Morphological, Molecular, and Biochemical Characterization of a Unique Lentil (Lens culinaris Medik.) Genotype Showing Seed-Coat Color Anomalies Due to Altered Anthocyanin Pathway
Abstract
:1. Introduction
2. Results
2.1. Seed-Coat Color Anomalies
2.2. Molecular Marker Analysis
2.3. Biochemical Studies
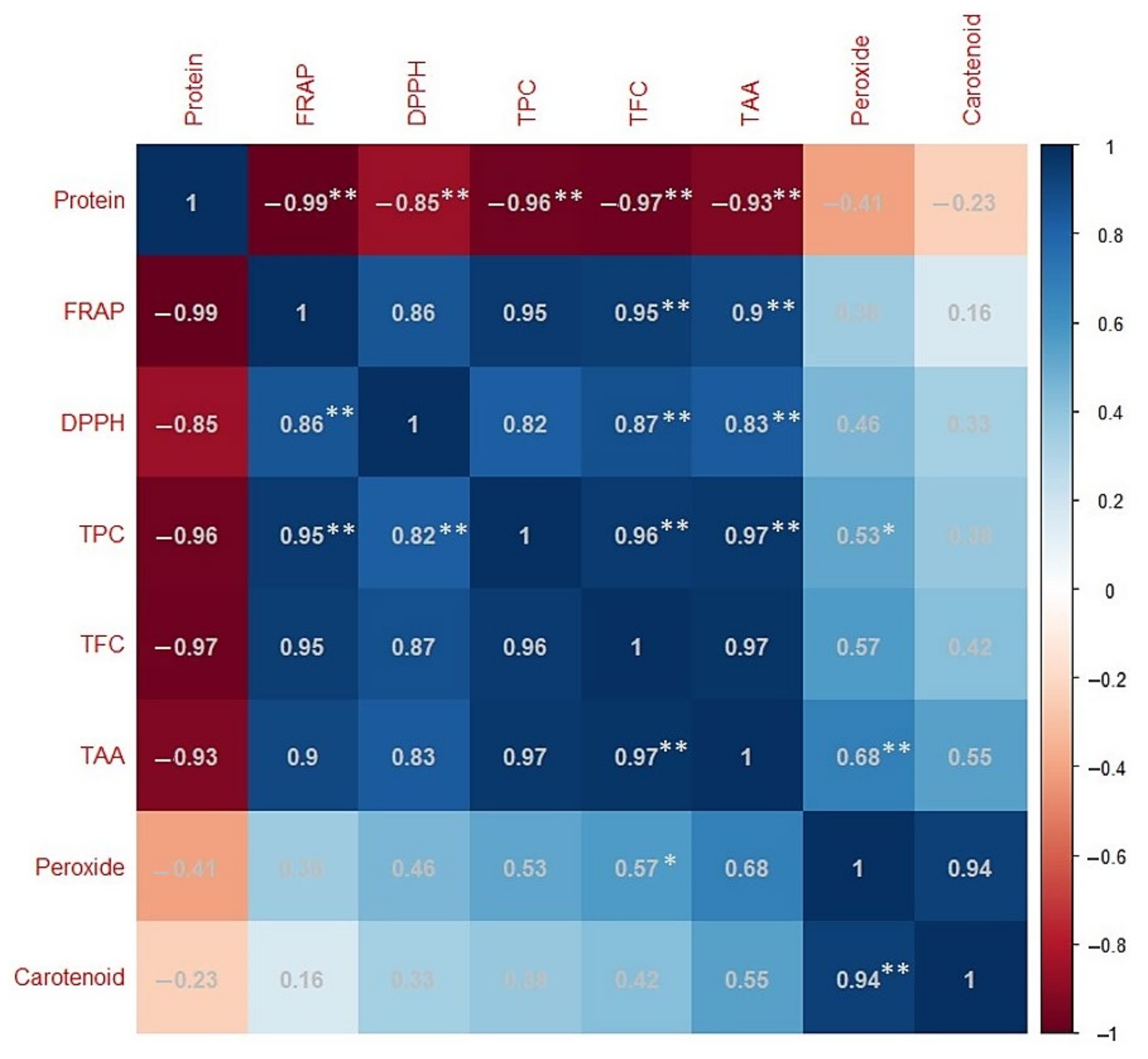
2.4. Correlation Analysis
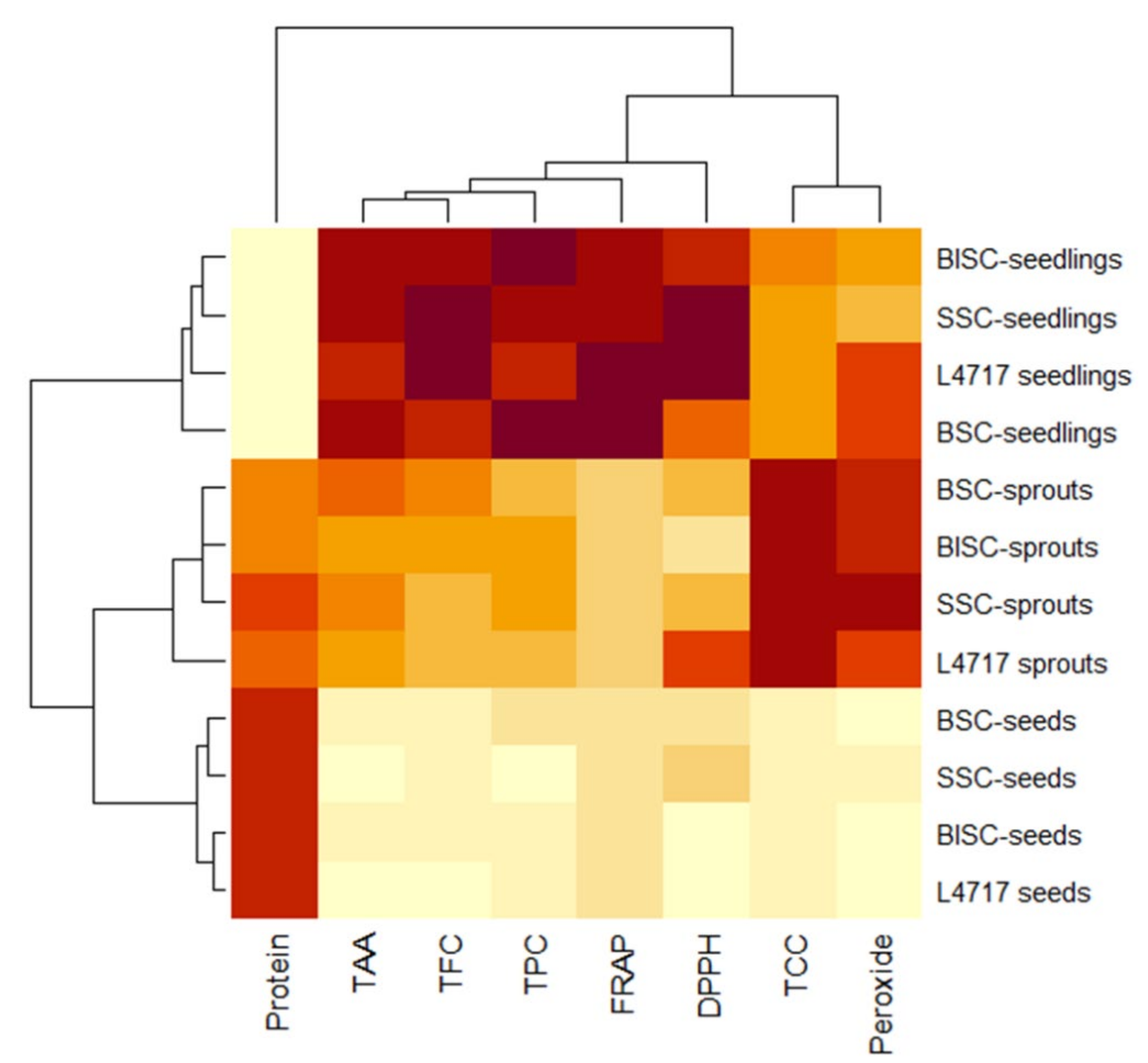
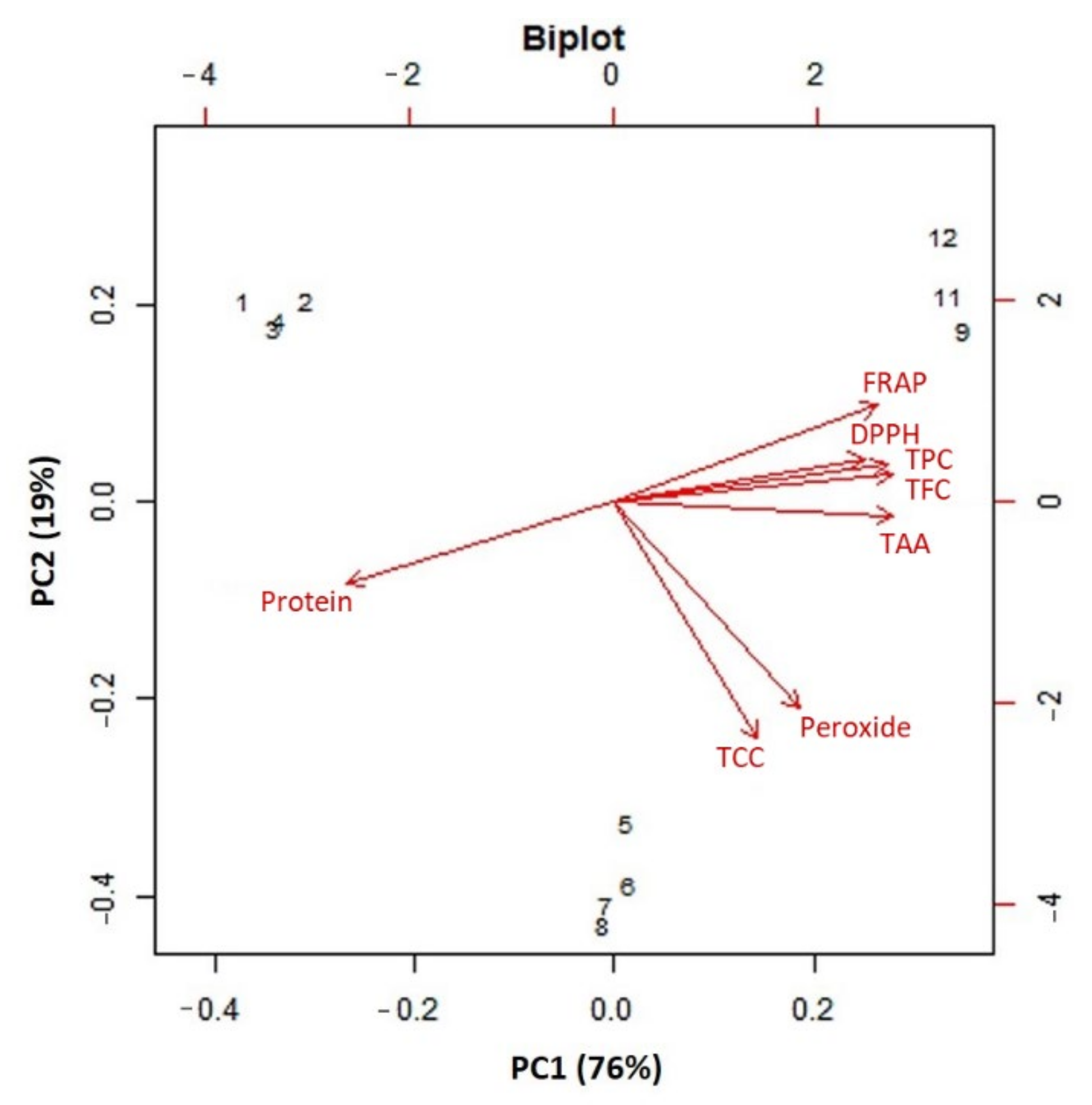
2.5. Cluster and PCA Analysis of Functional Features of Lentil Genotypes
3. Discussion
3.1. Morphological Characterization
