Effects of Zinc, Copper and Iron Oxide Nanoparticles on Induced DNA Methylation, Genomic Instability and LTR Retrotransposon Polymorphism in Wheat (Triticum aestivum L.)
Abstract
:1. Introduction
2. Results
2.1. Characterization of Nanoparticles
2.2. iPBS Analysis
2.3. CRED-iPBS Analysis
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, W.; Tan, W.; Peralta-Videa, J.R.; Gardea-Torresdey, J.L.; Ji, R.; Yin, Y.; Guo, H. Interaction of metal oxide nanoparticles with higher terrestrial plants: Physiological and biochemical aspects. Plant Physiol. Biochem. 2017, 110, 210–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolaymat, T.; Genaidy, A.; Abdelraheem, W.; Dionysiou, D.; Andersen, C. The effects of metallic engineered nanoparticles upon plant systems: An analytic examination of scientific evidence. Sci. Total Environ. 2017, 579, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Servin, A.; Elmer, W.; Mukherjee, A.; De la Torre-Roche, R.; Hamdi, H.; White, J.C.; Bindraban, P.; Dimkpa, C. A review of the use of engineered nanomaterials to suppress plant disease and enhance crop yield. J. Nanoparticle Res. 2015, 17, 1–21. [Google Scholar] [CrossRef]
- Zhang, L.; Webster, T.J. Nanotechnology and nanomaterials: Promises for improved tissue regeneration. Nano Today 2009, 4, 66–80. [Google Scholar] [CrossRef]
- Khan, A.; Zinta, G. Drought stress and chromatin: An epigenetic perspective. In Drought Stress Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2016; Volume 2, pp. 571–586. [Google Scholar]
- Kumar, S.; Beena, A.; Awana, M.; Singh, A. Physiological, biochemical, epigenetic and molecular analyses of wheat (Triticum aestivum) genotypes with contrasting salt tolerance. Front. Plant Sci. 2017, 8, 1151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, S. Factors Influencing Agricultural Professionals’ Attitudes towards Organic Agriculture and Biotechnology; ANU: Canberra, Australia, 2005. [Google Scholar]
- Galbraith, D.W. Silica breaks through in plants. Nat. Nanotechnol. 2007, 2, 272–273. [Google Scholar] [CrossRef]
- Torney, F.; Trewyn, B.G.; Lin, V.S.-Y.; Wang, K. Mesoporous silica nanoparticles deliver DNA and chemicals into plants. Nat. Nanotechnol. 2007, 2, 295–300. [Google Scholar] [CrossRef]
- Erturk, F.A.; Aydin, M.; Sigmaz, B.; Taspinar, M.S.; Arslan, E.; Agar, G.; Yagci, S. Effects of As2O3 on DNA methylation, genomic instability, and LTR retrotransposon polymorphism in Zea mays. Environ. Sci. Pollut. Res. 2015, 22, 18601–18606. [Google Scholar] [CrossRef]
- Zulfiqar, F.; Navarro, M.; Ashraf, M.; Akram, N.A.; Munné-Bosch, S. Nanofertilizer use for sustainable agriculture: Advantages and limitations. Plant Sci. 2019, 289, 110270. [Google Scholar] [CrossRef]
- Goldberg, A.D.; Allis, C.D.; Bernstein, E. Epigenetics: A landscape takes shape. Cell 2007, 128, 635–638. [Google Scholar] [CrossRef] [Green Version]
- Hauser, M.T.; Aufsatz, W.; Jonak, C.; Luschnig, C. Transgenerational epigenetic inheritance in plants. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 459–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef] [Green Version]
- Xue-Lin, L.; Zhong-Xu, L.; Yi-Chun, N.; Xiao-Ping, G.; Zhang, X.-L. Methylation-sensitive amplification polymorphism of epigenetic changes in cotton under salt stress. Acta Agron. Sin. 2009, 35, 588–596. [Google Scholar]
- Pereira, W.J.; Pappas, M.D.C.R.; Grattapaglia, D.; Pappas, G.J., Jr. A cost-effective approach to DNA methylation detection by Methyl Sensitive DArT sequencing. PLoS ONE 2020, 15, e0233800. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, P.T.; Orłowska, R.; Niedziela, A. A relative quantitative methylation-sensitive amplified polymorphism (MSAP) method for the analysis of abiotic stress. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Lv, J.; Zhang, L.; Dou, J.; Sun, Y.; Li, X.; Fu, X.; Dou, H.; Mao, J.; Hu, X. MethylRAD: A simple and scalable method for genome-wide DNA methylation profiling using methylation-dependent restriction enzymes. Open Biol. 2015, 5, 150130. [Google Scholar] [CrossRef] [Green Version]
- Brunner, A.L.; Johnson, D.S.; Kim, S.W.; Valouev, A.; Reddy, T.E.; Neff, N.F.; Anton, E.; Medina, C.; Nguyen, L.; Chiao, E. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009, 19, 1044–1056. [Google Scholar] [CrossRef] [Green Version]
