Genome-Wide Association Study of Zinc Toxicity Tolerance within a Rice Core Collection (Oryza sativa L.)
Abstract
1. Introduction
2. Results
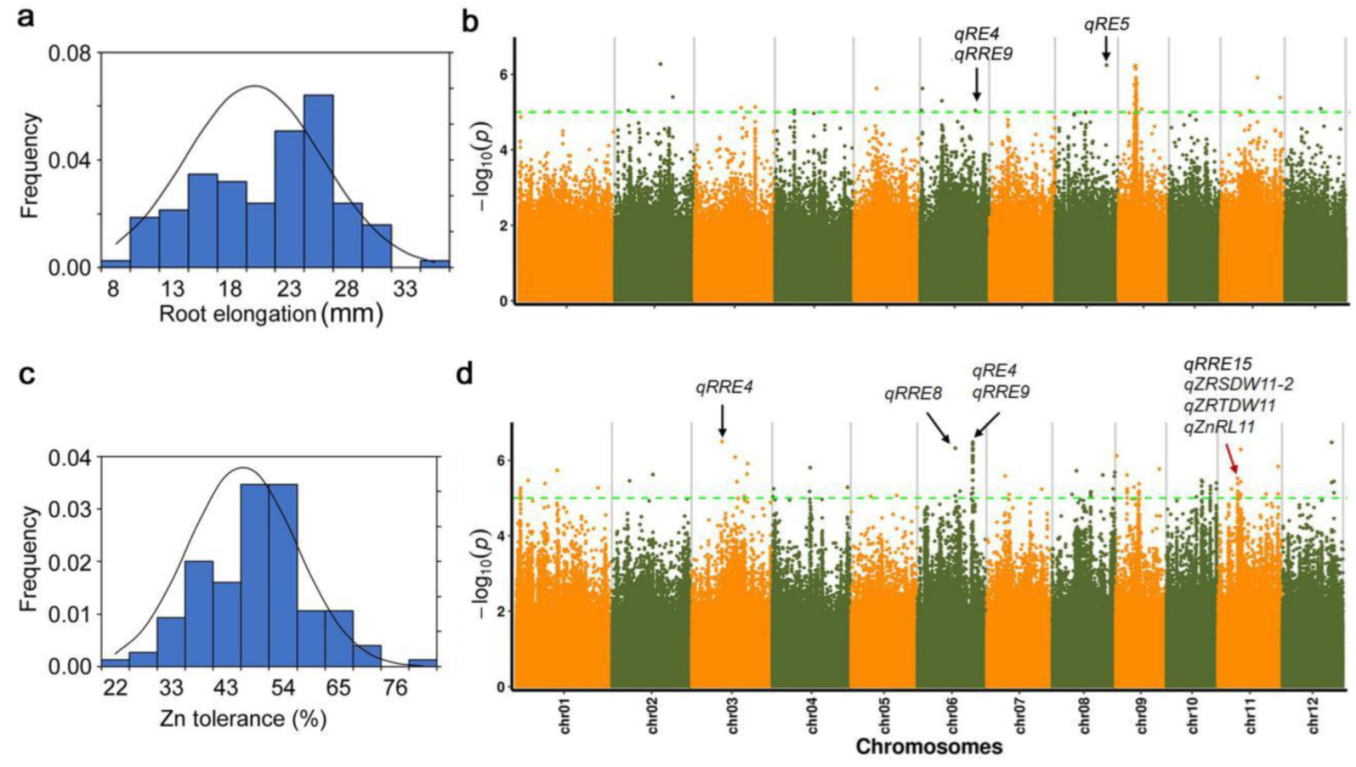
2.1. Identification of Phenotypic Variations for Zn Tolerance within Ting’s Core Collection
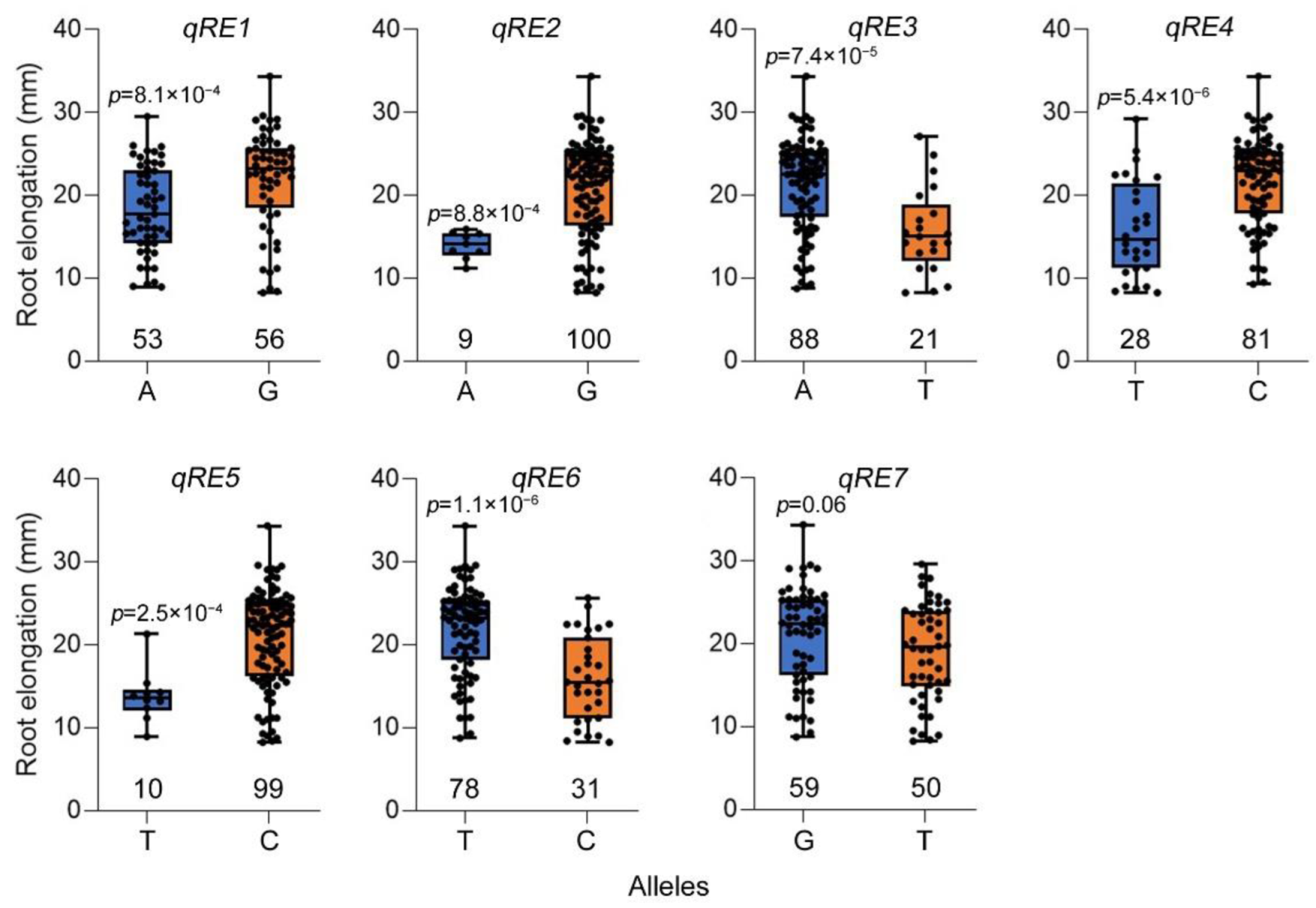
2.2. GWAS for Root Elongation under High Zn Toxicity within Ting’s Core Collection
2.3. GWAS for Relative Root Elongation under High Zn Toxicity within Ting’s Core Collection
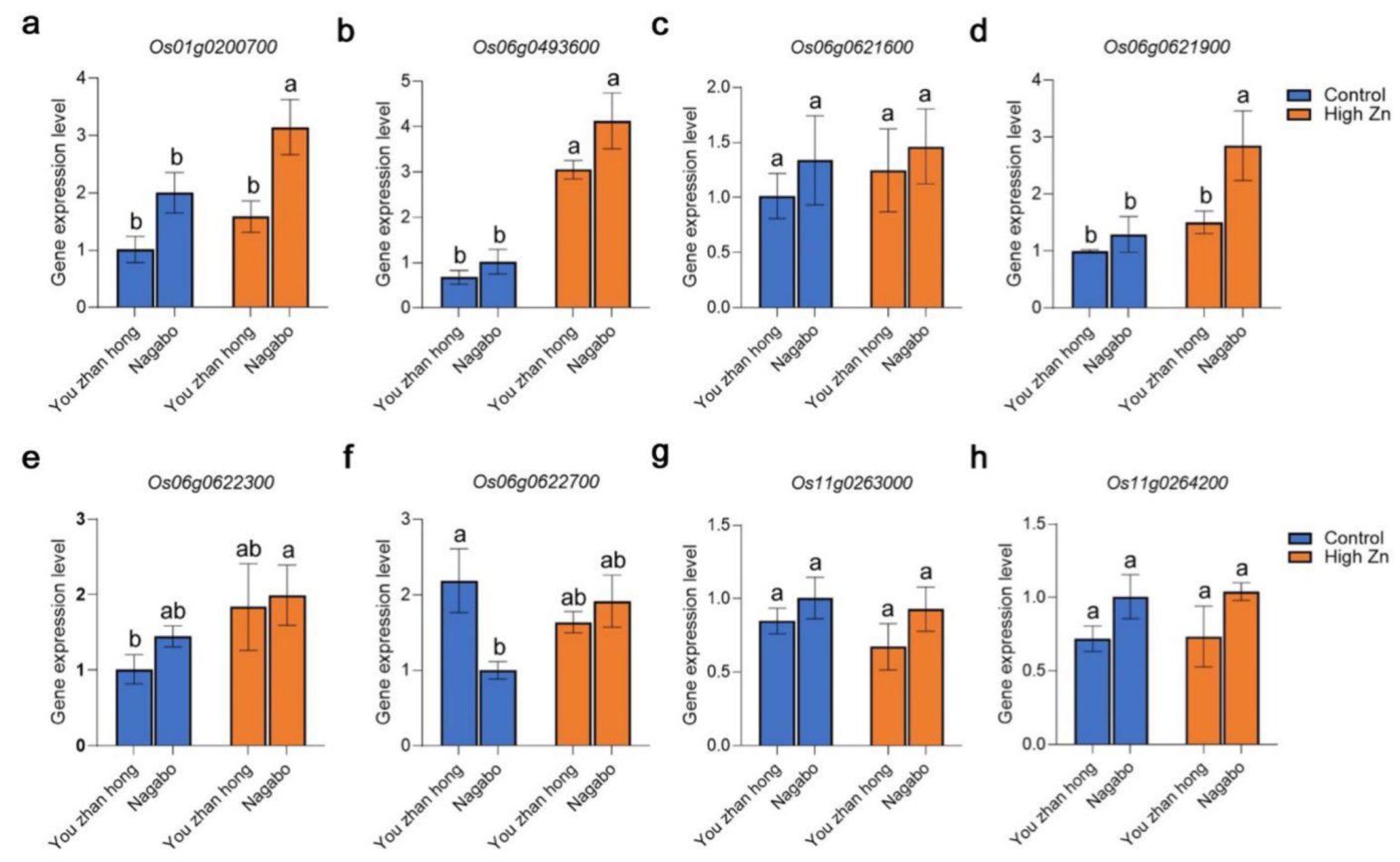
2.4. Candidate Genes Analysis of Detected QTL Responding to Zn Toxicity in the Present Study
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Phenotyping for Zn Toxicity Tolerance
4.3. SNPs Calling
4.4. GWAS Analysis
4.5. qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Broadley, M.R.; White, P.J.; Hammond, J.P.; Zelko, I.; Lux, A. Zinc in plants. New Phytol. 2012, 173, 677–702. [Google Scholar] [CrossRef] [PubMed]
- Chaney, R.L. Zinc phytotoxicity. In Zinc in Soil and Plants; Robson, A.D., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1993; pp. 135–150. [Google Scholar]
- Hussain, D.; Haydon, M.J.; Wang, Y.W.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-Type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef] [PubMed]
- Verret, F.; Gravot, A.; Auroy, P.; Leonhardt, N.; David, P.; Nussaume, L.; Vavasseur, A.; Richaud, P. Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 2004, 576, 306–312. [Google Scholar] [CrossRef]
- Hanikenne, M.; Talke, I.N.; Haydon, M.J.; Lanz, C.; Nolte, A.; Motte, P.; Kroymann, J.; Weigel, D.; Krämer, U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 2008, 453, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Choi, K.S.; Kim, D.Y.; Geisler, M.; Park, J.Y.; Vincenzetti, V.; Schellenberg, M.; Kim, S.H.; Lim, Y.P.; Noh, E.W.; et al. Arabidopsis PCR2 is a zinc exporter involved in both zinc extrusion and long-distance zinc transport. Plant Cell 2010, 22, 2237–2252. [Google Scholar] [CrossRef] [PubMed]
