Somatic embryogenesis and plants from zygotic embryos of coconut (Cocos nucifera L.) in vitro. Plant Cell Rep. 1984, 3, 222–225. [Google Scholar] [CrossRef]
- Chan, J.L.; Saénz, L.; Talavera, C.; Hornung, R.; Robert, M.; Oropeza, C. Regeneration of coconut (Cocos nucifera L.) from plumule explants through somatic embryogenesis. Plant Cell Rep. 1998, 17, 515–521. [Google Scholar] [CrossRef]
- Fernando, S.; Verdeil, J.; Hocher, V.; Weerakoon, L.; Hirimburegama, K. Histological analysis of plant regeneration from plumule explants of Cocos nucifera. Plant Cell Tissue Organ Cult. 2003, 72, 281–283. [Google Scholar] [CrossRef]
- Verdeil, J.-L.; Hornung, R.; Jacobsen, H.-J.; Rillo, E.; Oropeza, C.; Bourdeix, R.; N’Cho, Y.-P.; Hocher, V.; Hamon, S.; Sangare, A. Recent progress on coconut micropropagation through a joined effort involving different countries. In Current Advances in Coconut Biotechnology; Oropeza, C., Cardeña, R., Santamaría, J.M., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 391–405. [Google Scholar]
- Pérez-Núñez, M.T.; Chan, J.L.; Sáenz, L.; González, T.; Verdeil, J.L.; Oropeza, C. Improved somatic embryogenesis from Cocos nucifera (L.) plumule explants. Vitr. Cell. Dev. Biol.-Plant 2006, 42, 37–43. [Google Scholar] [CrossRef]
- Montero-Cortés, M.; Rodríguez-Paredes, F.; Burgeff, C.; Pérez-Nuñez, T.; Cordova, I.; Oropeza, C.; Verdeil, J.-L.; Saenz, L. Characterisation of a cyclin-dependent kinase (CDKA) gene expressed during somatic embryogenesis of coconut palm. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 102, 251–258. [Google Scholar] [CrossRef]
- Lakshmi, J.; Bhavyashree, U.; Fayas, T.P.; Sajini, K.K.; Rajesh, M.K.; Karun, A. Histological studies of cellular differentiation during somatic embryogenesis of coconut plumule-derived calli. J. Plant. Crops 2015, 43, 196–203. [Google Scholar]
- Sáenz, L.; Chan, J.L.; Narvaez, M.; Oropeza, C. Protocol for the Micropropagation of Coconut from Plumule Explants. In Plant Cell Culture Protocols; Loyola-Vargas, V.M., Ochoa-Alejo, N., Eds.; Springer: New York, NY, USA, 2018; pp. 161–170. [Google Scholar]
- Oropeza, C.; Sandoval-Cancino, G.; Sáenz, L.; Narváez, M.; Rodríguez, G.; Chan, J.L. Coconut (Cocos nucifera L.) Somatic Embryogenesis Using Immature Inflorescence Explants. In Step Wise Protocols for Somatic Embryogenesis of Important Woody Plants: Volume II; Jain, S.M., Gupta, P., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 103–111. [Google Scholar]
- Sandoval-Cancino, G.; Sáenz, L.; Chan, J.L.; Oropeza, C. Improved formation of embryogenic callus from coconut immature inflorescence explants. Vitr. Cell. Dev. Biol.-Plant 2016, 52, 367–378. [Google Scholar] [CrossRef]
- Perera, P.I.P.; Hocher, V.; Verdeil, J.L.; Doulbeau, S.; Yakandawala, D.M.D.; Weerakoon, L.K. Unfertilized ovary: A novel explant for coconut (Cocos nucifera L.) somatic embryogenesis. Plant Cell Rep. 2007, 26, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Fernando, S.C.; Vidhanaarachchi, V.R.M.; Weerakoon, L.K.; Santha, E.S. What makes clonal propagation of coconut difficult? Asia-Pac. J. Mol. Biol. Biotechnol. 2010, 18, 163–165. [Google Scholar]
- Nguyen, Q.T.; Bandupriya, H.D.D.; López-Villalobos, A.; Sisunandar, S.; Foale, M.; Adkins, S.W. Tissue culture and associated biotechnological interventions for the improvement of coconut (Cocos nucifera L.): A review. Planta 2015, 242, 1059–1076. [Google Scholar] [CrossRef]