- Orłowska, R.; Pachota, K.A.; Machczyńska, J.; Niedziela, A.; Makowska, K.; Zimny, J.; Bednarek, P.T. Improvement of anther cultures conditions using the Taguchi method in three cereal crops. Electron. J. Biotechnol. 2020, 43, 8–15. [Google Scholar] [CrossRef]
- Lu, Y.; Rong, T.; Cao, M. Analysis of DNA methylation in different maize tissues. J. Genet. Genom. 2008, 35, 41–48. [Google Scholar] [CrossRef]
- Hosseinpour, A.; Ozkan, G.; Nalci, O.; Haliloğlu, K. Estimation of genomic instability and DNA methylation due to aluminum (Al) stress in wheat (Triticum aestivum L.) using iPBS and CRED-iPBS analyses. Turk J. Bot. 2019, 43, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Hosseinpour, A.; Ilhan, E.; Özkan, G.; Öztürk, H.İ.; Haliloglu, K.; Cinisli, K.T. Plant growth-promoting bacteria (PGPBs) and copper (II) oxide (CuO) nanoparticle ameliorates DNA damage and DNA Methylation in wheat (Triticum aestivum L.) exposed to NaCl stress. J. Plant Biochem. Biotechnol. 2021, 1, 1–14. [Google Scholar] [CrossRef]
- Nalci, O.B.; Nadaroglu, H.; Pour, A.H.; Gungor, A.A.; Haliloglu, K. Effects of ZnO, CuO and γ-Fe3O4 nanoparticles on mature embryo culture of wheat (Triticum aestivum L.). Plant Cell Tissue Organ. Cult. 2019, 136, 269–277. [Google Scholar] [CrossRef]
- Nazli, A.; Baig, M.W.; Zia, M.; Ali, M.; Shinwari, Z.K.; Haq, I.U. Plant-based metallic nanoparticles as potential theranostics agents: Bioinspired tool for imaging and treatment. IET NanoBiotechnol. 2018, 12, 869–878. [Google Scholar] [CrossRef]
- Nadaroglu, H.; Cicek, S.; Gungor, A.A. Removing Trypan blue dye using nano-Zn modified Luffa sponge. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017, 172, 2–8. [Google Scholar] [CrossRef]
- Kuzma, J. Moving forward responsibly: Oversight for the nanotechnology-biology interface. J. Nanoparticle Res. 2007, 9, 165–182. [Google Scholar] [CrossRef]
- Shankramma, K.; Yallappa, S.; Shivanna, M.; Manjanna, J. Fe2O3 magnetic nanoparticles to enhance S. lycopersicum (tomato) plant growth and their biomineralization. Appl. Nanosci. 2016, 6, 983–990. [Google Scholar]
- Lengke, M.F.; Fleet, M.E.; Southam, G. Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver (I) nitrate complex. Langmuir 2007, 23, 2694–2699. [Google Scholar] [CrossRef] [PubMed]
- Vafaie Moghadam, A.; Iranbakhsh, A.; Saadatmand, S.; Ebadi, M.; Oraghi Ardebili, Z. New insights into the transcriptional, epigenetic, and physiological responses to zinc oxide nanoparticles in datura stramonium; potential species for phytoremediation. J. Plant Growth Regul. 2021, 41, 271–281. [Google Scholar] [CrossRef]
- Safari, M.; Oraghi Ardebili, Z.; Iranbakhsh, A. Selenium nanoparticle induced alterations in expression patterns of heat shock factor A4A (HSFA4A), and high molecular weight glutenin subunit 1Bx (Glu-1Bx) and enhanced nitrate reductase activity in wheat (Triticum aestivum L.). Acta Physiol. Plant. 2018, 40, 1–8. [Google Scholar] [CrossRef]
- Gao, X.; Zou, C.; Wang, L.; Zhang, F. Silicon decreases transpiration rate and conductance from stomata of maize plants. J. Plant Nutr. 2006, 29, 1637–1647. [Google Scholar] [CrossRef]
- Feizi, H.; Moghaddam, P.R.; Shahtahmassebi, N.; Fotovat, A. Impact of bulk and nano-sized titanium dioxide (TiO2) on wheat seed germination and seedling growth. Biol. Trace Elem. Res. 2012, 146, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Javed, R.; Usman, M.; Yücesan, B.; Zia, M.; Gürel, E. Effect of zinc oxide (ZnO) nanoparticles on physiology and steviol glycosides production in micropropagated shoots of Stevia rebaudiana Bertoni. Plant Physiol. Biochem. 2017, 110, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Tarafdar, J.; Raliya, R.; Mahawar, H.; Rathore, I. Development of zinc nanofertilizer to enhance crop production in pearl millet (Pennisetum americanum). Agric. Res. 2014, 3, 257–262. [Google Scholar] [CrossRef]
- Pagano, L.; Servin, A.D.; De La Torre-Roche, R.; Mukherjee, A.; Majumdar, S.; Hawthorne, J.; Marmiroli, M.; Maestri, E.; Marra, R.E.; Isch, S.M. Molecular response of crop plants to engineered nanomaterials. Environ. Sci. Technol. 2016, 50, 7198–7207. [Google Scholar] [CrossRef]
- Ruotolo, R.; Maestri, E.; Pagano, L.; Marmiroli, M.; White, J.C.; Marmiroli, N. Plant response to metal-containing engineered nanomaterials: An omics-based perspective. Environ. Sci. Technol. 2018, 52, 2451–2467. [Google Scholar] [CrossRef]
