Genome-Wide Identification and Expression Analysis of eIF Family Genes from Brassica rapa in Response to TuMV Resistance
Abstract
1. Introduction
2. Results
2.1. Identification of eIF Family Genes in B. rapa and Arabidopsis thaliana
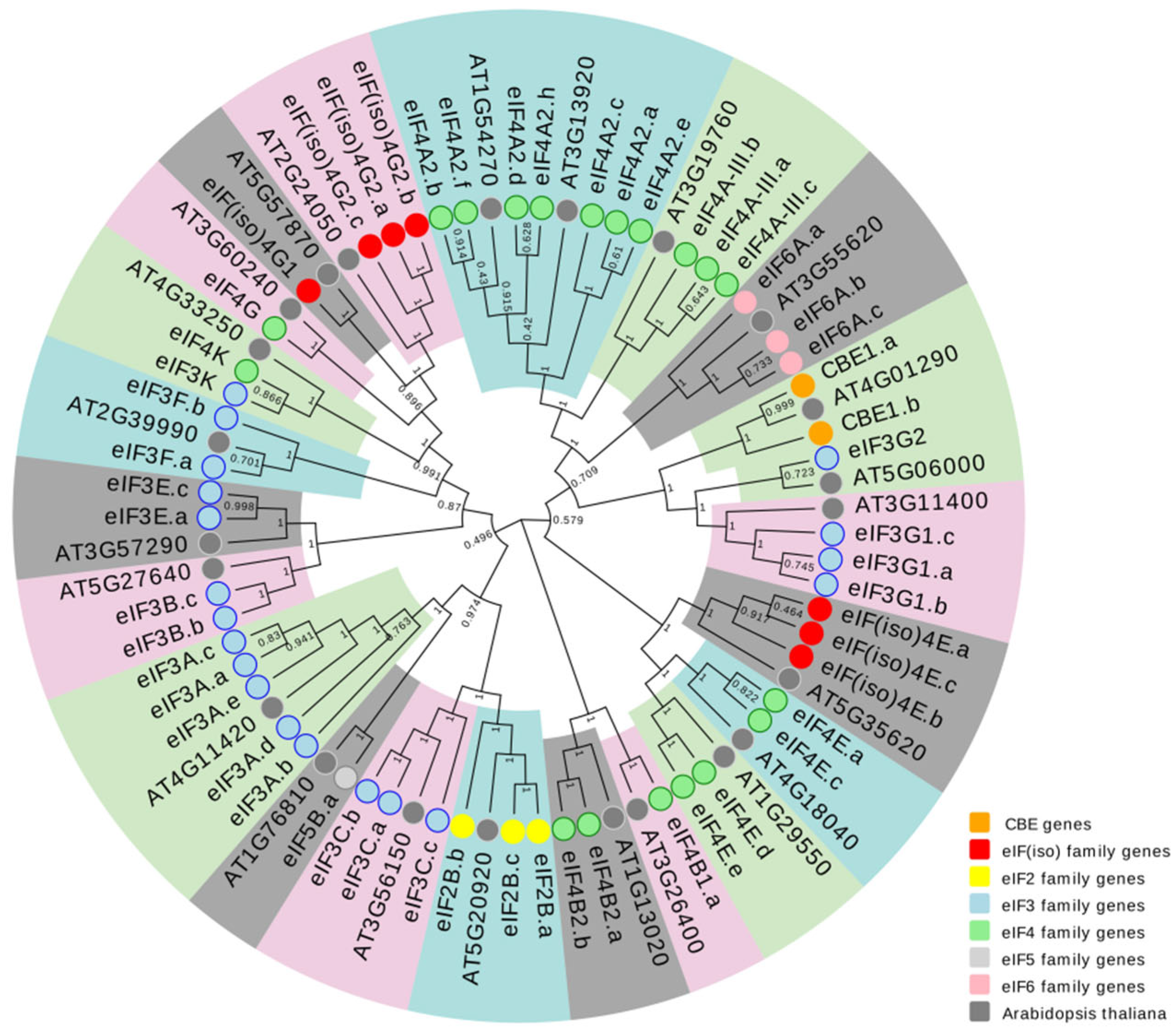
2.2. Phylogenetic Analysis of eIF Family Genes Sequences
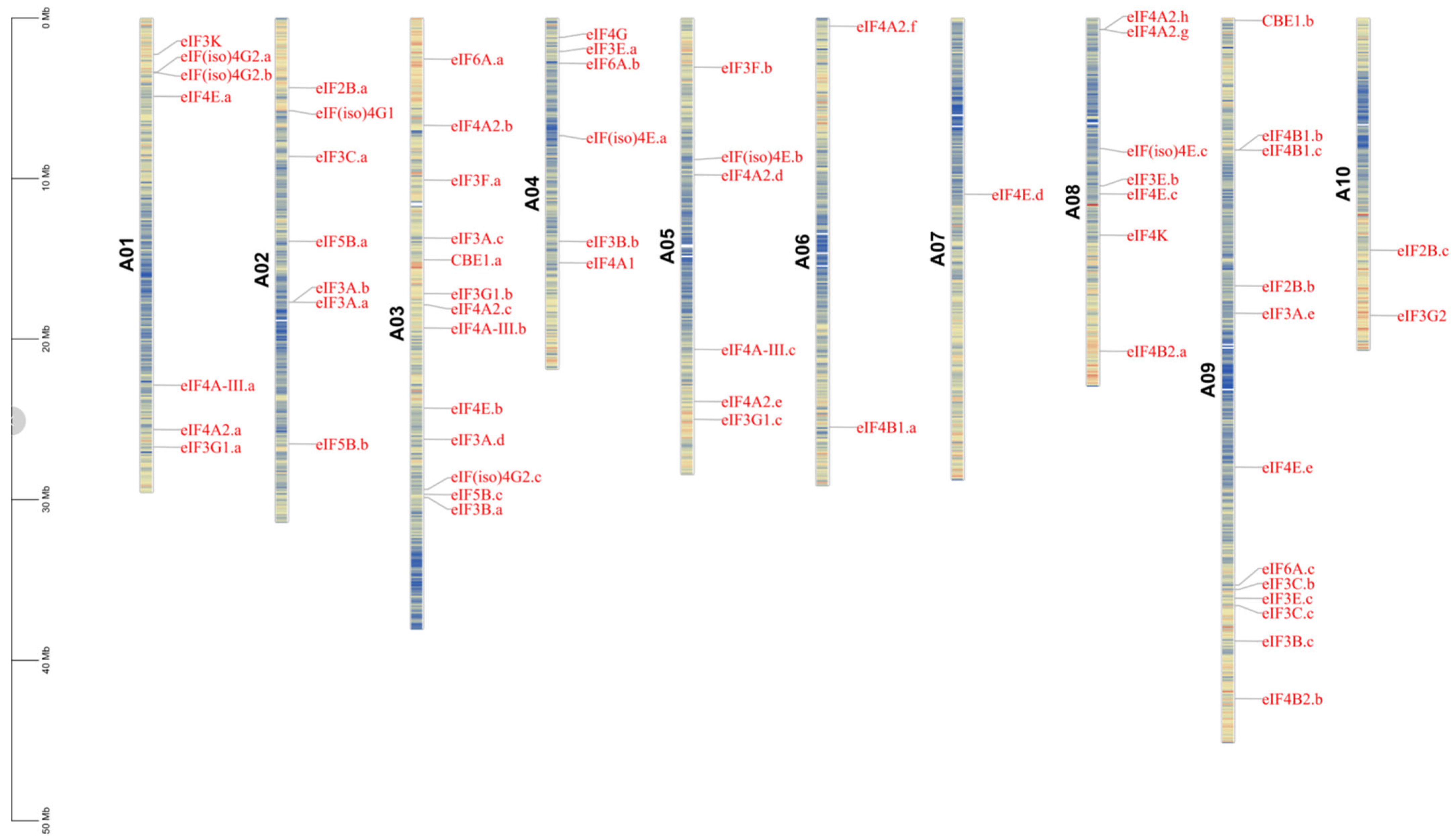
2.3. Chromosomal Distribution of eIF Family Genes in B. rapa
2.4. Identification of the Physicochemical Properties of eIF Family Genes
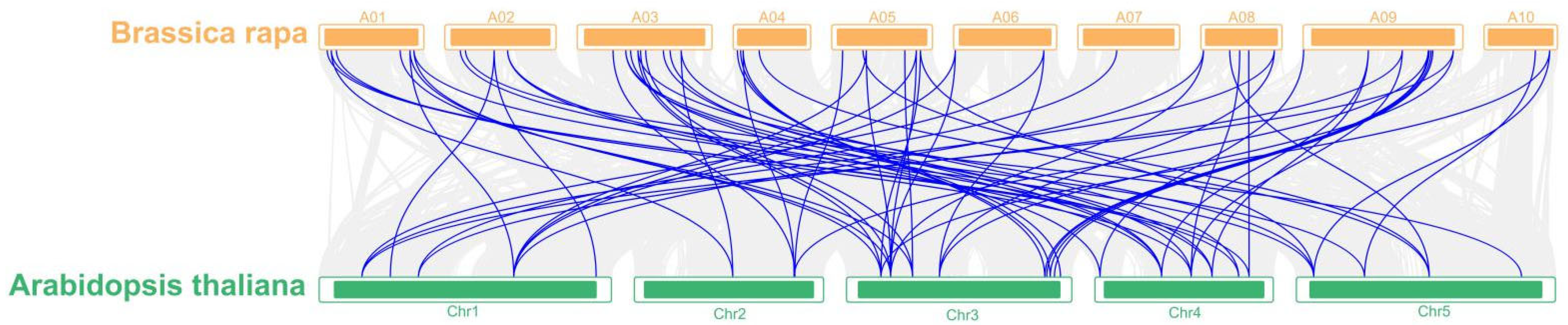
2.5. Evolutionary Analysis of eIF Family Genes in B. rapa
2.6. Conserved Analysis of the Gene Sequences and Gene Structures
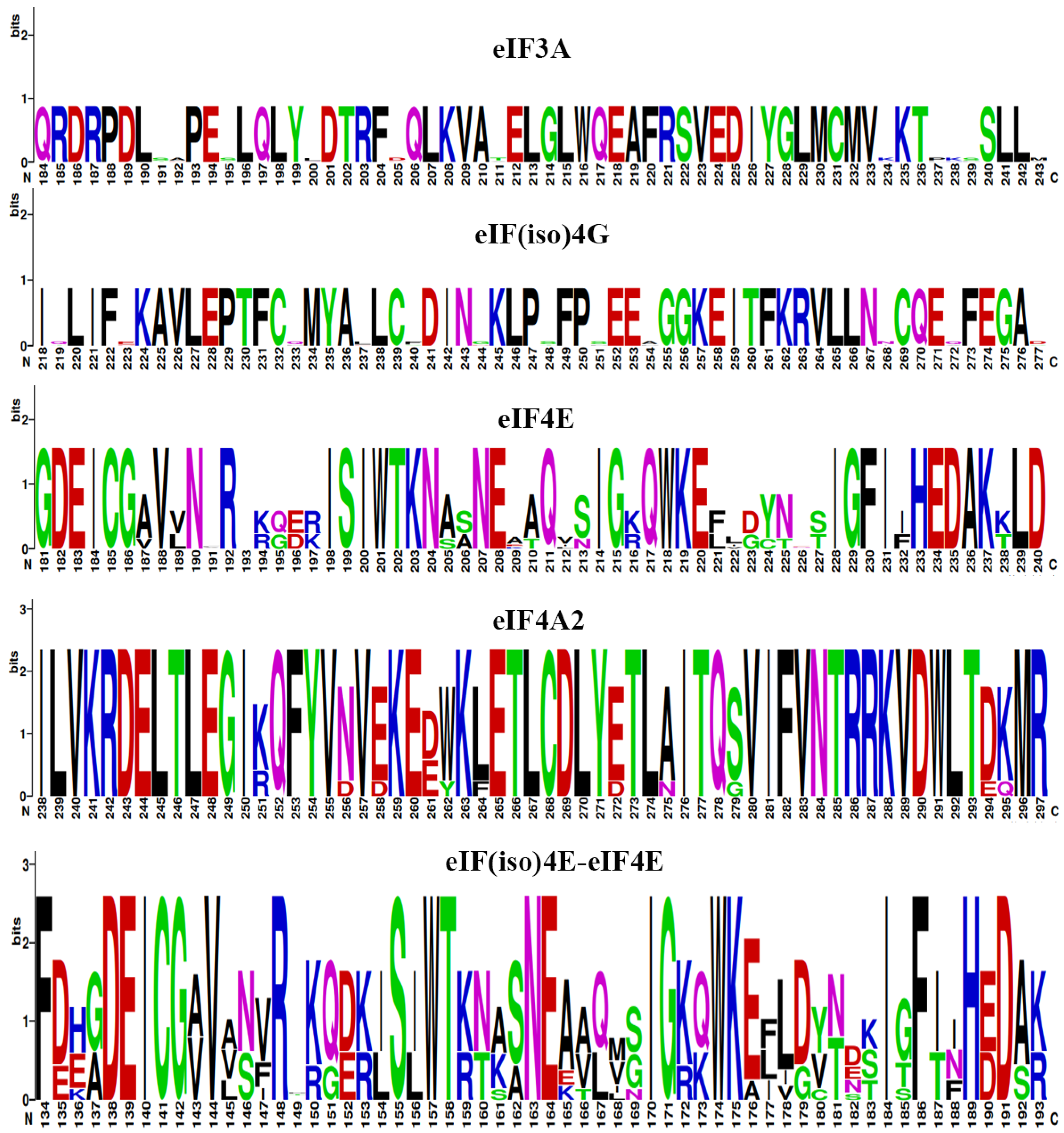
2.7. Three-Dimensional (3D) Structure and Sequence Logo Analyses of eIF Proteins
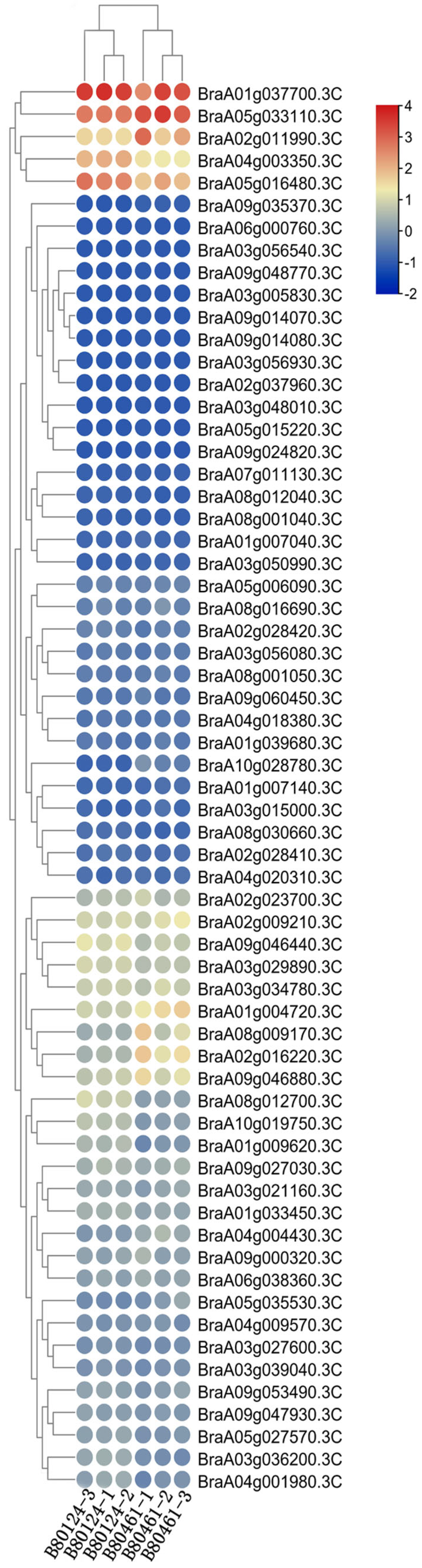
2.8. Expression Characterization of eIF Family Genes in Resistant/Susceptible Brassica rapa Lines
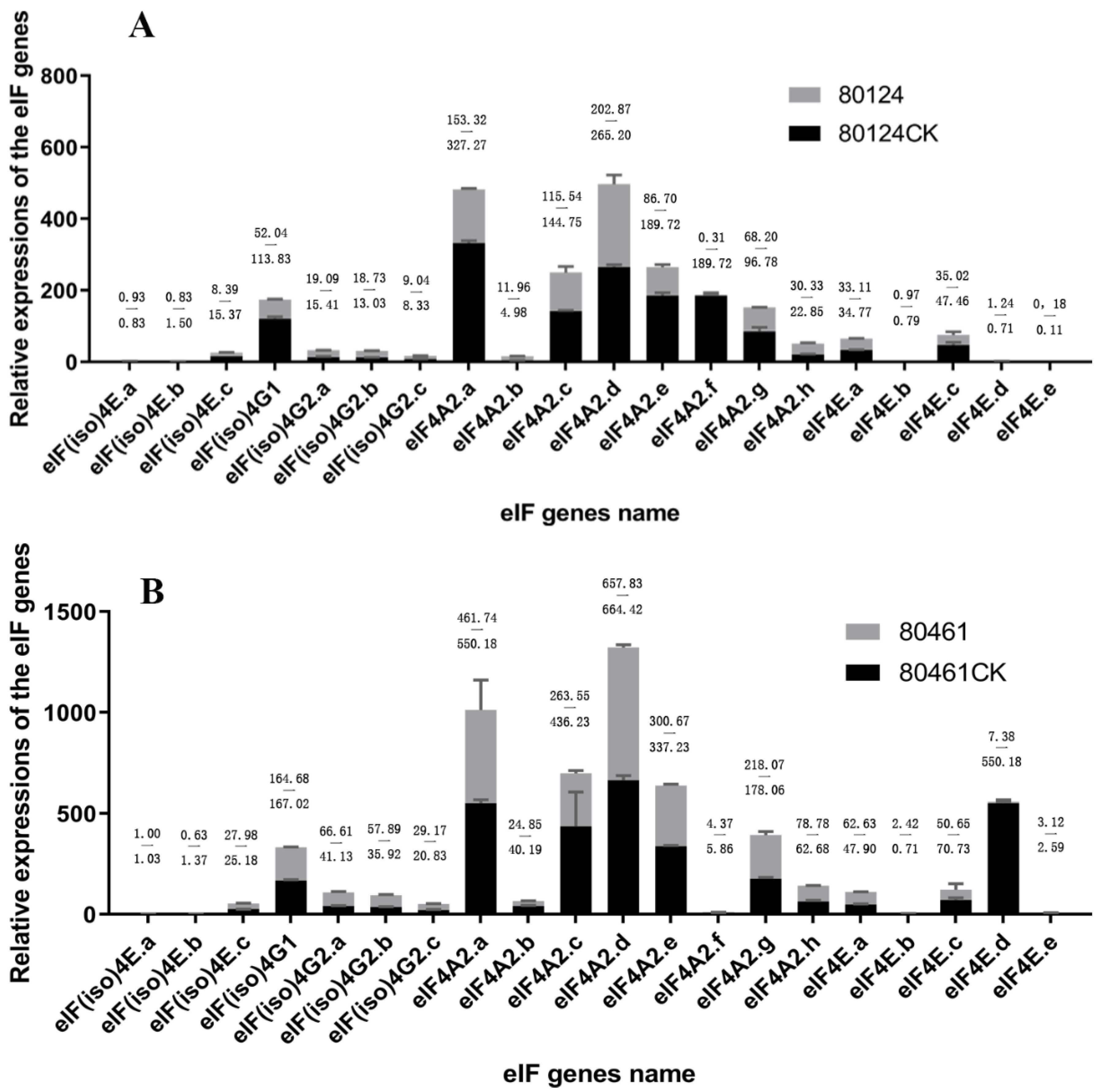
2.9. qRT-PCR Analyses of eIF Family Genes in Resistant/Susceptible B. rapa Lines
2.10. Differential Metabolites Analysis between the Susceptible and Resistant B. rapa Lines
3. Discussion
3.1. Evolution of eIF Family Genes
3.2. eIF Family Genes as Important Resistance Factors against Viruses in Plants
3.3. Effects of the Key Amino Acids at 3D Structure on Resistance
4. Materials and Methods
4.1. Plant Materials and TuMV Inoculation
4.2. Identification and Characterization of eIF Family Genes in A. thaliana and B. rapa
4.3. Phylogenetic Analyses of eIF Family Genes
4.4. Chromosome Distribution of eIF Family Genes
4.5. Analysis of Conserved Motifs, Gene Structures, and Cis-Acting Elements
4.6. Transcriptome Data Analysis, Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR), and Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Browning, K.S.; Bailey-Serres, J. Mechanism of cytoplasmic mRNA translation. Arabidopsis Book 2015, 13, e0176. [Google Scholar] [CrossRef] [PubMed]
- Joshi, B.; Lee, K.; Maeder, D.L.; Jagus, R. Phylogenetic analysis of eIF4E-family members. BMC Evol. Biol. 2005, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Mayberry, L.K.; Allen, M.L.; Dennis, M.D.; Browning, K.S. Evidence for variation in the optimal translation initiation complex: Plant eIF4B, eIF4F, and eIF(iso)4F differentially promote translation of mRNAs. Plant Physiol. 2009, 150, 1844–1854. [Google Scholar] [CrossRef] [PubMed]
- Kaye, N.M.; Emmett, K.J.; Merrick, W.C.; Jankowsky, E. Intrinsic RNA binding by the eukaryotic initiation factor 4F depends on a minimal RNA length but not on the m7G cap. J. Biol. Chem. 2009, 284, 17742–17750. [Google Scholar] [CrossRef]