- Kobae, Y.; Uemura, T.; Sato, M.S.; Ohnishi, M.; Mimura, T.; Nakagawa, T.; Maeshima, M. Zinc transporter of Arabidopsis thaliana AtMTP1 is localized to vacuolar membranes and implicated in zinc homeostasis. Plant Cell Physiol. 2004, 45, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Desbrosses-Fonrouge, A.G.; Voigt, K.; Schröder, A.; Arrivault, S.; Thomine, S.; Krämer, U. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005, 579, 4165–4174. [Google Scholar] [CrossRef]
- Arrivault, S.; Senger, T.; Krämer, U. The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 2006, 46, 861–879. [Google Scholar] [CrossRef]
- Haydon, M.J.; Cobbett, C.S. A novel major facilitator superfamily protein at the tonoplast influence zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 2007, 143, 1705–1719. [Google Scholar] [CrossRef]
- Pineau, C.; Loubet, S.; Lefoulon, C.; Chalies, C.; Fizames, C.; Lacombe, B.; Ferrand, M.; Loudet, O.; Berthomieu, P.; Richard, O. Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn Tolerance in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1003120. [Google Scholar] [CrossRef]
- Dong, Y.J.; Ogawa, T.; Lin, D.Z.; Koh, H.J.; Kamiunten, H.; Matsuo, M.; Cheng, S.H. Molecular mapping of quantitative trait loci for zinc toxicity tolerance in rice seedling (Oryza sativa L.). Field Crops Res. 2006, 95, 420–425. [Google Scholar] [CrossRef]
- Song, A.L.; Li, P.; Li, Z.J.; Fan, F.L.; Nikolic, M.; Liang, Y.C. The alleviation of zinc toxicity by silicon is related to zinc transport and antioxidative reactions in rice. Plant Soil 2011, 344, 319–333. [Google Scholar] [CrossRef]