- Ullah, S.; Ullah, R. Beneficial effects of several nanoparticles on the growth of different plants species. Curr. Nanosci. 2019, 15, 460–470. [Google Scholar]
- Mota, M.B.S.; Carvalho, M.A.; Monteiro, A.N.; Mesquita, R.D. DNA damage response and repair in perspective: Aedes aegypti, Drosophila melanogaster and Homo sapiens. Parasites Vectors 2019, 12, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Sundaria, N.; Singh, M.; Upreti, P.; Chauhan, R.P.; Jaiswal, J.; Kumar, A. Seed priming with Iron oxide nanoparticles triggers Iron acquisition and biofortification in wheat (Triticum aestivum L.) grains. J. Plant Growth Regul. 2019, 38, 122–131. [Google Scholar] [CrossRef]
- Zhang, Z.; Ke, M.; Qu, Q.; Peijnenburg, W.; Lu, T.; Zhang, Q.; Ye, Y.; Xu, P.; Du, B.; Sun, L. Impact of copper nanoparticles and ionic copper exposure on wheat (Triticum aestivum L.) root morphology and antioxidant response. Environ. Pollut. 2018, 239, 689–697. [Google Scholar] [CrossRef]
- Ravet, K.; Pilon, M. Copper and iron homeostasis in plants: The challenges of oxidative stress. Antioxid. Redox Signal. 2013, 19, 919–932. [Google Scholar] [CrossRef] [Green Version]
- Rafique, M.; Shaikh, A.J.; Rasheed, R.; Tahir, M.B.; Bakhat, H.F.; Rafique, M.S.; Rabbani, F. A review on synthesis, characterization and applications of copper nanoparticles using green method. Nano 2017, 12, 1750043. [Google Scholar] [CrossRef]
- Szőllősi, R. Superoxide dismutase (SOD) and abiotic stress tolerance in plants: An overview. Oxidative Damage Plants 2014, 3, 89–129. [Google Scholar]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Alegría-Torres, J.A.; Baccarelli, A.; Bollati, V. Epigenetics and lifestyle. Epigenomics 2011, 3, 267–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajput, V.; Minkina, T.; Suskova, S.; Mandzhieva, S.; Tsitsuashvili, V.; Chapligin, V.; Fedorenko, A. Effects of copper nanoparticles (CuO NPs) on crop plants: A mini review. BioNano Sci. 2018, 8, 36–42. [Google Scholar] [CrossRef]
- Yuan, P.; Peng, C.; Shi, J.; Liu, J.; Cai, D.; Wang, D.; Shen, Y. Ferrous ions inhibit Cu uptake and accumulation via inducing iron plaque and regulating the metabolism of rice plants exposed to CuO nanoparticles. Environ. Sci. Nano 2021, 8, 1456–1468. [Google Scholar] [CrossRef]
- Mimmo, T.; Del Buono, D.; Terzano, R.; Tomasi, N.; Vigani, G.; Crecchio, C.; Pinton, R.; Zocchi, G.; Cesco, S. Rhizospheric organic compounds in the soil–microorganism–plant system: Their role in iron availability. Eur. J. Soil Sci. 2014, 65, 629–642. [Google Scholar] [CrossRef]
- Martínez-Fernández, D.; Barroso, D.; Komárek, M. Root water transport of Helianthus annuus L. under iron oxide nanoparticle exposure. Environ. Sci. Pollut. Res. 2016, 23, 1732–1741. [Google Scholar] [CrossRef]
- Alidoust, D.; Isoda, A. Effect of γFe 2 O 3 nanoparticles on photosynthetic characteristic of soybean (Glycine max (L.) Merr.): Foliar spray versus soil amendment. Acta Physiol. Plant 2013, 35, 3365–3375. [Google Scholar] [CrossRef]
- Bakhtiari, M.; Moaveni, P.; Sani, B. The effect of iron nanoparticles spraying time and concentration on wheat. BFIJ 2015, 7, 679–683. [Google Scholar]
- Rui, M.; Ma, C.; Hao, Y.; Guo, J.; Rui, Y.; Tang, X.; Zhao, Q.; Fan, X.; Zhang, Z.; Hou, T. Iron oxide nanoparticles as a potential iron fertilizer for peanut (Arachis hypogaea). Front. Plant Sci. 2016, 7, 815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egger, G.; Liang, G.; Aparicio, A.; Jones, P.A. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004, 429, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Smolkova, B.; El Yamani, N.; Collins, A.R.; Gutleb, A.C.; Dusinska, M. Nanoparticles in food. Epigenetic changes induced by nanomaterials and possible impact on health. Food Chem. Toxicol. 2015, 77, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Simkó, M.; Gazsó, A.; Fiedeler, U.; Nentwich, M. Nanoparticles, Free Radicals and Oxidative Stress (NanoTrust Dossier No. 012en–January 2011). 2011. Austrian Academy of Sciences; Legal Person under Public Law (BGBl 569/1921; BGBl I 130/2003); Dr. Ignaz Seipel-Platz 2, A-1010 Vienna. Available online: epub.oeaw.ac.at/ita/nanotrust-dossiers/dossier012en.pdf (accessed on 28 July 2022).
