Distribution, Polymorphism and Function Characteristics of the GST-Encoding Fhb7 in Triticeae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Sequence Amplification and Cloning of Fhb7 Homologs
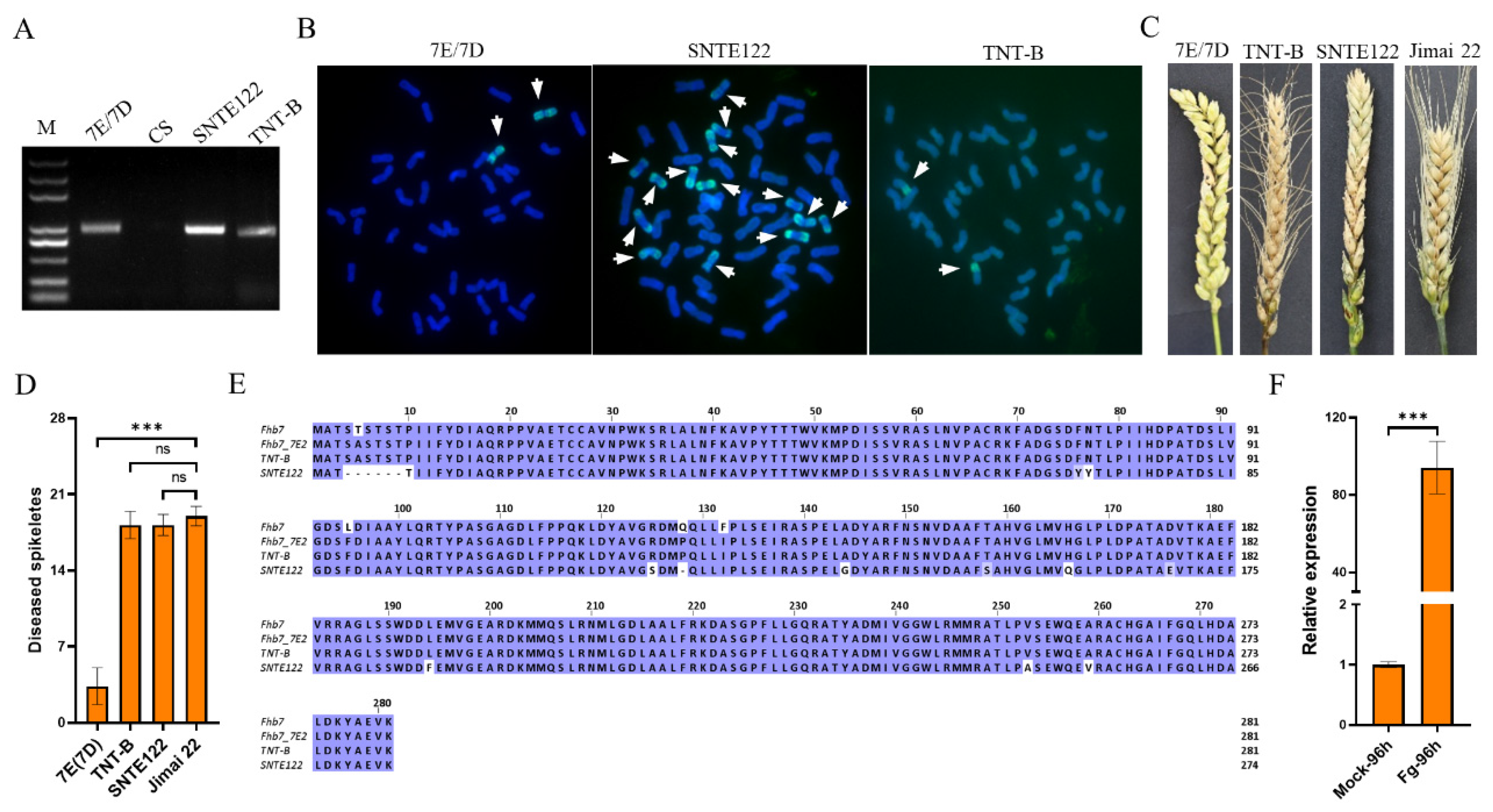
2.2.2. Chromosome Preparation and Fluorescence In Situ Hybridization (FISH)
2.2.3. FHB Resistance Evaluation on Wheat-Thinopyrum Derivatives
2.2.4. The Expression Analysis of the Fhb7 Homolog in the Line TNT-B
3. Results
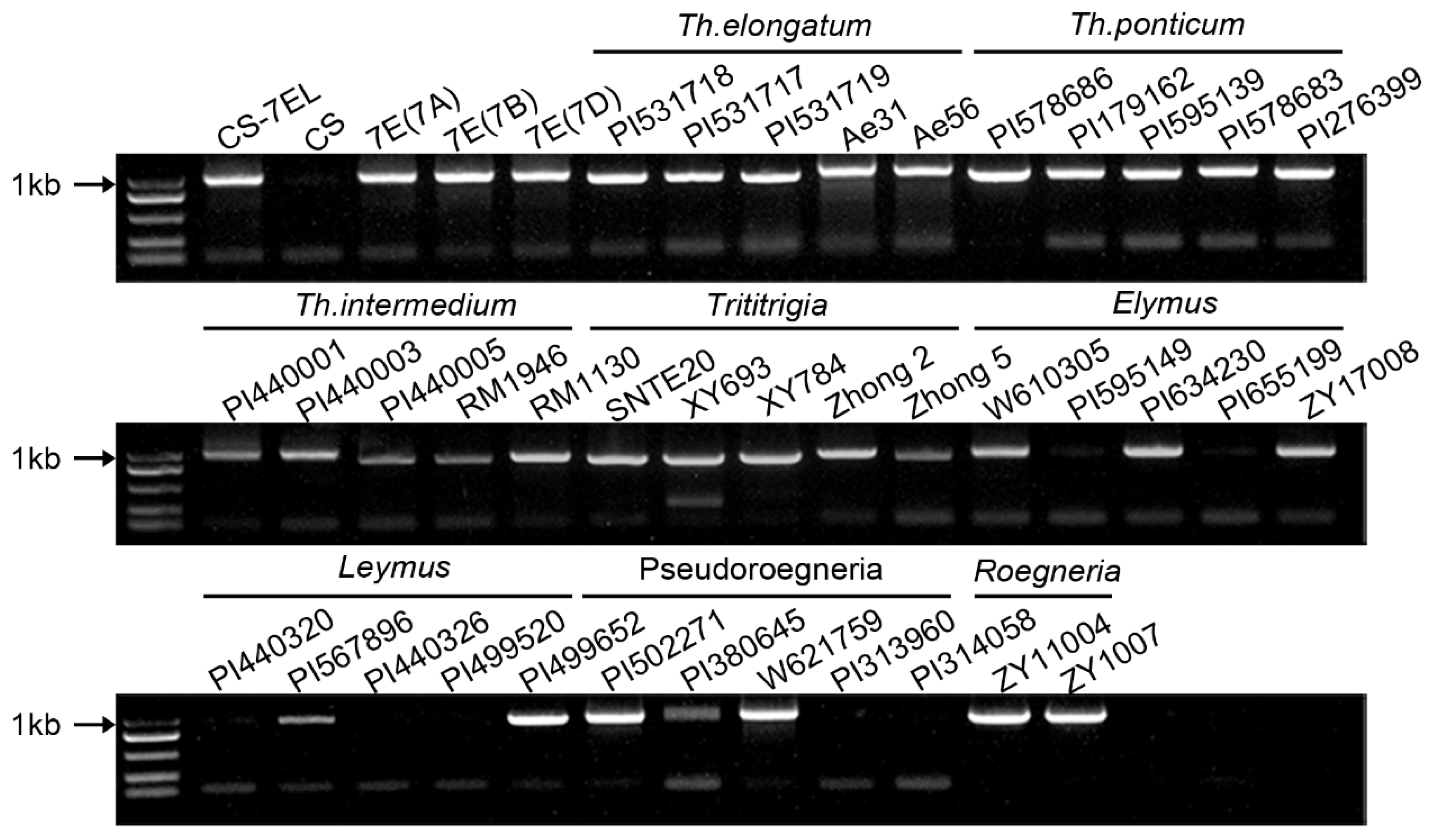
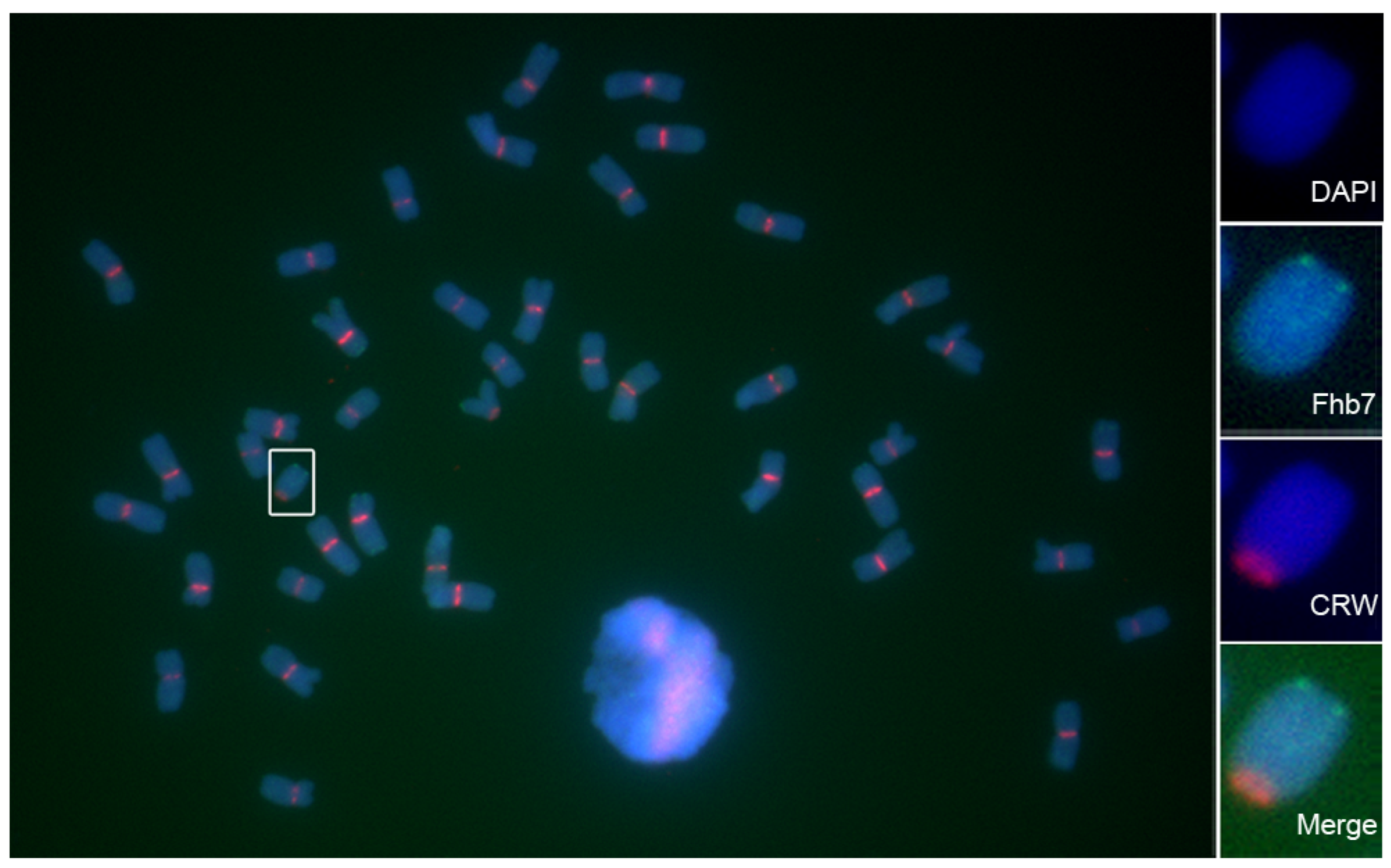
3.1. Fhb7 Is Not Unique to Thinopyrum in Triticeae
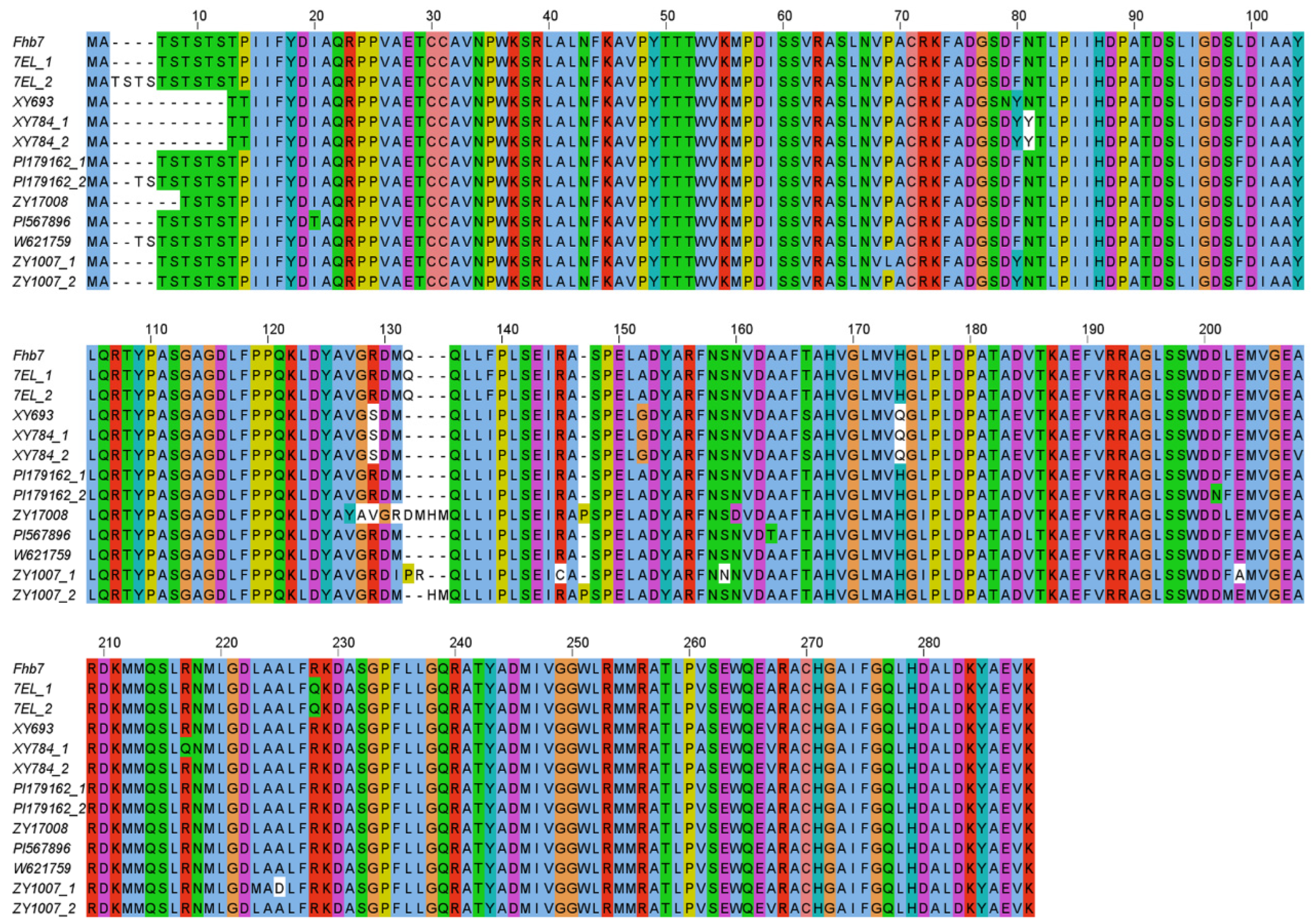
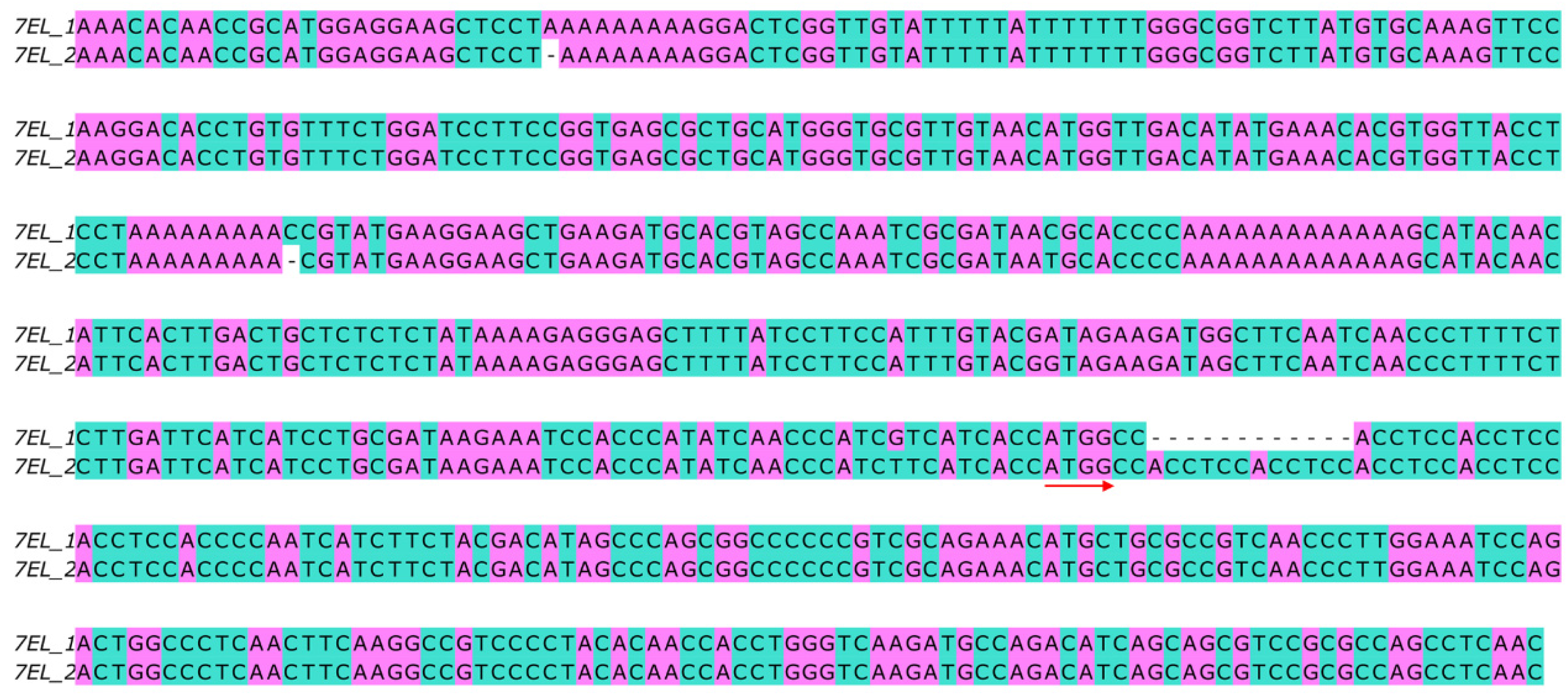
3.2. Polymorphism of Fhb7 Homologs in Triticeae
3.3. Contrast Reactions of Wheat-Thinopyrum Derivatives Carrying Fhb7 Homologs to FHB
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Angulo, V.; Beriot, N.; Garcia-Hernandez, E.; Li, E.; Masteling, R.; Lau, J.A. Plant-microbe eco-evolutionary dynamics in a changing world. New Phytol. 2022, 234, 1919–1928. [Google Scholar] [CrossRef] [PubMed]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: Genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, P.H. The Endosphere Microbiome of Ginseng. Plants 2022, 11, 415. [Google Scholar] [CrossRef] [PubMed]