- Liu, H.; Soomro, A.; Zhu, Y.J.; Qiu, X.J.; Chen, K.; Zheng, T.Q.; Yang, L.W.; Xing, D.Y.; Xu, J.L. QTL underlying iron and zinc toxicity tolerances at seedling stage revealed by two sets of reciprocal introgression populations of rice (Oryza sativa L.). Crop. J. 2016, 4, 280–289. [Google Scholar] [CrossRef]
- Meng, L.J.; Wang, B.X.; Zhao, X.Q.; Ponce, K.; Qian, Q.; Ye, G.Y. Association mapping of ferrous, zinc, and aluminum tolerance at the seedling stage in indica rice using MAGIC populations. Front. Plant Sci. 2017, 8, 1822. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, K.; Pang, Y.L.; Naveed, S.A.; Zhao, X.Q.; Wang, X.Q.; Wang, Y.; Dingkuhn, M.; Pasuquin, J.; Li, Z.K.; et al. QTL mapping and candidate gene analysis of ferrous iron and zinc toxicity tolerance at seedling stage in rice by genome-wide association study. BMC Genom. 2017, 18, 828. [Google Scholar] [CrossRef]
- Wilkins, D.A. The measurement of tolerance to edaphic factors by means of root growth. New Phytol. 1978, 80, 623–633. [Google Scholar] [CrossRef]
- Platre, M.P.; Satbhai, S.B.; Brent, L.; Gleason, M.F.; Cao, M.; Grison, M.; Glavier, M.; Zhang, L.; Gaillochet, C.; Goeschl, C.; et al. The receptor kinase SRF3 coordinates iron-level and flagellin dependent defense and growth responses in plants. Nat. Commun. 2022, 13, 4445. [Google Scholar] [CrossRef]
- Li, B.H.; Sun, L.; Huang, J.Y.; Göschl, C.; Shi, W.M.; Chory, J.; Busch, W. GSNOR provides plant tolerance to iron toxicity via preventing iron-dependent nitrosative and oxidative cytotoxicity. Nat. Commun. 2019, 10, 3896. [Google Scholar] [CrossRef]
- Satbhai, S.; Setzer, C.; Freynschlag, F.; Slovak, R.; Kerdaffrec, E.; Busch, W. Natural allelic variation of FRO2 modulates Arabidopsis root growth under iron deficiency. Nat. Commun. 2017, 8, 15603. [Google Scholar] [CrossRef]
- Li, X.L.; Lu, Y.G.; Li, J.Q.; Muhammad, Q. Strategies on sample size determination and qualitative and quantitative traits integration to construct core collection of rice (Oryza sativa). Rice Sci. 2011, 18, 46–55. [Google Scholar] [CrossRef]