- Rhoads, R.E. eIF4E: New family members, new binding partners, new roles. J. Biol. Chem. 2009, 284, 16711–16715. [Google Scholar] [CrossRef]
- Hernandez, G.; Vazquez-Pianzola, P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 2005, 122, 865–876. [Google Scholar] [CrossRef] [PubMed]
- Syntichaki, P.; Troulinaki, K.; Tavernarakis, N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 2007, 445, 922–926. [Google Scholar] [CrossRef]
- Sanfacon, H. Plant Translation Factors and Virus Resistance. Viruses 2015, 7, 3392–3419. [Google Scholar] [CrossRef]
- Patrick, R.M.; Browning, K.S. The eIF4F and eIFiso4F Complexes of Plants: An Evolutionary Perspective. Comp. Func.t Genom. 2012, 2012, 287812–287814. [Google Scholar] [CrossRef]
- Rhoads, R.E.; Dinkova, T.D.; Jagus, R. Approaches for Analyzing the Differential Activities and Functions of eIF4E Family Members. Methods Enzymol. 2007, 429, 261–297. [Google Scholar] [CrossRef]
- Sato, M.; Nakahara, K.; Yoshii, M.; Ishikawa, M.; Uyeda, I. Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. Febs Lett. 2005, 579, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Jenner, C.E.; Nellist, C.F.; Barker, G.C.; Walsh, J.A. Turnip mosaic virus (TuMV) Is Able to Use Alleles of Both eIF4E and eIF(iso)4E from Multiple Loci of the Diploid Brassica rapa. Mol. Plant-Microbe Interact. 2010, 23, 1498–1505. [Google Scholar] [CrossRef] [PubMed]
- Lellis, A.D.; Kasschau, K.D.; Whitham, S.A.; Carrington, J.C. Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr. Biol. 2002, 12, 1046–1051. [Google Scholar] [CrossRef]
- Yoshii, M.; Nishikiori, M.; Tomita, K.; Yoshioka, N.; Kozuka, R.; Naito, S.; Ishikawa, M. The Arabidopsis Cucumovirus multiplication 1 and 2 loci encode tip translation initiation factors 4E and 4G. J. Virol. 2004, 78, 6102–6111. [Google Scholar] [CrossRef]
- Kim, J.; Kang, W.H.; Hwang, J.; Yang, H.B.; Dosun, K.; Oh, C.S.; Kang, B.C. Transgenic Brassica rapa plants over-expressing eIF(iso)4E variants show broad-spectrum Turnip mosaic virus (TuMV) resistance. Mol. Plant Pathol. 2014, 15, 615–626. [Google Scholar] [CrossRef]
- Nellist, C.F.; Qian, W.; Jenner, C.E.; Moore, J.D.; Zhang, S.J.; Wang, X.W.; Briggs, W.H.; Barker, G.C.; Sun, R.F.; Walsh, J.A. Multiple copies of eukaryotic translation initiation factors in Brassica rapa facilitate redundancy, enabling diversification through variation in splicing and broad-spectrum virus resistance. Plant J. 2014, 77, 261–268. [Google Scholar] [CrossRef]
- Rusholme, R.L.; Higgins, E.E.; Walsh, J.A.; Lydiate, D.J. Genetic control of broad-spectrum resistance to turnip mosaic virus in Brassica rapa (Chinese cabbage). J. General Virol. 2007, 88, 3177–3186. [Google Scholar] [CrossRef]