- Mejia, L.C.; Herre, E.A.; Sparks, J.P.; Winter, K.; Garcia, M.N.; Van Bael, S.A.; Stitt, J.; Shi, Z.; Zhang, Y.; Guiltinan, M.J.; et al. Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Front. Microbiol. 2014, 5, 479. [Google Scholar] [CrossRef]
- Huang, J. Horizontal gene transfer in eukaryotes: The weak-link model. Bioessays 2013, 35, 868–875. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, P.; Bae, H. Horizontal Gene Transfer and Endophytes: An Implication for the Acquisition of Novel Traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef] [Green Version]
- Ricard, G.; McEwan, N.R.; Dutilh, B.E.; Jouany, J.P.; Macheboeuf, D.; Mitsumori, M.; McIntosh, F.M.; Michalowski, T.; Nagamine, T.; Nelson, N.; et al. Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genom. 2006, 7, 22. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Zhao, J.; Tang, N.; Sun, H.; Huang, J. Horizontal gene transfer from bacteria and plants to the Arbuscular Mycorrhizal Fungus Rhizophagus irregularis. Front. Plant Sci. 2018, 9, 701. [Google Scholar] [CrossRef]
- Shinozuka, H.; Hettiarachchige, I.K.; Shinozuka, M.; Cogan, N.O.I.; Spangenberg, G.C.; Cocks, B.G.; Forster, J.W.; Sawbridge, T.I. Horizontal transfer of a ss-1,6-glucanase gene from an ancestral species of fungal endophyte to a cool-season grass host. Sci. Rep. 2017, 7, 9024. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.; Hu, X.; Sun, H.; Yang, Y.; Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 2012, 3, 1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Makarova, K.S.; Aravind, L. Horizontal gene transfer in prokaryotes: Quantification and classification. Annu. Rev. Microbiol. 2001, 55, 709–742. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Lawrence, J.G.; Groisman, E.A. Lateral gene transfer and the nature of bacterial innovation. Nature 2000, 405, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Nan, Z.; Song, Q.; Xia, C.; Li, X.; Yao, X.; Xu, W.; Kuang, Y.; Tian, P.; Zhang, Q. Advances in research on Epichloë endophytes in Chinese native grasses. Front. Microbiol. 2016, 7, 1399. [Google Scholar] [CrossRef] [Green Version]
- Timmusk, S.; Nevo, E.; Ayele, F.; Noe, S.; Niinemets, Y. Fighting Fusarium pathogens in the era of climate change: A conceptual approach. Pathogens 2020, 9, 419. [Google Scholar] [CrossRef]