3.2. Molecular Characterization
3.3. Biochemical Characterization
3.4. Correlation, Cluster and PCA Analysis
3.5. Probable Effect on the Anthocyanin Biosynthesis Pathway
4. Materials and Methods
4.1. Sowing and Generation-Based Seed-Coat Color Analysis
4.2. SSR Marker Analysis
4.3. Biochemical Studies
4.3.1. Total Phenolic Content (TPC)
4.3.2. Total Flavonoids Content (TFC)
4.3.3. DPPH (1,1-Diphenyl-2-picrylhydrazyl) Scavenging Activity
4.3.4. FRAP (Ferric Reducing Antioxidant Power) Assay
4.3.5. Peroxide Quantification
4.3.6. Total Carotenoid Content (TCC)
4.3.7. Total Ascorbic Acid (TAA)
4.3.8. Total (Crude) Soluble Protein
4.3.9. Total Anthocyanin Content (TAC)
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mishra, G.P.; Aski, M.S.; Bosamia, T.; Chaurasia, S.; Mishra, D.C.; Bhati, J.; Kumar, A.; Javeria, S.; Tripathi, K.; Kohli, M.; et al. Insights into the host-pathogen interaction pathways through RNA-Seq analysis of Lens culinaris Medik. in response to Rhizoctonia bataticola infection. Genes 2022, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- FAOSTAT. 2021. Available online: https://www.fao.org/faostat/en/#home (accessed on 31 December 2021).