- Fratelli, M.; Goodwin, L.O.; Ørom, U.A.; Lombardi, S.; Tonelli, R.; Mengozzi, M.; Ghezzi, P. Gene expression profiling reveals a signaling role of glutathione in redox regulation. Proc. Natl. Acad. Sci. USA 2005, 102, 13998–14003. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Lee, S.; Lee, I. Alteration of phytotoxicity and oxidant stress potential by metal oxide nanoparticles in Cucumis sativus. Water Air Soil Pollut. 2012, 223, 2799–2806. [Google Scholar] [CrossRef]
- Dusinska, M.; Magdolenova, Z.; Fjellsbø, L.M. Toxicological aspects for nanomaterial in humans. Nanotechnol. Nucleic Acid Deliv. 2013, 948, 1–12. [Google Scholar]
- Yao, Y.; Costa, M. Genetic and epigenetic effects of nanoparticles. J. Mol. Genet. Med. 2013, 7, 1–6. [Google Scholar]
- Georgantzopoulou, A.; Serchi, T.; Cambier, S.; Leclercq, C.C.; Renaut, J.; Shao, J.; Kruszewski, M.; Lentzen, E.; Grysan, P.; Eswara, S. Effects of silver nanoparticles and ions on a co-culture model for the gastrointestinal epithelium. Part. Fibre Toxicol. 2015, 13, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Dietz, K.J.; Herth, S. Plant nanotoxicology. Trends Plant Sci. 2011, 16, 582–589. [Google Scholar] [CrossRef]
- Zeinalzadehtabrizi, H.; Hosseinpour, A.; Aydin, M.; Haliloglu, K. A modified genomic DNA extraction method from leaves of sunflower for PCR based analyzes. J. Biodivers. Environ. Sci. 2015, 7, 222–225. [Google Scholar]
- Kalendar, R.; Antonius, K.; Smýkal, P.; Schulman, A.H. iPBS: A universal method for DNA fingerprinting and retrotransposon isolation. Theor. Appl. Genet. 2010, 121, 1419–1430. [Google Scholar] [CrossRef]
- Türkoğlu, A.; Haliloğlu, K.; Balpinar, Ö.; Öztürk, H.I.; Özkan, G.; Poczai, P. The Effect of Mammalian Sex Hormones on Polymorphism and Genomic Instability in the Common Bean (Phaseolus vulgaris L.). Plants 2022, 11, 2071. [Google Scholar] [CrossRef]
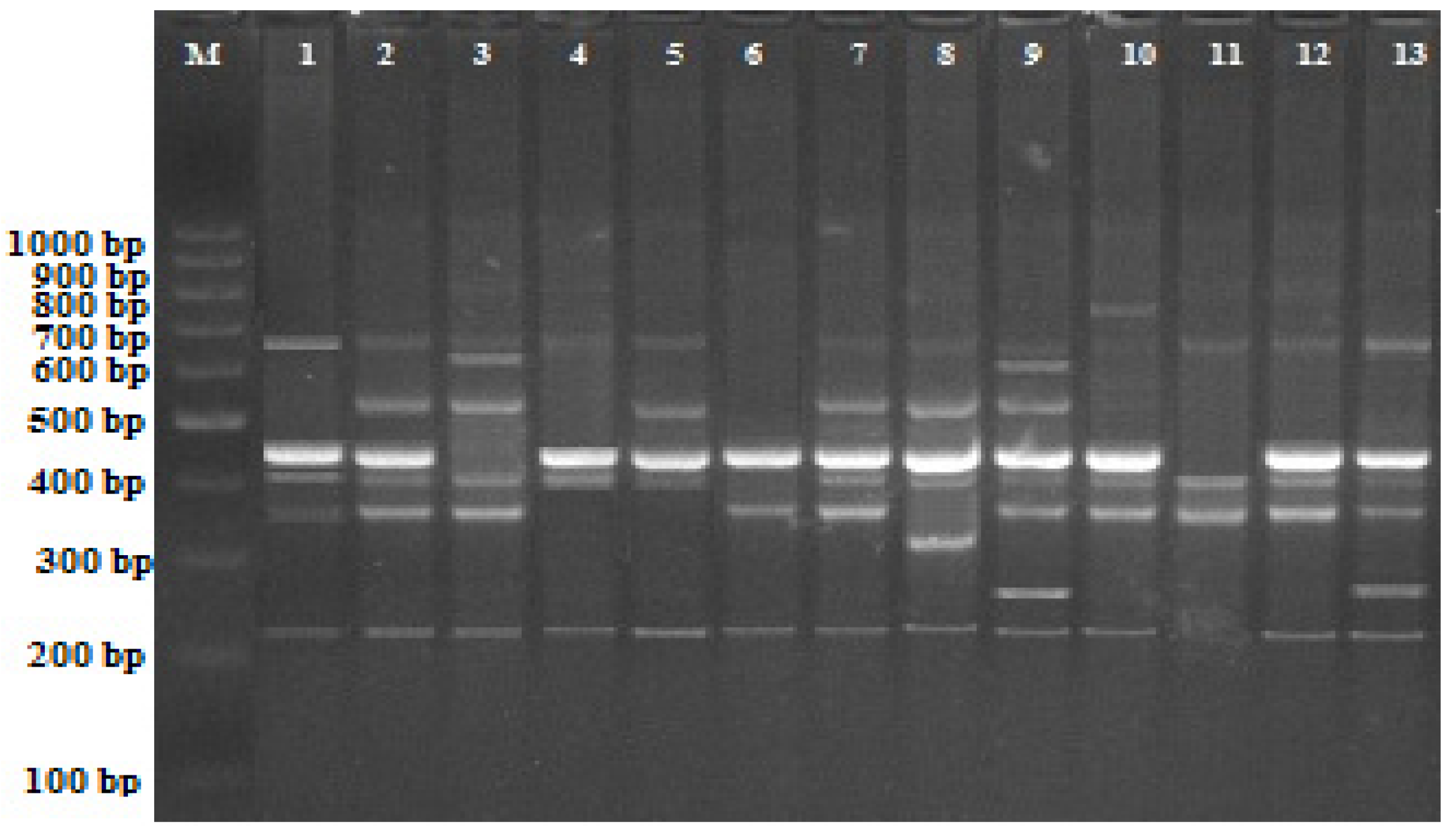
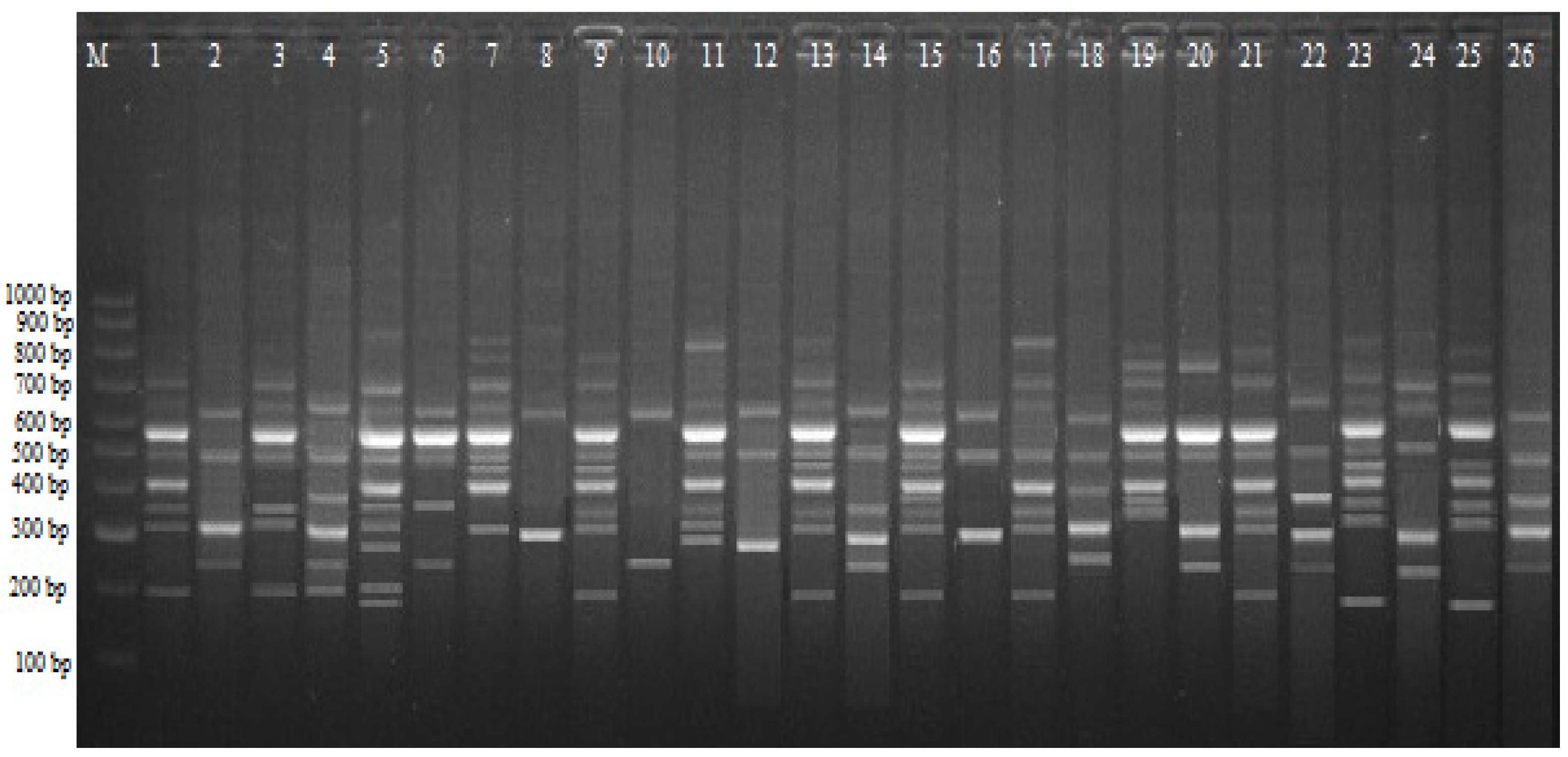
| Primer | * +/− ** | Control *** | Experimental Groups | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZnO NPs | CuO NPs | γ-Fe3O4 NPs | ||||||||||||
| 0 a | 1X a | 2X b | 3X c | 0 b | 1X b | 2X b | 3X b | 0 c | 1X c | 2X c | 3X c | |||