- Ma, Z.; Xie, Q.; Li, G.; Jia, H.; Zhou, J.; Kong, Z.; Li, N.; Yuan, Y. Germplasms, genetics and genomics for better control of disastrous wheat Fusarium head blight. Theor. Appl. Genet. 2020, 133, 1541–1568. [Google Scholar] [CrossRef]
- Cuthbert, P.A.; Somers, D.J.; Thomas, J.; Cloutier, S.; Brulé-Babel, A. Fine mapping Fhb1, a major gene controlling Fusarium head blight resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2006, 112, 1465. [Google Scholar] [CrossRef]
- Cuthbert, P.A.; Somers, D.J.; Brulé-Babel, A. Mapping of Fhb2 on chromosome 6BS: A gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 114, 429–437. [Google Scholar] [CrossRef]
- Qi, L.L.; Pumphrey, M.O.; Friebe, B.; Chen, P.D.; Gill, B.S. Molecular cytogenetic characterization of alien introgressions with gene Fhb3 for resistance to Fusarium head blight disease of wheat. Theor. Appl. Genet. 2008, 117, 1155–1166. [Google Scholar] [CrossRef]
- Xue, S.; Li, G.; Jia, H.; Xu, F.; Lin, F.; Tang, M.; Wang, Y.; An, X.; Xu, H.; Zhang, L.; et al. Fine mapping Fhb4, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2010, 121, 147–156. [Google Scholar] [CrossRef]