- Mishra, G.P.; Dikshit, H.K.; Kumari, J.; Priti; Tripathi, K.; Devi, J.; Aski, M.; Mehra, R.; Sarker, A.; Kumar, S. Identification and characterization of novel Penta-podded genotypes in the cultivated lentil (Lens culinaris Medik.). Crop Sci. 2020, 60, 1974–1985. [Google Scholar] [CrossRef]
- Tschermak-Seysenegg, E. Lentil and Field bean crosses. Sityringsber Akad. Wiss. Wein Math. Nat. KI.I. Abt. 1928, 137, 171–181. [Google Scholar]
- Ladizinsky, G. The genetics of several morphological traits in the lentil. J. Hered. 1979, 70, 135–137. [Google Scholar] [CrossRef]
- Erskine, W.; Witcomb, J.R. Lentil Germplasm Catalogue; ICARDA: Allepo, Syria, 1984; p. 45. [Google Scholar]
- Emami, M.K.; Sharma, B. Inheritance of black testa color in lentil (Lens culinaris Medik). Euphytica 2000, 115, 43–47. [Google Scholar] [CrossRef]
- Wilson, V.E.; Hudson, L.W. Inheritance of lentil flower color. J. Hered. 1978, 69, 129–130. [Google Scholar] [CrossRef]
- Vandenberg, A.; Slinkard, A.E. Genetics of seed coat colour and pattern in lentil. J. Hered. 1990, 81, 484–488. [Google Scholar] [CrossRef]
- Slinkard, A.E. Inheritance of cotyledon color in lentils. J. Hered. 1978, 69, 139–140. [Google Scholar] [CrossRef]
- Sharma, B.; Emami, M.K. Discovery of a new gene causing dark green cotyledon and pathway of pigment synthesis in lentil (Lens culinaris Medil). Euphytica 2002, 124, 349–353. [Google Scholar] [CrossRef]
- Subedi, M.; Tabil, L.G. Vandenberg A Influence of seed coat color genes on milling qualities of red lentil (Lens culinaris Medik). J. Agril. Sci. 2018, 10, 88–97. [Google Scholar] [CrossRef]
- Mirali, M.; Purves, R.W.; Stonehouse, R.; Song, R.; Bett, K.; Vandenberg, A. Genetics and biochemistry of zero-tannin lentils. PLoS ONE 2016, 11, e0164624. [Google Scholar] [CrossRef]
- Vaillancourt, R.; Slinkard, A.E.; Reichert, R.D. The inheritance of condensed tannin concentration in lentil. Can. J. Plant Sci. 1986, 66, 241–246. [Google Scholar] [CrossRef]
- Miyamoto, T.; Everson, E.H. Biochemical and physiological studies of wheat seed pigmentation. Agron. J. 1958, 50, 733–734. [Google Scholar] [CrossRef]
- Styles, E.D.; Ceska, O. The genetic control of flavonoid synthesis in maize. Can J. Genet. Cytol. 1977, 19, 289–302. [Google Scholar] [CrossRef]
- McCallum, J.A.; Walker, J.R.L. Proanthocyanidins in Wheat Bran. Cereal Chem. 1990, 67, 282–285. Available online: https://www.cerealsgrains.org/publications/cc/backissues/1990/Documents/67_282.pdf (accessed on 23 May 2022).
- Finocchiaro, F.; Ferrari, B.; Gianinetti, A. A study of biodiversity of flavonoid content in the rice caryopsis evidencing simultaneous accumulation of anthocyanins and proanthocyanidins in a black-grained genotype. J. Cereal Sci. 2010, 51, 28–34. [Google Scholar] [CrossRef]
- Kohyama, N.; Chono, M.; Nakagawa, H.; Matsuo, Y.; Ono, H.; Matsunaka, H. Flavonoid compounds related to seed coat color of wheat. Biosci. Biotechnol. Biochem. 2017, 81, 2112–2118. [Google Scholar] [CrossRef] [Green Version]
- Mannino, G.; Gentile, C.; Ertani, A.; Serio, G.; Bertea, C.M. Anthocyanins: Biosynthesis, Distribution, Ecological Role, and Use of Biostimulants to Increase Their Content in Plant Foods—A Review. Agriculture 2021, 11, 212. [Google Scholar] [CrossRef]
- Yue, E.; Huang, Y.; Qian, L.; Lu, Q.; Wang, X.; Qian, H.; Yan, J.; Ruan, S. Comparative Analysis of Proanthocyanidin Metabolism and Genes Regulatory Network in Fresh Leaves of Two Different Ecotypes of Tetrastigma hemsleyanum. Plants 2022, 11, 211. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef]
- Mannino, G.; Chinigò, G.; Serio, G.; Genova, T.; Gentile, C.; Munaron, L.; Bertea, C.M. Proanthocyanidins and Where to Find Them: A Meta-Analytic Approach to Investigate Their Chemistry, Biosynthesis, Distribution, and Effect on Human Health. Antioxidants 2021, 10, 1229. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, H.-M.; Ma, Y.-X.; Wang, X.-D. Developments in extraction, purification, and structural elucidation of proanthocyanidins (2000–2019). Stud. Nat. Prod. Chem. 2021, 68, 347–391. [Google Scholar] [CrossRef]