| iPBS 2382 | + | 9 | 627; 166 | 239 | 850; 520 | - | 458 | - | 560; 265 | - | 625; 300 | 580 | 750; 536 | 625; 245 |
| − | 605 | 287 | 485 | 698 | 308 | 146 | 335 | 420 | 352 | - | 485; 146 | 335 | ||
| iPBS 2383 | + | 6 | 430; 307 | 440; 207 | 520 | 330; 207 | - | - | 610 | 327 | 188 | 350 | - | 218 |
| − | - | 350 | 400; 258 | 350 | 515; 285 | 400 | 258 | 515; 400 | 651 | 285 | 651 | 400 | ||
| iPBS 2384 | + | 7 | 608 | 670; 505 | 682; 426 | 285 | 300 | 300 | 483; 322 | 584 | 480; 265 | 185 | 465 | - |
| − | - | 359 | 315; 105 | - | 655; 250 | - | - | 359 | 450 | 550; 450 | 315; 105 | 655 | ||
| iPBS 2385 | + | 8 | 397 | - | 100 | 274 | 350 | - | - | 428; 125 | 385 | - | - | 505 |
| − | - | 305; 158 | 571; 409 | - | - | 550; 350 | 675 | - | 450; 158 | 158 | - | - | ||
| iPBS 2386 | + | 6 | 524 | 440 | 350; 205 | 450 | 287 | 405 | 620 | 355 | - | - | 450; 366 | - |
| − | 352 | - | 600 | - | - | 750 | 450 | - | 450; 305;205 | 450; 205 | 600 | - | ||
| iPBS 2387 | + | 5 | 650 | 392 | - | 450; 205 | - | 650; 258 | 715 | 115 | 320 | - | 452 | 502 |
| − | 560; 367 | - | 367; 239 | 600 | 239 | - | - | 367 | 560; 153 | 239 | 285 | 153 | ||
| iPBS 2388 | + | 6 | - | 342 | 467 | 565; 295 | 487 | 378 | 350; 258 | 405 | - | 482 | 489; 326 | - |
| − | 221 | 285 | 362 | 160 | 160 | - | 221; 160 | - | 450 | 650; 362 | 285 | 362 | ||
| iPBS 2389 | + | 8 | 118 | - | 404 | - | 858; 465 | 157 | 248 | - | 580 | 571 | 535; 228 | 776 |
| − | - | 450 | - | 285 | 550 | 360 | - | 517 | 600; 350; 140 | - | - | - | ||
| iPBS 2390 | + | 7 | 657 | 267 | - | - | - | 850; 609 | 615 | - | 305; 127 | 422 | - | 115 |
| − | 632; 362 | - | 605 | 495 | 362 | 405 | - | - | - | - | 605 | 503 | ||
| iPBS 2391 | + | 5 | 547 | 620; 552 | - | 507 | - | 535 | 310; 539 | 618; 250 | 778 | - | - | 250 |
| − | - | 450 | 350 | 350 | 650; 407 | - | 350 | - | - | 209; 450 | - | - | ||
| Total band | 67 | 17 | 19 | 23 | 17 | 18 | 16 | 20 | 15 | 25 | 17 | 19 | 14 | |
| Polymorphism (%) | 25.37 | 28.35 | 34.32 | 25.37 | 26.86 | 23.88 | 29.85 | 22.38 | 37.31 | 25.37 | 28.35 | 20.89 | ||
| GTS value | 74.62 | 71.64 | 65.67 | 74.62 | 73.13 | 76.11 | 70.14 | 77.61 | 62.68 | 74.62 | 71.64 | 79.10 | ||