- Rubio, M.; Nicolai, M.; Caranta, C.; Palloix, A. Allele mining in the pepper gene pool provided new complementation effects between pvr2-eIF4E and pvr6-eIF(iso)4E alleles for resistance to pepper veinal mottle virus. J. General Virol. 2009, 90, 2808–2814. [Google Scholar] [CrossRef]
- Ayme, V.; Petit-Pierre, J.; Souche, S.; Palloix, A.; Moury, B. Molecular dissection of the potato virus YVPg virulence factor reveals complex adaptations to the pvr2 resistance allelic series in pepper. J. General Virol. 2007, 88, 1594–1601. [Google Scholar] [CrossRef]
- Ayme, V.; Souche, S.; Caranta, C.; Jacquemond, M.; Chadoeuf, J.; Palloix, A.; Moury, B. Different mutations in the genome-linked protein VPg of Potato virus Y confer virulence on the pvr2(3) resistance in pepper. Mol. Plant-Microbe Interact. 2006, 19, 557–563. [Google Scholar] [CrossRef]
- Moury, B.; Morel, C.; Johansen, E.; Guilbaud, L.; Souche, S.; Ayme, V.; Caranta, C.; Palloix, A.; Jacquemond, M. Mutations in Potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant-Microbe Interact. 2004, 17, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Ling, K.S.; Harris, K.R.; Meyer, J.D.F.; Levi, A.; Guner, N.; Wehner, T.C.; Bendahmane, A.; Havey, M.J. Non-synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to Zucchini yellow mosaic virus. Theor. Appl. Genet. 2009, 120, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Perovic, D.; Kramer, I.; Habekuss, A.; Perner, K.; Pickering, R.; Proeseler, G.; Kanyuka, K.; Ordon, F. Genetic analyses of BaMMV/BaYMV resistance in barley accession HOR4224 result in the identification of an allele of the translation initiation factor 4e (Hv-eIF4E) exclusively effective against Barley mild mosaic virus (BaMMV). Theor. Appl. Genet. 2014, 127, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- German-Retana, S.; Walter, J.; Doublet, B.; Roudet-Tavert, G.; Nicaise, V.; Lecampion, C.; Houvenaghel, M.C.; Robaglia, C.; Michon, T.; Le Gall, O. Mutational analysis of plant cap-binding protein eIF4E reveals key amino acids involved in biochemical functions and potyvirus infection. J. Virol. 2008, 82, 7601–7612. [Google Scholar] [CrossRef]
- Lee, J.H.; Muhsin, M.; Atienza, G.A.; Kwak, D.Y.; Kim, S.M.; De Leon, T.B.; Angeles, E.R.; Coloquio, E.; Kondoh, H.; Satoh, K.; et al. Single Nucleotide Polymorphisms in a Gene for Translation Initiation Factor (eIF4G) of Rice (Oryza sativa) Associated with Resistance to Rice tungro spherical virus. Mol. Plant-Microbe Interact. 2010, 23, 29–38. [Google Scholar] [CrossRef]