- Zhu, F. Proanthocyanidins in cereals and pseudocereals. Crit. Rev. Food Sci. Nutr. 2019, 59, 1521–1533. [Google Scholar] [CrossRef]
- Jende-Strid, B. Genetic control of flavonoid biosynthesis in barley. Hereditas 1993, 119, 187–204. [Google Scholar] [CrossRef]
- Trojan, V.; Musilova, M.; Vyhnanek, T.; Klejdus, B.; Hanadek, P.; Havel, L. Chalcone synthase expression and pigment deposition in wheat with purple and blue colored caryopsis. J. Cereal. Sci. 2014, 59, 48–55. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Mock, H.P.; Kukoeva, T.V.; Börner, A.; Khlestkina, E.K. Regulation of the Flavonoid Biosynthesis Pathway Genes in Purple and Black Grains of Hordeum vulgare. PLoS ONE 2016, 11, e0163782. [Google Scholar] [CrossRef] [Green Version]
- Nozzolillo, C.; Ricciardi, L. Proanthocyanidin Content of Broad Bean Seeds: Relationship of Seed Coat Color. In Plant Polyphenols, Basic Life Sciences; Hemingway, R.W., Laks, P.E., Eds.; Springer: Boston, MA, USA, 1992; Volume 59. [Google Scholar] [CrossRef]
- Todd, J.J.; Vodkin, L.O. Pigmented Soybean (Glycine max) Seed Coats Accumulate Proanthocyanidins during Development. Plant Physiol. 1993, 102, 663–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirali, M.; Purves, R.W.; Vandenberg, A. Profiling the phenolic compounds of the four major seed coat types and their relation to color genes in lentil. J. Nat. Prod. 2017, 80, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Wilson, V.E.; Hudson, L.W. Seedcoat color anomalies in early generations of lentils. J. Hered. 1978, 69, 205–206. [Google Scholar] [CrossRef]
- Wilson, V.E.; Hudson, L.W. Lentil seedcoat background color inheritance. J. Hered. 1980, 71, 149–150. [Google Scholar] [CrossRef]
- Narendra, V.G.; Abdorrazzaghi, M. An Intelligent system for identification of Indian Lentil types using Artificial Neural Network (BPNN). IOSR J. Comp. Engg. 2013, 15, 54–60. Available online: https://www.iosrjournals.org/iosr-jce/papers/Vol15-issue5/J01555460.pdf?id=8505 (accessed on 31 December 2021).
- Duenas, M.; Sun, B.; Hernaändez, T.; Estrella, I.; Spranger, M.I. Proanthocyanidin composition in the seed coat of lentils (Lens culinaris L.). J. Agric. Food Chem. 2003, 51, 7999–8004. [Google Scholar] [CrossRef]
- Bakhsh, A.; Iqbal, S.M.; Cheema, N.M. Inheritance of morphological characters associated with plant and dried seeds in lentil (Lens culinaris Medik.). Pak. J. Bot. 2013, 45, 1497–1502. Available online: https://www.pakbs.org/pjbot/PDFs/45(5)/02.pdf (accessed on 31 December 2021).
- Prakken, R. Inheritance of colour in Phaseolus vulgaris L. II. A critical review. Meded. Landbouwhogesch Wagening. 1970, 70–23, 1–38. Available online: https://edepot.wur.nl/290008 (accessed on 31 December 2021).
- Harland, S.C. Inheritance of certain characters in the cowpea (Vigna sinensis). J. Genet. 1919, 8, 101–131. [Google Scholar] [CrossRef]
- Fruwirth, C. Selection in pure lines. J. Hered. 1917, 8, 90–94. [Google Scholar] [CrossRef]
- Imai, Y. Recurrent Pseudomutation. Am. Nat. 1935, 69, 456–459. [Google Scholar] [CrossRef]
- Jambhale, N.D.; Nerkar, Y.S. An unstable gene controlling developmental variegation in okra (Abelmoschus esculentus (L.) Moench). Theoret. Appl. Genet. 1985, 71, 122–125. [Google Scholar] [CrossRef]
- Punia, S.S.; Ram, B.; Dheer, M.; Jain, N.K.; Koli, N.R.; Khedar, O.P. Hyper-variable spontaneous genetic variation for earliness, seed characters and other yield-contributing traits in lentil (Lens culinaris Med.). Curr. Sci. 2014, 106, 75–83. Available online: https://www.currentscience.ac.in/Volumes/106/01/0075.pdf (accessed on 31 December 2021).
- Wilson, V.E. Morphology and technique for crossing Lens esculenta Moench. Crop Sci. 1972, 12, 231–232. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, D.P. Hybridization technique in lentil under field conditions. Lens Newslett. 1998, 25, 1–3. Available online: https://agris.fao.org/agris-search/search.do?recordID=QV1999000032 (accessed on 31 December 2021).