| Primer | M */H ** | + ***/− **** | Control ***** | Experimental Groups | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZnO NPs | CuO NPs | γ-Fe3O4 NPs | |||||||||||||
| 0 a | 1X a | 2X b | 3X c | 0 b | 1X b | 2X b | 3X b | 0 c | 1X c | 2X c | 3X c | ||||
| iPBS 2382 | M | + | 8 | 658; 434 | 420; 156 | 590; | 660 | 645 | 450 | 660; 450; 305 | - | 785; 475; 325; 120 | - | 650; 450 | - |
| − | - | 600 | 250; 165 | - | - | 250 | - | 485; 375 | 298 | 205 | 165 | 345 | |||
| H | + | 6 | 575 | 497 | 320; 139 | 365 | 335 | 150; 216 | 685; 455 | - | 168; 102 | 605 | 335 | ||
| − | 415 | 305 | - | - | 180 | - | - | 350 | 254; 128 | - | 128 | - | |||
| iPBS 2383 | M | + | 5 | 754; 515 | 555;450 | 550; 362; 125 | 538 | 360 | 780 | 785; 650 | 375 | - | 605; 515 | 785 | 645; 360 |
| − | - | 245 | 405; 350 | - | - | - | - | - | 350; 245; 165 | - | 525 | - | |||
| H | + | 4 | - | 450; 325 | 667; 240 | - | 455; 300 | 409; 125 | 600 | - | 480 | 170 | - | - | |
| − | 405 | - | - | 405 | - | - | 395 | 540 | 305 | 155 | 305; 155 | 305 | |||
| iPBS 2384 | M | + | 7 | - | 604; 425 | 475; 282 | 378 | 655; 509; 430 | - | 708; 506 | - | 360; 270; 235 | - | 505; 337; 125 | 475; 345 |
| − | 390; 240 | 450 | 350 | - | - | 625; 120 | - | 390 | 350; 205 | 450; 120 | - | - | |||
| H | + | 6 | 285 | - | 125 | 465 | 495 | - | - | 275 | 760; 605; 375 | 260 | 550 | 262 | |
| − | - | 350 | 425; 350 | - | 250 | 165 | 450 | 450 | 165 | 568; 425 | 425; 350 | - | |||
| iPBS 2385 | M | + | 11 | 450 | 808; 745; 625 | 350; 128 | 825; 486 | - | 760; 456 | 785; 345 | 625 | 458; 325; 124 | - | 850; 650; 485; 120 | 625 |
| − | 340 | - | 785 | - | 325; 410 | 325 | 440 | - | 705; 650 | 605 | - | - | |||
| H | + | 10 | - | 695 | 374; 125 | 560; 393 | 260 | - | 755; 550; 469 | 745 | 710; 650 | 330; 280 | 355 | - | |
| − | 425 | 525; 365 | - | - | 300; 170 | 751; 636, 225 | - | 225 | 345 | - | 450; 345 | 525 | |||
| iPBS 2386 | M | + | 4 | 340; 115 | - | 405; 300 | 807; 485 | 300; 220 | - | 615; 430 | 405 | 345; 262 | 600 | 565; 450 | 475 |
| − | - | 255; 458 | 654 | - | 458; 355 | 355 | - | - | 255 | - | - | ||||
| H | + | 8 | - | - | 505; 118 | 715 | 450 | 650 | 685 | - | 600; 450 | 605 | 655 | 305 | |