- Xue, S.; Xu, F.; Tang, M.; Zhou, Y.; Li, G.; An, X.; Lin, F.; Xu, H.; Jia, H.; Zhang, L.; et al. Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2011, 123, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Cainong, J.C.; Bockus, W.W.; Feng, Y.; Chen, P.; Qi, L.; Sehgal, S.K.; Danilova, T.V.; Koo, D.-H.; Friebe, B.; Gill, B.S. Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor. Appl. Genet. 2015, 128, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Zhang, X.; Hou, Y.; Cai, J.; Shen, X.; Zhou, T.; Xu, H.; Ohm, H.W.; Wang, H.; Li, A. High-density mapping of the major FHB resistance gene Fhb7 derived from Thinopyrum ponticum and its pyramiding with Fhb1 by marker-assisted selection. Theor. Appl. Genet. 2015, 128, 2301–2316. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Ohm, H. Molecular mapping of Thinopyrum-derived Fusarium head blight resistance in common wheat. Mol. Breed. 2007, 20, 131–140. [Google Scholar] [CrossRef]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Kong, L. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, J. Fungal genes in plants: Impact and potential applications. Trends Plant Sci. 2020, 25, 1064–1067. [Google Scholar] [CrossRef]
- Fu, S.; Lv, Z.; Qi, B.; Guo, X.; Li, J.; Liu, B.; Han, F. Molecular cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium head blight. J. Genet. Genom. 2012, 39, 103–110. [Google Scholar] [CrossRef]