- Wilson, V.E.; Law, A.G. Natural crossing in Lens esculenta Moench. J. Am. Soc. Hortic. Sci. 1972, 97, 142–143. [Google Scholar]
- Chandlee, J.M.; Vodkin, L.O. Unstable expression of a soybean gene during seed coat development. Theoret. Appl. Genet. 1989, 77, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Groose, R.W.; Schulte, S.M.; Palmer, R.G. Germinal reversion of an unstable mutation for anthocyanin pigmentation in soybean. Theoret. Appl. Genet. 1990, 79, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Zabala, G.; Vodkin, L.O. Methylation Affects Transposition and Splicing of a Large CACTA Transposon from a MYB Transcription Factor Regulating Anthocyanin Synthase Genes in Soybean Seed Coats. PLoS ONE 2014, 9, e111959. [Google Scholar] [CrossRef] [Green Version]
- Prior, R.L.; Wu, X.; Schaich, K. Standardized methods for the determination of antioxidant capacity and phenolics in foods and dietary supplements. J. Agric. Food Chem. 2005, 53, 4290–4302. [Google Scholar] [CrossRef] [PubMed]
- Heim, K.E.; Tagliaferro, A.R.; Bobilya, D.J. Flavonoid antioxidants: Chemistry, metabolism and structure-activity relationships. J. Nutr. Biochem. 2002, 13, 572–584. [Google Scholar] [CrossRef]
- Aron, P.M.; Kennedy, J.A. Flavan-3-ols: Nature, occurrence and biological activity. Mol. Nutr. Food Res. 2008, 52, 79–104. [Google Scholar] [CrossRef]
- Swieca, M.; Surdyka, M.; Gawlik-Dziki, U.; Złotek, U.; Baraniak, B. Antioxidant potential of fresh and stored lentil sprouts affected by elicitation with temperature stresses. Int. J. Food Sci. Technol. 2014, 49, 1811–1817. [Google Scholar] [CrossRef]
- Fouad, A.A.; Rehab, F.M.A. Effect of germination time on proximate analysis, bioactive compounds and antioxidant activity of lentil (Lens culinaris Medik.) sprouts. Acta Sci. Pol. Technol. Aliment. 2015, 14, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Altuner, F. Determination of biochemical composition and pigment content in legume and cereal microgreens. Legume Res. 2021, 44, 1018–1025. [Google Scholar] [CrossRef]
- Fuente, B.D.L.; López-García, G.; Máñez, V.; Alegría, A.; Barberá, R.; Cilla, A. Evaluation of the bioaccessibility of antioxidant bioactive compounds and minerals of four genotypes of brassicaceae microgreens. Foods 2019, 8, 250. [Google Scholar] [CrossRef] [Green Version]
- Priti; Mishra, G.P.; Dikshit, H.K.; Vinutha, T.; Tontang, M.T.; Stobdan, T.; Sangwan, S.; Aski, M.; Singh, A.; Kumar, R.R.; et al. Diversity in phytochemical composition, antioxidant capacities, and nutrient contents among mungbean and lentil microgreens when grown at plain-altitude region (Delhi) and high-altitude region (Leh-Ladakh), India. Front. Plant Sci. 2021, 12, 710812. [Google Scholar] [CrossRef]
- Swieca, M.; Gwalik-Dziki, U. Effects of sprouting and postharvest storage under cool temperature conditions on starch content and antioxidant capacity of green pea, lentil and young mung bean sprouts. Food Chem. 2015, 185, 99–105. [Google Scholar] [CrossRef]
- Ahuja, H.; Kaur, S.; Gupta, A.K.; Singh, S.; Kaur, J. Biochemical mapping of lentil (Lens culinaris Medik) genotypes for quality traits. Acta Physiol. Plant. 2015, 37, 179. [Google Scholar] [CrossRef]
- Wojdylo, A.; Nowicka, P.; Tkacz, K.; Turkiewicz, I.P. Sprouts vs. microgreens as novel functional foods: Variation of nutritional and phytochemical profiles and their in vitro bioactive properties. Molecules 2020, 25, 4648. [Google Scholar] [CrossRef]
- Cheeseman, J.M. Hydrogen peroxide concentrations in leaves under natural conditions. J. Exp. Bot. 2006, 57, 2435–2444. [Google Scholar] [CrossRef] [Green Version]
- Ozden, M.; Demirel, U.; Kahraman, A. Effects of proline on antioxidant system in leaves of grapevine (Vitis vinifera L.) exposed to oxidative stress by H2O2. Sci. Hortic. 2009, 119, 163–168. [Google Scholar] [CrossRef]
- Biju, S.; Fuentes, S.; Gupta, G. Physiological and Biochemical Responses of Lentils to Silicon Mediated Drought Tolerance. 2018. Available online: https://grdc.com.au/resources-and-publications/grdc-update-papers/tab-content/grdc-update-papers/2018/02/physiological-and-biochemical-responses-of-lentils-to-silicon-mediated-drought-tolerance (accessed on 31 December 2021).