| − | 525 | 400; 225 | 150 | 605; 150 | 360; 320 | - | - | 225; 150 | 375; 225 | 400; 320 | - | ||||
| iPBS 2387 | M | + | 3 | 285 | - | 605 | - | 425 | 785; 250 | - | - | 680; 510 | 670; 466 | 485 | 689 |
| − | 185 | 185 | - | 650; 580 | 580 | - | 650; 185 | 580; 185 | - | 580 | 650 | - | |||
| H | + | 9 | - | 420;285 | 605; 225 | 450 | 500 | - | 550 | 780 | 450; 245;125 | - | 885; 745; 600 | - | |
| − | 850; 405; 365 | 405;160 | - | 715 | 125 | 365 | 365; 125 | 160 | 345; 205 | 125 | - | 550; 205 | |||
| iPBS 2388 | M | + | 7 | 450; 226 | 250; 115 | 510; 258 | 388 | 520; 450 | 487 | 650; 255; 125 | 425 | 360; 220 | - | 655; 505 | 650 |
| − | - | - | 405; 100 | - | - | - | - | - | 185; 100 | 575; 345 | 405 | - | |||
| H | + | 8 | 250 | 397; 285 | 440; 338; 112 | 746; 350; 119 | 380 | 455; 350 | 580; 375; 120 | - | 498 | 624 | 404; 348 | - | |
| − | 535; 453 | 308 | - | - | - | - | 405 | 355; 105 | 500; 462 | - | - | 500 | |||
| iPBS 2389 | M | + | 11 | 375; 198 | 785; 625; 320 | 625; 405 | 550 | 680; 465; 345 | 460; 128 | 645; 460; 355 | - | 605; 485; 325 | 652 | 496; 320 | 128 |
| − | - | 490 | - | - | - | - | 105 | 447 | - | 520 | |||||
| H | + | 8 | 285; 159 | 153 | 750; 418 | - | 785; 560; 245 | - | 424; 335 | - | 450; 355; 265 | 600; 505; 485 | 525 | - | |
| − | 255 | 350; 228 | 402; 350 | 228;125 | 450 | 350 | 320 | 350 | 255; 125 | - | 350 | 320 | |||
| iPBS 2390 | M | + | 5 | 490; 350 | 485; 309 | 365; 263 | 785; 460; 340 | - | 750 | 464; 128 | - | 560; 258 | 365 | 775 | - |
| − | - | 180 | 450; 180 | - | 654; 350 | - | 450 | 350; 180 | 350;180 | - | - | 245 | |||
| H | + | 4 | 365 | - | 582; 278; 145 | 525;410 | 669; 285 | 680; 490 | 325 | - | 458 | 635 | - | 605 | |
| − | 125 | 600;450 | - | - | - | 125 | 600 | 345 | 125 | 600; 450 | 125 | ||||
| iPBS 2391 | M | + | 8 | - | 850; 290;185 | 758; 800; 450 | 750; 450 | 800; 285 | 810; 450 | 456; 378 | 830 | 805; 753; 385 | 800 | 850; 450 | 750; 330 |
| − | 400 | - | 350; 200 | - | 705; 200 | - | - | 555 | 315; 200 | 645 | - | 495 | |||
| H | + | 4 | 350; 200 | 550; 360 | - | - | 346 | - | 345 | 775; 555 | 390 | 650 | 350 | ||