- Kalaipandian, S.; Mu, Z.; Kong, E.Y.Y.; Biddle, J.; Cave, R.; Bazrafshan, A.; Wijayabandara, K.; Beveridge, F.C.; Nguyen, Q.; Adkins, S.W. Cloning Coconut via Somatic Embryogenesis: A Review of the Current Status and Future Prospects. Plants 2021, 10, 2050. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Sakaguchi, K. DNA repair in plants. Chem. Rev. 2006, 106, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Jasrotia, R.S.; Iquebal, M.; Jaiswal, S.; Angadi, U.; Rai, A.; Kumar, D. Deciphering genes associated with root wilt disease of coconut and development of its transcriptomic database (CnTDB). Physiol. Mol. Plant Pathol. 2017, 100, 255–263. [Google Scholar] [CrossRef]
- Andrade-Torres, A.; Oropeza, C.; Sáenz, L.; González-Estrada, T.; Ramírez-Benítez, J.E.; Becerril, K.; Chan, J.L.; Rodríguez-Zapata, L.C. Transient genetic transformation of embryogenic callus of Cocos nucifera. Biologia 2011, 66, 790–800. [Google Scholar] [CrossRef]
- Knutzon, D.S.; Hayes, T.R.; Wyrick, A.; Xiong, H.; Davies, H.M.; Voelker, T.A. Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant Physiol. 1999, 120, 739–746. [Google Scholar] [CrossRef]
- Davies, H.M.; Hawkins, D.J.; Nelsen, J.S. Lysophosphatidic acid acyltransferase from immature coconut endosperm having medium chain length substrate specificity. Phytochemistry 1995, 39, 989–996. [Google Scholar] [CrossRef]
- Sun, R.; Ye, R.; Gao, L.; Zhang, L.; Wang, R.; Mao, T.; Zheng, Y.; Li, D.; Lin, Y. Characterization and Ectopic Expression of CoWRI1, an AP2/EREBP Domain-Containing Transcription Factor from Coconut (Cocos nucifera L.) Endosperm, Changes the Seeds Oil Content in Transgenic Arabidopsis thaliana and Rice (Oryza sativa L.). Front. Plant Sci. 2017, 8, 63. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, K.B.; Taylor, M.; Zhou, X.-R.; Vanhercke, T.; Wood, C.C.; Blanchard, C.L.; Singh, S.; Petrie, J.R. Metabolic engineering of medium-chain fatty acid biosynthesis in Nicotiana benthamiana plant leaf lipids. Front. Plant Sci. 2015, 6, 164. [Google Scholar] [CrossRef] [PubMed]
- Bhatta, M.; Sandro, P.; Smith, M.R.; Delaney, O.; Voss-Fels, K.P.; Gutierrez, L.; Hickey, L.T. Need for speed: Manipulating plant growth to accelerate breeding cycles. Curr. Opin. Plant Biol. 2021, 60, 101986. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Hatta, M.A.M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Watson, A.; Gonzalez-Navarro, O.E.; Ramirez-Gonzalez, R.H.; Yanes, L.; Mendoza-Suárez, M.; Simmonds, J.; Wells, R.; Rayner, T.; Green, P.; et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018, 13, 2944–2963. [Google Scholar] [CrossRef]
- Alahmad, S.; Dinglasan, E.; Leung, K.M.; Riaz, A.; Derbal, N.; Voss-Fels, K.P.; Able, J.A.; Bassi, F.M.; Christopher, J.; Hickey, L.T. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods 2018, 14, 36. [Google Scholar] [CrossRef]
- Watson, A.; Hickey, L.T.; Christopher, J.; Rutkoski, J.; Poland, J.; Hayes, B.J. Multivariate Genomic Selection and Potential of Rapid Indirect Selection with Speed Breeding in Spring Wheat. Crop Sci. 2019, 59, 1945–1959. [Google Scholar] [CrossRef]
- Zheng, Z.; Wang, H.B.; Chen, G.D.; Yan, G.; Liu, C.J. A procedure allowing up to eight generations of wheat and nine generations of barley per annum. Euphytica 2013, 191, 311–316. [Google Scholar] [CrossRef]