- Yang, C.; Shahidi, F.; Tsao, R. Biomarkers of oxidative stress and cellular-based assays of indirect antioxidant measurement. In Measurement of Antioxidant Activity and Capacity: Recent Trends and Applications; Apak, R., Capanoglu, E., Shahidi, F., Hoboken, N.J., Eds.; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 2018; pp. 165–186. [Google Scholar] [CrossRef]
- Zhang, B.; Deng, Z.; Tang, Y.; Chen, P.; Liu, R.; Ramdath, D.D.; Liu, Q.; Hernandez, M.; Tsao, R. Fatty acid, carotenoid and tocopherol compositions of 20 Canadian lentil cultivars and synergistic contribution to antioxidant activities. Food Chem. 2014, 161, 296–304. [Google Scholar] [CrossRef]
- Ashokkumar, K.; Diapari, M.; Jha, A.B.; Tar’an, B.; Arganosa, G.; Warkentin, T.D. Genetic diversity of nutritionally important carotenoids in 94 pea and 121 chickpea accessions. J. Food Compos. Anal. 2015, 43, 49–60. [Google Scholar] [CrossRef]
- Kowitcharoen, L.; Phornvillay, S.; Lekkham, P.; Pongprasert, N.; Srilaong, V. Bioactive composition and nutritional profile of microgreens cultivated in Thailand. Appl. Sci. 2021, 11, 7981. [Google Scholar] [CrossRef]
- Swieca, M.; Baraniak, B. Nutritional and Antioxidant Potential of Lentil Sprouts Affected by Elicitation with Temperature Stress. J. Agric. Food Chem. 2014, 62, 3306–3313. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Wheeler, G.L. Ascorbic acid in plants: Biosynthesis and function. Critical. Rev. Plant Sci. 2000, 19, 267–290. [Google Scholar] [CrossRef]
- Paciolla, C.; Fortunato, S.; Dipierro, N.; Paradiso, A.; Leonardis, S.D.; Mastropasqua, L. Pinto MCD Vitamin C in plants: From functions to biofortification. Antioxidants 2019, 8, 519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fidrianny, I.; Suhendy, H.; Insanu, M. Correlation of phytochemical content with antioxidant potential of various sweet potato (Ipomoea batatas) in West Java, Indonesia. Asian Pac. J. Trop. Biomed. 2018, 8, 25–30. [Google Scholar] [CrossRef]
- Bhoyar, M.S.; Mishra, G.P.; Naik, P.K.; Srivastava, R.B. Estimation of antioxidant activity and total phenolics among natural populations of Caper (Capparis spinosa) leaves collected from cold arid desert of trans-Himalayas. Aust. J. Crop Sci. 2011, 5, 912–919. Available online: http://www.cropj.com/mishra_5_7_2011_912_919.pdf (accessed on 31 December 2021).
- Bhoyar, M.S.; Mishra, G.P.; Naik, P.K.; Singh, S.B. Evaluation of antioxidant capacities and total polyphenols in various edible parts of Capparis spinosa L. collected from trans-Himalayas. Def. Life Sci. J. 2018, 3, 30–36. [Google Scholar] [CrossRef]
- Patel, K.G.; Mandaliya, V.B.; Mishra, G.P.; Dobaria, J.R.; Radhakrishnan, T. Transgenic peanut overexpressing mtlD gene confers enhanced salinitystress tolerance via mannitol accumulation and differential antioxidative responses. Acta Physiol. Plant 2016, 38, 181. [Google Scholar] [CrossRef]
- Bhalani, H.; Mishra, G.P.; Sarkar, T.; Radhakrishnan, T. Regulation of antioxidant mechanisms by AtDREB1A improves soil-moisture deficit stress tolerance in transgenic peanut (Arachis hypogaea L.). PLoS ONE 2019, 14, e0216706. [Google Scholar] [CrossRef] [Green Version]
- Devi, J.; Sanwal, S.K.; Koley, T.K.; Mishra, G.P.; Karmakar, P.; Singh, P.; Singh, B. Variations in the total phenolics and antioxidant activities among garden pea (Pisum sativum L.) genotypes differing for maturity duration, seed and flower traits and their association with the yield. Sci. Hortic. 2019, 244, 141–150. [Google Scholar] [CrossRef]
- Pereira, D.M.; Valentao, P.; Pereira, J.A.; Andrade, P.B. Phenolics: From chemistry to biology. Molecules 2009, 14, 2202–2211. [Google Scholar] [CrossRef]
- Oktay, M.; Gulcin, I.; Kufrevioglu, O.I. Determination of in vitro antioxidant activity of fennel (Foeniculum vulgare) seed extracts. LebensmWiss Techol. 2003, 36, 263–271. [Google Scholar] [CrossRef]
- El-Nakhel, C.; Pannico, A.; Kyriacou, M.C.; Giordano, M.; De Pascale, S.; Rouphael, Y. Macronutrient deprivation eustress elicits differential secondary metabolites in red and green-pigmented butterhead lettuce grown in a closed soilless system. J. Sci. Food Agric. 2019, 99, 6962–6972. [Google Scholar] [CrossRef]
- El-Nakhel, C.; Giordano, M.; Pannico, A.; Carillo, P.; Fusco, G.M.; De Pascale, S.; Rouphael, Y. Cultivar-specific performance and qualitative descriptors for butterhead Salanova lettuce produced in closed soilless cultivation as a candidate salad crop for human life support in space. Life 2019, 9, 61. [Google Scholar] [CrossRef] [Green Version]
- Kyriacou, M.C.; El-Nakhel, C.; Pannico, A.; Graziani, G.; Soteriou, G.A.; Giordano, M.; Zarrelli, A.; Ritieni, A.; De Pascale, S.; Rouphael, Y. Genotype-specific modulatory effects of select spectral bandwidths on the nutritive and phytochemical composition of microgreens. Front. Plant Sci. 2019, 10, 1501. [Google Scholar] [CrossRef]