- Ceoloni, C.; Forte, P.; Kuzmanovic, L.; Tundo, S.; Moscetti, I.; De Vita, P.; Virili, M.E.; D’Ovidio, R. Cytogenetic mapping of a major locus for resistance to Fusarium head blight and crown rot of wheat on Thinopyrum elongatum 7EL and its pyramiding with valuable genes from a Th. ponticum homoeologous arm onto bread wheat 7DL. Theor. Appl. Genet. 2017, 130, 2005–2024. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Hu, Y.; Qin, L.; Wang, D.; Jia, J.; Jiao, Y. A recent burst of gene duplications in Triticeae. Plant Com. 2021, 3, 100268. [Google Scholar] [CrossRef]
- Falke, K.C.; Susic, Z.; Wilde, P.; Wortmann, H.; Mohring, J.; Piepho, H.P.; Geiger, H.H.; Miedaner, T. Testcross performance of rye introgression lines developed by marker-assisted backcrossing using an Iranian accession as donor. Theor. Appl. Genet. 2009, 118, 1225–1238. [Google Scholar] [CrossRef]
- Jiang, J.; Friebe, B.; Gill, B.S. Recent advances in alien gene transfer in wheat. Euphytica 1993, 73, 199–212. [Google Scholar] [CrossRef]
| Primer Name | Sequence (5′-3′) | Tm (°C) | Amplified Region |
|---|---|---|---|
| 26102F | CGATAGAAGATAGCTTCAATCAACCCTTT | 60 | CDS |