- Hart, J.P.; Griffiths, P.D. A series of eIF4E alleles at the Bc-3 locus are associated with recessive resistance to Clover yellow vein virus in common bean. Theor. Appl. Genet. 2013, 126, 2849–2863. [Google Scholar] [CrossRef]
- Konecna, E.; Safarova, D.; Navratil, M.; Hanacek, P.; Coyne, C.; Flavell, A.; Vishnyakova, M.; Ambrose, M.; Redden, R.; Smykal, P. Geographical Gradient of the eIF4E Alleles Conferring Resistance to Potyviruses in Pea (Pisum) Germplasm. PLoS ONE 2014, 9, 90394. [Google Scholar] [CrossRef]
- Marandel, G.; Salava, J.; Abbott, A.; Candresse, T.; Decroocq, V. Quantitative trait loci meta-analysis of Plum pox virus resistance in apricot (Prunus armeniaca L.): New insights on the organization and the identification of genomic resistance factors. Mol. Plant Pathol. 2009, 10, 347–360. [Google Scholar] [CrossRef]
- Wang, X.H.; Kohalmi, S.E.; Svircev, A.; Wang, A.M.; Sanfacon, H.; Tian, L.N. Silencing of the Host Factor eIF(iso)4E Gene Confers Plum Pox Virus Resistance in Plum. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Poulicard, N.; Pinel-Galzi, A.; Fargette, D.; Hebrard, E. Alternative mutational pathways, outside the VPg, of rice yellow mottle virus to overcome eIF(iso)4G-mediated rice resistance under strong genetic constraints. J. General Virol. 2014, 95, 219–224. [Google Scholar] [CrossRef]
- Thiemele, D.; Boisnard, A.; Ndjiondjop, M.N.; Cheron, S.; Sere, Y.; Ake, S.; Ghesquiere, A.; Albar, L. Identification of a second major resistance gene to Rice yellow mottle virus, RYMV2, in the African cultivated rice species, O. glaberrima. Theor. Appl. Genet. 2010, 121, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Traore, O.; Pinel-Galzi, A.; Issaka, S.; Poulicard, N.; Aribi, J.; Ake, S.; Ghesquiere, A.; Sere, Y.; Konate, G.; Hebrard, E.; et al. The adaptation of Rice yellow mottle virus to the eIF(iso)4G-mediated rice resistance. Virology 2010, 408, 103–108. [Google Scholar] [CrossRef]
- Ruffel, S.; Gallois, J.L.; Lesage, M.L.; Caranta, C. The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genom. 2005, 274, 346–353. [Google Scholar] [CrossRef] [PubMed]
- Gallie, D.R.; Browning, K.S. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J. Biol. Chem. 2001, 276, 36951–36960. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.A.; Sharpe, A.G.; Jenner, C.E.; Lydiate, D.J. Characterisation of resistance to turnip mosaic virus in oilseed rape (Brassica napus) and genetic mapping of TuRB01. Theor. Appl. Genet. 1999, 99, 1149–1154. [Google Scholar] [CrossRef]