- González-Barrios, P.; Bhatta, M.; Halley, M.; Sandro, P.; Gutiérrez, L. Speed breeding and early panicle harvest accelerates oat (Avena sativa L.) breeding cycles. Crop Sci. 2021, 61, 320–330. [Google Scholar] [CrossRef]
- Yao, Y.; Zhang, P.; Wang, H.B.; Lu, Z.Y.; Liu, C.J.; Liu, H.; Yan, G.J. How to advance up to seven generations of canola (Brassica napus L.) per annum for the production of pure line populations? Euphytica 2016, 209, 113–119. [Google Scholar] [CrossRef]
- Bandupriya, H.D.D.; Perera, C.; Pereira, M.G.; Bourdeix, R. Towards Innovative Coconut Breeding Programs; Springer: Cham, Switzerland, 2020; pp. 241–272. [Google Scholar]
- Naresh Kumar, S.; Bai, K. Photo-oxidative stress in coconut seedlings: Early events to leaf scorching and seedling death. Braz. J. Plant Physiol. 2008, 21, 223–232. [Google Scholar] [CrossRef]
- Corley, R.H.V. Potential Productivity of Tropical Perennial Crops. Exp. Agric. 1983, 19, 217–237. [Google Scholar] [CrossRef]
- Gjuvsland, A.B.; Vik, J.O.; Beard, D.A.; Hunter, P.J.; Omholt, S.W. Bridging the genotype-phenotype gap: What does it take? J. Physiol. 2013, 591, 2055–2066. [Google Scholar] [CrossRef] [PubMed]
- N’Cho, Y.P.; Sangare, A.; Bourdeix, R.; Bonnot, F.; Baudouin, L. Assessment of a Few Coconut Ecotypes a Biometric Approach. 1. Study of Tall Populations. Oleagineux 1993, 48, 121–132. [Google Scholar]
- Natarajan, C.K.; Ganesamurthy, K.; Kavitha, M. Genetic variability in coconut (Cocos nucifera). Electron. J. Plant Breed. 2010, 1, 1367–1370. [Google Scholar]
- Friend, D.; Corley, R. Measuring Coconut Palm Dry Matter Production. Exp. Agric. 1994, 30, 223–235. [Google Scholar] [CrossRef]
- Hardon, J.J.; Williams, C.N.; Watson, I.A. Leaf Area and Yield in the Oil Palm in Malaya. Exp. Agric. 1969, 5, 25–32. [Google Scholar] [CrossRef]
- Müller-Linow, M.; Wilhelm, J.; Briese, C.; Wojciechowski, T.; Schurr, U.; Fiorani, F. Plant Screen Mobile: An open-source mobile device app for plant trait analysis. Plant Methods 2019, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Valiaparambil Sebastian, J.S.; Hebbar, K.; Prasad, P.V.V. Phenotyping Tools to Understand Effects of Climate Change; Astral Internati onal Pvt. Ltd.: New Delhi, India, 2016; pp. 169–188. [Google Scholar]
- Nampoothiri, K.U.K.; Parthasarathy, V.A. Varietal Improvement. In The Coconut Palm (Cocos nucifera L.)—Research and Development Perspectives; Krishnakumar, V., Thampan, P.K., Nair, M.A., Eds.; Springer: Singapore, 2018; pp. 113–156. [Google Scholar]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Isabel, N.; Holliday, J.A.; Aitken, S.N. Forest genomics: Advancing climate adaptation, forest health, productivity, and conservation. Evol. Appl. 2020, 13, 3–10. [Google Scholar] [CrossRef]
- Cortés, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern Strategies to Assess and Breed Forest Tree Adaptation to Changing Climate. Front. Plant Sci. 2020, 11, 583323. [Google Scholar] [CrossRef]
- Kumar, S.N.; Kasturi Bai, K.V.; Rajagopal, V.; Aggarwal, P.K. Simulating coconut growth, development and yield with the InfoCrop-coconut model. Tree Physiol. 2008, 28, 1049–1058. [Google Scholar] [CrossRef]