| 26102R | CTACTTCACCTCGGCATACTTGTC | ||
| 26102ProF | TCCGCATTTCCCTTGCAGAT | 60 | Promoter and CDS |
| 26102RT-F | GGACTTCCCTTGGATCCTGC | 60 | CDS |
| 26102RT-R | ACCGACAATCATGTCCGCAT | ||
| Actin-F | CAACGAGCTCCGTGTCGCA | 60 | CDS |
| Actin-R | GAGGAAGCGTGTATCCCTCATAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Wang, M.; Kang, H.; Zhou, Y.; Han, F. Distribution, Polymorphism and Function Characteristics of the GST-Encoding Fhb7 in Triticeae. Plants 2022, 11, 2074. https://doi.org/10.3390/plants11162074
Guo X, Wang M, Kang H, Zhou Y, Han F. Distribution, Polymorphism and Function Characteristics of the GST-Encoding Fhb7 in Triticeae. Plants. 2022; 11(16):2074. https://doi.org/10.3390/plants11162074
Chicago/Turabian StyleGuo, Xianrui, Mian Wang, Houyang Kang, Yonghong Zhou, and Fangpu Han. 2022. "Distribution, Polymorphism and Function Characteristics of the GST-Encoding Fhb7 in Triticeae" Plants 11, no. 16: 2074. https://doi.org/10.3390/plants11162074
APA StyleGuo, X., Wang, M., Kang, H., Zhou, Y., & Han, F. (2022). Distribution, Polymorphism and Function Characteristics of the GST-Encoding Fhb7 in Triticeae. Plants, 11(16), 2074. https://doi.org/10.3390/plants11162074