- Hughes, S.L.; Hunter, P.J.; Sharpe, A.G.; Kearsey, M.J.; Lydiate, D.J.; Walsh, J.A. Genetic mapping of the novel Turnip mosaic virus resistance gene TuRB03 in Brassica napus. Theor. Appl. Genet. 2003, 107, 1169–1173. [Google Scholar] [CrossRef]
- Jenner, C.E.; Tomimura, K.; Ohshima, K.; Hughes, S.L.; Walsh, J.A. Mutations in Turnip mosaic virus P3 and Cylindrical Inclusion Proteins Are Separately Required to Overcome Two Brassica napus Resistance Genes. Virology 2002, 300, 50–59. [Google Scholar] [CrossRef][Green Version]
- Jenner, C.E.; Xiaowu, W.; Tomimura, K.; Ohshima, K.; Ponz, F.; Walsh, J.A. The dual role of the potyvirus P3 protein of Turnip mosaic virus as a symptom and avirulence determinant in brassicas. Mol. Plant Microbe Interact. 2003, 16, 777–784. [Google Scholar] [CrossRef]
- Qian, W.; Zhang, S.; Zhang, S.; Li, F.; Zhang, H.; Wu, J.; Wang, X.; Walsh, J.A.; Sun, R. Mapping and candidate-gene screening of the novel Turnip mosaic virus resistance gene retr02 in Chinese cabbage (Brassica rapa L.). Theor. Appl. Genet. 2012, 126, 179–188. [Google Scholar] [CrossRef]
- Shopan, J.; Mou, H.; Zhang, L.; Zhang, C.; Ma, W.; Walsh, J.A.; Hu, Z.; Yang, J.; Zhang, M. Eukaryotic translation initiation factor 2B-beta (eIF2Bβ), a new class of plant virus resistance gene. Plant J. 2017, 90, 929–940. [Google Scholar] [CrossRef]
- Li, G.; Zhang, S.; Li, F.; Zhang, H.; Zhang, S.; Zhao, J.; Sun, R. Variability in the Viral Protein Linked to the Genome of Turnip Mosaic Virus Influences Interactions with eIF(iso)4Es in Brassica rapa. The Plant Pathol. J. 2021, 37, 47–56. [Google Scholar] [CrossRef]
- Li, G.L.; Qian, W.; Zhang, S.J.; Zhang, S.F.; Li, F.; Zhang, H.; Fang, Z.Y.; Wu, J.; Wang, X.W.; Sun, R.F. Variability in eukaryotic initiation factor iso4E in Brassica rapa influences interactions with the viral protein linked to the genome of Turnip mosaic virus. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Sun, R.; Bonnema, A.B. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- des Georges, A.; Dhote, V.; Kuhn, L.; Hellen, C.U.T.; Pestova, T.V.; Frank, J.; Hashem, Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 2015, 525, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Kuhle, B.; Ficner, R. e IF 5 B employs a novel domain release mechanism to catalyze ribosomal subunit joining. EMBO J. 2014, 33, 1177–1191. [Google Scholar] [CrossRef]
- Mössner, E.; Huber-Wunderlich, M.; Rietsch, A.; Beckwith, J.; Glockshuber, R.; Åslund, F. Importance of Redox Potential for the in Vivo Function of the Cytoplasmic Disulfide Reductant Thioredoxin from Escherichia coli. J. Biol. Chem. 1999, 274, 25254–25259. [Google Scholar] [CrossRef]