- Zhang, P.; Zhong, K.Z.; Zhong, Z.Z.; Tong, H.H. Mining candidate gene for rice aluminum tolerance through genome wide association study and transcriptomic analysis. BMC Plant Biol. 2019, 19, 490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhong, K.Z.; Tong, H.H.; Qasim, M.; Li, J.Q. Association mapping for aluminum tolerance in a core collection of rice landraces. Front. Plant Sci. 2016, 7, 1415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhong, K.Z.; Zhong, Z.Z.; Tong, H.H. Genome-wide associated study of important agronomic traits within a core collection of rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 259. [Google Scholar]
- Fu, D.; Zhong, K.Z.; Zhong, Z.Z.; Hu, G.C.; Zhang, P.; Tong, H.H. Genome-wide association study of sheath blight resistance within a core collection of rice (Oryza sativa L.). Agronomy 2022, 12, 1493. [Google Scholar] [CrossRef]
- Lee, S.; Chiecko, J.C.; Kim, S.A.; Walker, E.L.; Lee, Y.S.; Guerinot, M.L.; An, G. Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 2009, 150, 786–800. [Google Scholar] [CrossRef]
- Inoue, H.; Kobayashi, T.; Nozoye, T.; Takahashi, M.; Kakei, Y.; Suzuki, K.; Nakazono, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 2009, 284, 3470–3479. [Google Scholar] [CrossRef]
- Shahpiri, A.; Soleimanifard, I.; Asadollahi, M.A. Functional characterization of a type 3 methallolthionein isoform (OsMTI-3a) from rice. Int. J. Biol. Macromol. 2015, 73, 154–159. [Google Scholar] [CrossRef]
- Ai, P.H.; Sun, S.B.; Zhao, J.N.; Fan, X.R.; Xin, W.J.; Guo, Q.; Yu, L.; Shen, Q.R.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6 have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef]
- Kaur, H.; Garg, N. Zinc toxicity in plants: A review. Planta 2021, 253, 129. [Google Scholar] [CrossRef]