- Rajagopal, V.; Shivashankar, S.; Kasturibai, K.V.; Voleti, S.R. Leaf Water Potential as an Index of Drought Tolerance in Coconut (Cocos nucifera L.). Plant Physiol. Biochem. 1988, 15, 80–86. [Google Scholar]
- Repellin, A.; Daniel, C.; Zuily-Fodil, Y. Merits of physiological tests for characterizing the performance of different coconut varieties subjected to drought. Oleagineux 1994, 49, 155–169. [Google Scholar]
- Gomes, F.; Prado, C.H.B.A. Ecophysiology of coconut palm under water stress. Braz. J. Plant Physiol. 2007, 19, 377–391. [Google Scholar] [CrossRef]
- Cintra, F.L.D.; Passos, E.E.M.; Leal, S. Evaluation of root system distribution in tall coconut cultivars. Oleagineux 1993, 48, 453–461. [Google Scholar]
- Kasturi Bai, K.V.; Rajagopal, V.; Arunachalam, V. Assessment of diversity in coconut varieties for drought responsive physiological traits. J. Plant. Crops 2006, 34, 118–120. [Google Scholar]
- V Rajagopal, K.V.; Bai, K.; Kumar, N. Breeding for drought tolerance in coconut: Status and potentials. In Coconut Genetic Resources; International Plant Genetic Resources Institute-Regional Office for Asia, the Pacific and Oceania (IPGRI-APO): Serdang, Malaysia, 2005. [Google Scholar]
- EPPN, European Plant Phenotyping Network Report. 2016. Available online: https://cordis.europa.eu/project/id/284443/reporting:FORSCHUNGSZENTRUMJULICHGMBH (accessed on 9 March 2022).
- Jonker, F. Next Generation Phenotyping with Uavs. In Laboratory of Geo-Information Science and Remore Sensing; Wageningen University & Research Centre: Wageningen, The Netherlands, 2019. [Google Scholar]
- Barh, D.; Khan, M.S.; Davies, E. PlantOmics: The Omics of Plant Science; Springer: New Delhi, India, 2015. [Google Scholar]
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Entekhabi, D.; Njoku, E.G.; O’Neill, P.E.; Kellogg, K.H.; Crow, W.T.; Edelstein, W.N.; Entin, J.K.; Goodman, S.D.; Jackson, T.J.; Johnson, J.; et al. The Soil Moisture Active Passive (SMAP) Mission. Proc. IEEE 2010, 98, 704–716. [Google Scholar] [CrossRef]
- Fisher, J.B.; Lee, B.; Purdy, A.J.; Halverson, G.H.; Dohlen, M.B.; Cawse-Nicholson, K.; Wang, A.; Anderson, R.G.; Aragon, B.; Arain, M.A.; et al. ECOSTRESS: NASA’s Next Generation Mission to Measure Evapotranspiration from the International Space Station. Water Resour. Res. 2020, 56, e2019WR026058. [Google Scholar] [CrossRef]
- Lee, C.M.; Cable, M.L.; Hook, S.J.; Green, R.O.; Ustin, S.L.; Mandl, D.J.; Middleton, E.M. An introduction to the NASA Hyperspectral InfraRed Imager (HyspIRI) mission and preparatory activities. Remote Sens. Environ. 2015, 167, 6–19. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arumugam, T.; Hatta, M.A.M. Improving Coconut Using Modern Breeding Technologies: Challenges and Opportunities. Plants 2022, 11, 3414. https://doi.org/10.3390/plants11243414
Arumugam T, Hatta MAM. Improving Coconut Using Modern Breeding Technologies: Challenges and Opportunities. Plants. 2022; 11(24):3414. https://doi.org/10.3390/plants11243414
Chicago/Turabian StyleArumugam, Thayalan, and Muhammad Asyraf Md Hatta. 2022. "Improving Coconut Using Modern Breeding Technologies: Challenges and Opportunities" Plants 11, no. 24: 3414. https://doi.org/10.3390/plants11243414
APA StyleArumugam, T., & Hatta, M. A. M. (2022). Improving Coconut Using Modern Breeding Technologies: Challenges and Opportunities. Plants, 11(24), 3414. https://doi.org/10.3390/plants11243414