- Chang, J.H.; Cho, Y.H.; Sohn, S.Y.; Choi, J.M.; Kim, A.; Kim, Y.C.; Jang, S.K.; Cho, Y. Crystal structure of the eIF4A–PDCD4 complex. Proc. Natl. Acad. Sci. USA 2009, 106, 3148–3153. [Google Scholar] [CrossRef]
- Dinkova, T.D.; Martinez-Castilla, L.; Cruz-Espíndola, M.A. The Diversification of eIF4E Family Members in Plants and Their Role in the Plant-Virus Interaction. In Evolution of the Protein Synthesis Machinery and Its Regulation; Hernández, G., Jagus, R., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 187–205. [Google Scholar]
- Gao, Z.; Johansen, E.; Eyers, S.; Thomas, C.L.; Noel Ellis, T.H.; Maule, A.J. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 2004, 40, 376–385. [Google Scholar] [CrossRef]
- Yeam, I.; Cavatorta, J.R.; Ripoll, D.R.; Kang, B.-C.; Jahn, M.M. Functional Dissection of Naturally Occurring Amino Acid Substitutions in eIF4E That Confers Recessive Potyvirus Resistance in Plants. Plant Cell 2007, 19, 2913–2928. [Google Scholar] [CrossRef]
- Nicaise, V.; German-Retana, S.; Sanjuan, R.; Dubrana, M.P.; Mazier, M.; Maisonneuve, B.; Candresse, T.; Caranta, C.; Le Gall, O. The eukaryotic translation initiation factor 4E controls lettuce susceptibility to the potyvirus Lettuce mosaic virus. Plant Physiol. 2003, 132, 1272–1282. [Google Scholar] [CrossRef]
- Duprat, A.; Caranta, C.; Revers, F.; Menand, B.; Browning, K.S.; Robaglia, C. The Arabidopsis eukaryotic initiation factor (iso)4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 2002, 32, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.N.; Li, J.J.; Liu, W.-Y.; An, S.J.; Cho, H.J.; Her, N.H.; Yeam, I.H.; Kim, D.S.; Kang, B.C.E.m.b.s.a.k. Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells 2009, 27, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Naderpour, M.; Lund, O.L.E.S.; Larsen, R.; Johansen, E. Potyviral resistance derived from cultivars of Phaseolus vulgaris carrying bc-3 is associated with the homozygotic presence of a mutated eIF4E allele. Mol. Plant Pathol. 2010, 11, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Hofinger, B.J.; Russell, J.R.; Bass, C.G.; Baldwin, T.; Dos Reis, M.; Hedley, P.E.; Li, Y.; Macaulay, M.; Waugh, R.; Hammond-Kosack, K.E.; et al. An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol. Ecol. 2011, 20, 3653–3668. [Google Scholar] [CrossRef]
- Piron, F.; Nicolaï, M.; Minoïa, S.; Piednoir, E.; Moretti, A.; Salgues, A.; Zamir, D.; Caranta, C.; Bendahmane, A. An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS ONE 2010, 5, e11313. [Google Scholar] [CrossRef]
- Duan, H.; Richael, C.; Rommens, C.M. Overexpression of the wild potato eIF4E-1 variant Eva1 elicits Potato virus Y resistance in plants silenced for native eIF4E-1. Transgenic Res. 2011, 21, 929–938. [Google Scholar] [CrossRef]