- Curie, C.; Cassin, G.; Couch, D.; Divol, F.; Higuchi, K.; Jean, M.L.; Misson, J.; Schikora, A.; Czernic, P.; Mari, S. Metal movement within the plant: Contribution of nicotianamine and yellow stripe 1-like transporter. Ann. Bot. 2009, 103, 1–11. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, Y.R.; Li, Y.; Ling, H.Q.; Chu, C.C. OsMT1a, a type 1 metallothionein, plays the pivotal role in zinc homeostasis and drought tolerance in rice. Plant Mol. Biol. 2009, 70, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ma, J.F. Silicon suppresses zinc uptake through down-regulating zinc transporter gene in rice. Physiol. Plantarum 2020, 170, 580–591. [Google Scholar] [CrossRef] [PubMed]


| Trait | Range | Mean ± SD |
|---|---|---|
| Root elongation (RE, control) | 16.71–56.12 (mm) | 43.54 ± 8.95 (mm) |
| Root elongation (RE, under 120 μM Zn) | 8.25–34.39 (mm) | 20.16 ± 5.91 (mm) |
| Relative root elongation (RRE, %) | 21.86–78.19 (%) | 46.42 ± 10.48 (%) |
| QTL | Chromosome | Position | p-Value | Allele (the Reference/the Alternative) |
|---|---|---|---|---|
| qRE1 | chr02 | 20,681,680 | 5.34 × 10−7 | A/G |
| qRE2 | chr02 | 26,288,978 | 3.98 × 10−6 | A/G |
| qRE3 | chr06 | 1,118,739 | 2.35 × 10−6 | A/T |
| qRE4 | chr06 | 25,006,144 | 9.09 × 10−6 | T/C |
| qRE5 | chr08 | 23,227,944 | 5.70 × 10−7 | T/C |
| qRE6 | chr09 | 7,341,101 | 8.07 × 10−8 | T/C |
| qRE7 | chr11 | 27,019,946 | 4.12 × 10−6 | G/T |
| QTL | Chromosome | Position | p-Value | Allele (the Reference/the Alternative) | Previously Reported QTL |
|---|---|---|---|---|---|
| qRRE1 | chr01 | 5,483,287 | 3.43 × 10−6 | A/G | OsMTI-3a [28] |
| qRRE2 | chr01 | 18,578,810 | 1.84 × 10−6 | A/G | |
| qRRE3 | chr02 | 7,969,392 | 3.50 × 10−6 | G/A | |
| qRRE4 | chr03 | 13,749,414 | 3.16 × 10−7 | G/A | |
| qRRE5 | chr03 | 19,713,560 | 8.19 × 10−7 | C/T | |
| qRRE6 | chr03 | 25,346,195 | 1.22 × 10−6 | T/C | |
| qRRE7 | chr04 | 17,157,142 | 1.56 × 10−6 | G/A | |