| − | - | 300 | 490; 245 | 490; 300 | 245 | - | 245 | - | 605 | - | - | - | |||
| Polymorphism % | M | 32.77 | 41.34 | 50.88 | 31.50 | 35.60 | 29.50 | 43.01 | 24.84 | 57.96 | 32.28 | 37.94 | 23.85 | ||
| H | 30.58 | 39.94 | 42.56 | 30.06 | 34.39 | 26.61 | 33.83 | 22.97 | 49.81 | 30.19 | 35.92 | 21.31 | |||
| No | Primer Name | Sequence (5′ to 3′) | Tm (°C) | CG (%) |
|---|---|---|---|---|
| 1 | iPBS 2382 | TGTTGGCTTCCA | 44.9 | 50 |
| 2 | iPBS 2383 | GCATGGCCTCCA | 50.5 | 66.7 |
| 3 | iPBS 2384 | GTAATGGGTCCA | 40.9 | 50 |
| 4 | iPBS 2385 | CCATTGGGTCCA | 45.7 | 58.3 |
| 5 | iPBS 2386 | CTGATCAACCCA | 41.4 | 50 |
| 6 | iPBS 2387 | GCGCAATACCCA | 47.3 | 58.3 |
| 7 | iPBS 2388 | TTGGAAGACCCA | 43.4 | 50 |
| 8 | iPBS 2389 | ACATCCTTCCCA | 43 | 50 |
| 9 | iPBS 2390 | GCAACAACCCCA | 47.6 | 58.3 |
| 10 | iPBS 2391 | ATCTGTCAGCCA | 43.6 | 50 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haliloğlu, K.; Türkoğlu, A.; Balpınar, Ö.; Nadaroğlu, H.; Alaylı, A.; Poczai, P. Effects of Zinc, Copper and Iron Oxide Nanoparticles on Induced DNA Methylation, Genomic Instability and LTR Retrotransposon Polymorphism in Wheat (Triticum aestivum L.). Plants 2022, 11, 2193. https://doi.org/10.3390/plants11172193
Haliloğlu K, Türkoğlu A, Balpınar Ö, Nadaroğlu H, Alaylı A, Poczai P. Effects of Zinc, Copper and Iron Oxide Nanoparticles on Induced DNA Methylation, Genomic Instability and LTR Retrotransposon Polymorphism in Wheat (Triticum aestivum L.). Plants. 2022; 11(17):2193. https://doi.org/10.3390/plants11172193
Chicago/Turabian StyleHaliloğlu, Kamil, Aras Türkoğlu, Özge Balpınar, Hayrunnisa Nadaroğlu, Azize Alaylı, and Peter Poczai. 2022. "Effects of Zinc, Copper and Iron Oxide Nanoparticles on Induced DNA Methylation, Genomic Instability and LTR Retrotransposon Polymorphism in Wheat (Triticum aestivum L.)" Plants 11, no. 17: 2193. https://doi.org/10.3390/plants11172193
APA StyleHaliloğlu, K., Türkoğlu, A., Balpınar, Ö., Nadaroğlu, H., Alaylı, A., & Poczai, P. (2022). Effects of Zinc, Copper and Iron Oxide Nanoparticles on Induced DNA Methylation, Genomic Instability and LTR Retrotransposon Polymorphism in Wheat (Triticum aestivum L.). Plants, 11(17), 2193. https://doi.org/10.3390/plants11172193