- Kang, B.C.; Yeam, I.; Frantz, J.D.; Murphy, J.F.; Jahn, M.M. The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virus VPg. Plant J. 2005, 42, 392–405. [Google Scholar] [CrossRef]
- Li, G.; Lv, H.; Zhang, S.; Zhang, S.; Li, F.; Zhang, H.; Qian, W.; Fang, Z.; Sun, R. TuMV management for brassica crops through host resistance: Retrospect and prospects. Plant Pathol. 2019, 68, 1035–1044. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Molecular Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]



| Plant Species | Genes | Effect | References |
|---|---|---|---|
| Arabidopsis thaliana | eIF4E | Resistance to C1YVV | [11] |
| Arabidopsis thaliana | eIF(iso)4E | Resistance to TuMV/LMV | [12,13] |
| Arabidopsis thaliana | eIF4G | Resistance to CMV/TCV | [14] |
| Brassica rapa | eIF(iso)4E | Resistance to PPV/TuMV | [15,16,17] |
| Capsicum spp. | eIF(iso)4E | Resistance to PVMV/ChiVMV | [18] |
| Capsicum spp. | eIF4E | Resistance to PVY/TEV/PepMoV | [19,20,21] |
| Citrullus lanatus | eIF4E | Resistance to ZYMV | [22] |
| Hordeum vulgare | eIF4E | Resistance to BaMMV/BaYMV | [23] |
| Lactuca sativa | eIF4E | Resistance to LMY | [24] |
| Oryza sativa | eIF4G | Resistance to RTSV | [25] |
| Phaseolus vulgaris | eIF4E | Resistance to BCMV/C1YVV | [26] |
| Pisum sativum | eIF4E | Resistance to PsBMV/BYMV/C1YVV | [27] |
| Prunus armeniaca | eIF4E | Resistance to PPV | [28] |
| Prunus domestica | eIF(iso)4E | Resistance to PPV | [29] |
| Ryza spp. | eIF(iso)4G | Resistance to RYMV | [30,31,32] |
| Solanum habrochaites | eIF4E | Resistance to PVY/TEV | [33] |
| Wheat | eIF4G and eIF(iso)4G | Interact with the tobacco etch virus (TEV) 5′ cap; initiate TEV RNA translation | [34] |
| Wheat | eIF4G | Resistance to TEV | [34] |
| Index | Compound | Formula | Class | Fold Change | Log2FC | Type |
|---|---|---|---|---|---|---|
| KMW0583 | Trans-beta-Ionone | C13H20O | Terpene | 0.4449 | −1.1685 | Down |
| KMW0504 | Benzene, 1-ethenyl-4-methoxy | C9H10O | Aromatic | 2.5819 | 1.3685 | Up |
| KMW0499 | 4-Methylthiazole | C4H5NS | Heterocyclic compound | 0.0067 | −7.2183 | Down |
| KMW0186 | Octanal | C8H16O | Aldehyde | 2.9778 | 1.5743 | Up |
| KMW0361 | Benzaldehyde, 4-ethyl | C9H10O | Aldehyde | 0.4638 | −1.1086 | Down |
| KMW0110 | Allyl Isothiocyanate | C4H5NS | Ester | 0.0000 | −16.8360 | Down |
| KMW0152 | Butane, 1-isothiocyanato | C5H9NS | Ester | 0.0329 | −4.9257 | Down |
| QWM0007 | Butane dioic acid, diethyl ester | C8H14O4 | Ester | 0.0373 | −4.7433 | Down |