| qRRE8 | chr06 | 17,185,339 | 4.76 × 10−7 | A/G | |
| qRRE9 | chr06 | 25,053,430 | 3.29 × 10−7 | G/A | |
| qRRE10 | chr07 | 8,451,115 | 2.59 × 10−6 | C/T | |
| qRRE11 | chr08 | 22,766,099 | 2.45 × 10−6 | T/C | |
| qRRE12 | chr08 | 28,240,531 | 2.06 × 10−6 | T/G | |
| qRRE13 | chr09 | 5,323,335 | 2.45 × 10−6 | C/T | |
| qRRE14 | chr10 | 22,657,213 | 3.96 × 10−6 | A/T | |
| qRRE15 | chr11 | 8,988,532 | 3.02 × 10−6 | C/G | qZRSDW11-2 qZRTDW11 [14]; qZnRL11 [15] |
| qRRE16 | chr11 | 10,391,796 | 5.15 × 10−7 | C/T | |
| qRRE17 | chr11 | 27,270,465 | 1.45 × 10−6 | C/T | |
| qRRE18 | chr12 | 22,393,995 | 3.33 × 10−7 | G/A | |
| qRRE19 | chr12 | 23,423,605 | 3.60 × 10−6 | A/G |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, K.; Xie, L.; Hu, S.; Shao, G.; Sheng, Z.; Jiao, G.; Wang, L.; Chen, Y.; Tang, S.; Wei, X.; et al. Genome-Wide Association Study of Zinc Toxicity Tolerance within a Rice Core Collection (Oryza sativa L.). Plants 2022, 11, 3138. https://doi.org/10.3390/plants11223138
Zhong K, Xie L, Hu S, Shao G, Sheng Z, Jiao G, Wang L, Chen Y, Tang S, Wei X, et al. Genome-Wide Association Study of Zinc Toxicity Tolerance within a Rice Core Collection (Oryza sativa L.). Plants. 2022; 11(22):3138. https://doi.org/10.3390/plants11223138
Chicago/Turabian StyleZhong, Kaizhen, Lihong Xie, Shikai Hu, Gaoneng Shao, Zhonghua Sheng, Guiai Jiao, Ling Wang, Ying Chen, Shaoqing Tang, Xiangjin Wei, and et al. 2022. "Genome-Wide Association Study of Zinc Toxicity Tolerance within a Rice Core Collection (Oryza sativa L.)" Plants 11, no. 22: 3138. https://doi.org/10.3390/plants11223138
APA StyleZhong, K., Xie, L., Hu, S., Shao, G., Sheng, Z., Jiao, G., Wang, L., Chen, Y., Tang, S., Wei, X., Zhang, P., & Hu, P. (2022). Genome-Wide Association Study of Zinc Toxicity Tolerance within a Rice Core Collection (Oryza sativa L.). Plants, 11(22), 3138. https://doi.org/10.3390/plants11223138