| WMW0081 | Cis-2-(2-Pentenyl) furan | C9H12O | Heterocyclic compound | 0.4219 | −1.2452 | Down |
| XMW0133 | 6-Methyl-6-(5-methylfuran-2-yl) heptan-2-one | C13H20O2 | Ketone | 0.4896 | −1.0303 | Down |
| XMW0048 | Phenol, 2-nitro- | C6H5NO3 | Phenol | 7.2955 | 2.8670 | Up |
| XMW0376 | 10-Methyltricyclo [4.3.1.1(2,5)] undecan-10-ol | C12H20O | Alcohol | 0.4898 | −1.0297 | Down |
| XMW0460 | 1,3-Cyclopentadiene, 5,5-dimethyl-1,2-Dipropyl | C13H22 | Olefin | 0.2946 | −1.7634 | Down |
| NMW0073 | Bicyclo-2-ene-2-carboxaldehyde, 6,6-dimethyl | C10H14O | Terpene | 0.4031 | −1.3107 | Down |
| NMW0135 | Thiocyanic acid, phenylmethyl ester | C8H7NS | Sulfide | 0.3163 | −1.6605 | Down |
| D27 | Benzene, (2-isothiocyanatoethyl) | C9H9NS | Sulfide | 0.0043 | −7.8497 | Down |
| D218 | Cyclohexanecarboxylic acid | C7H12O2 | Acid | 0.0614 | −4.0261 | Down |
| XMW0811 | Phenethyl isocyanate | C9H9NO | Ester | 0.2005 | −2.3183 | Down |
| XMW0812 | Benzene, (1-methyl-1-propylpentyl) | C15H24 | Aromatic | 0.3611 | −1.4697 | Down |
| XMW1102 | Vinyl trans-cinnamate | C11H10O2 | Ester | 2.1582 | 1.1099 | Up |
| XMW1245 | (+)-2-Carene, 2-isopropenyl | C13H20 | Olefin | 0.4683 | −1.0946 | Down |
| XMW1459 | Thiophene, 2-butyl-5-ethyl | C10H16S | Heterocyclic compound | 0.4348 | −1.2017 | Down |
| XMW1481 | 5′-Hydroxy-2′,3′,4′-trimethylacetophenone | C11H14O2 | Ketone | 0.3290 | −1.6037 | Down |
| D378 | 1-Butene, 4-isothiocyanato | C5H7NS | Sulfide | 0.0012 | −9.6749 | Down |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, W.; Wang, S.; Zhang, S.; Li, F.; Zhang, H.; Sun, R.; Zhang, S.; Li, G. Genome-Wide Identification and Expression Analysis of eIF Family Genes from Brassica rapa in Response to TuMV Resistance. Plants 2022, 11, 2248. https://doi.org/10.3390/plants11172248
Huang W, Wang S, Zhang S, Li F, Zhang H, Sun R, Zhang S, Li G. Genome-Wide Identification and Expression Analysis of eIF Family Genes from Brassica rapa in Response to TuMV Resistance. Plants. 2022; 11(17):2248. https://doi.org/10.3390/plants11172248
Chicago/Turabian StyleHuang, Wenyue, Shaoxing Wang, Shifan Zhang, Fei Li, Hui Zhang, Rifei Sun, Shujiang Zhang, and Guoliang Li. 2022. "Genome-Wide Identification and Expression Analysis of eIF Family Genes from Brassica rapa in Response to TuMV Resistance" Plants 11, no. 17: 2248. https://doi.org/10.3390/plants11172248
APA StyleHuang, W., Wang, S., Zhang, S., Li, F., Zhang, H., Sun, R., Zhang, S., & Li, G. (2022). Genome-Wide Identification and Expression Analysis of eIF Family Genes from Brassica rapa in Response to TuMV Resistance. Plants, 11(17), 2248. https://doi.org/10.3390/plants11172248