- El-Nakhel, C.; Pannico, A.; Graziani, G.; Kyriacou, M.C.; Giordano, M.; Ritieni, A.; De Pascale, S.; Rouphael, Y. Variation in macronutrient content, phytochemical constitution and in vitro antioxidant capacity of green and red butterhead lettuce dictated by different developmental stages of harvest maturity. Antioxidants 2020, 9, 300. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Jeong, N.; Moon, J.K.; Lee, Y.H.; Lee, S.H.; Kim, H.M.; Hwang, C.H.; Back, K.; Palmer, R.G.; Jeong, S.C. Genetic analysis of genes controlling natural variation of seed coat and flower colors in soybean. J. Hered. 2010, 101, 757–768. [Google Scholar] [CrossRef] [Green Version]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [Green Version]
- Quattrocchio, F.; Baudry, A.; Lepiniec, L.; Grotewold, E. The regulation of flavonoid biosynthesis. In The Science of Flavonoids; Grotewold, E., Ed.; Springer: New York, NY, USA, 2016; pp. 97–122. [Google Scholar] [CrossRef]
- Takeoka, G.R.; Dao, L.T.; Tamura, H.; Harden, L.A. Delphinidin 3-O-(2-O-â-D-Glucopyranosyl-r-L-arabinopyranoside): A novel anthocyanin identified in beluga black lentils. J. Agric. Food Chem. 2005, 53, 4932–4937. [Google Scholar] [CrossRef]
- Oomah, B.D.; Caspar, F.; Malcolmson, L.J.; Bellido, A.-S. Phenolics and antioxidant activity of lentil and pea hulls. Food Res. Int. 2011, 44, 436–441. [Google Scholar] [CrossRef]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Ichino, T.; Fuji, K.; Ueda, H.; Takahashi, H.; Koumoto, Y.; Takagi, J.; Tamura, K.; Sasaki, R.; Aoki, K.; Shimada, T.; et al. GFS9/TT9 contributes to intracellular membrane trafficking and flavonoid accumulation in Arabidopsis thaliana. Plant J. 2014, 80, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; He, Q.; Ma, X.; Zhang, L. Characteristics of Color Development in Seeds of Brownand Yellow-Seeded Heading Chinese Cabbage and Molecular Analysis of Brsc, the Candidate Gene Controlling Seed Coat Color. Front. Plant Sci. 2017, 8, 1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.; Thompson, W. The isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, N.; Dikshit, H.K.; Singh, D.; Singh, A.; Kumar, H. Discovery of EST-derived microsatellite primers in the legume Lens culinaris (Fabaceae). Appl. Plant Sci. 2013, 1, apps.1200539. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, U.; Mishra, G.P.; Dikshit, H.K.; Mishra, D.C.; Bosamia, T.; Roy, A.; Bhati, J.; Priti; Aski, M.; Kumar, R.R.; et al. Comparative RNA-Seq analysis unfolds a complex regulatory network imparting yellow mosaic disease resistance in mungbean [Vigna radiata (L.) R. Wilczek]. PLoS ONE 2021, 16, e0244593. [Google Scholar] [CrossRef]
- Kumar, H.; Singh, A.; Dikshit, H.K.; Mishra, G.P.; Aski, M.; Meena, M.C.; Kumar, S. Genetic dissection of grain iron and zinc concentrations in lentil (Lens culinaris Medik.). J. Genet. 2019, 98, 66. [Google Scholar] [CrossRef]
- Singleton, V.L.; Orthofer, R.; Lamuela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Ciocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar] [CrossRef]
- Woisky, R.G.; Salatino, A. Analysis of propolis: Some parameters and procedure for chemical quality control. J. Apicultural. Res. 1998, 37, 99–105. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method. LWT-Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Benzie, F.F.; Strain, J.J. Ferric Reducing/antioxidant power assay: Direct measure of total antioxidant activity of biological fluids and modified version for simultaneous measurement of total antioxidant power and ascorbic acid concentration. Methods Enzymol. 1999, 299, 15–23. [Google Scholar] [CrossRef]
- Loreto, F.; Velikova, V. Isoprene produced by leaves protects the photosynthetic apparatus against ozone damage, quenches ozone products, and reduces lipid peroxidation of cellular membranes. Plant Physiol. 2001, 127, 1781–1787. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Wellburn, A.R. Determination of total carotenoids and chlorophylls A and B of leaf in different solvents. Biol. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, S.S.; Vitamin, C. Determination by Indophenol method. In Food Analysis Laboratory Manual; Springer: Cham, Switzerland, 2017; pp. 143–146. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative Study. J AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef] [Green Version]
- Giusti, M.M.; Luis, E.; Saona, R.; Wrolstad, R.E. Molar absorptivity and color characteristics of acylated and non-acylated pelargonidin-based anthocyanins. J. Agric. Food Chem. 1999, 47, 4631–4637. [Google Scholar] [CrossRef]

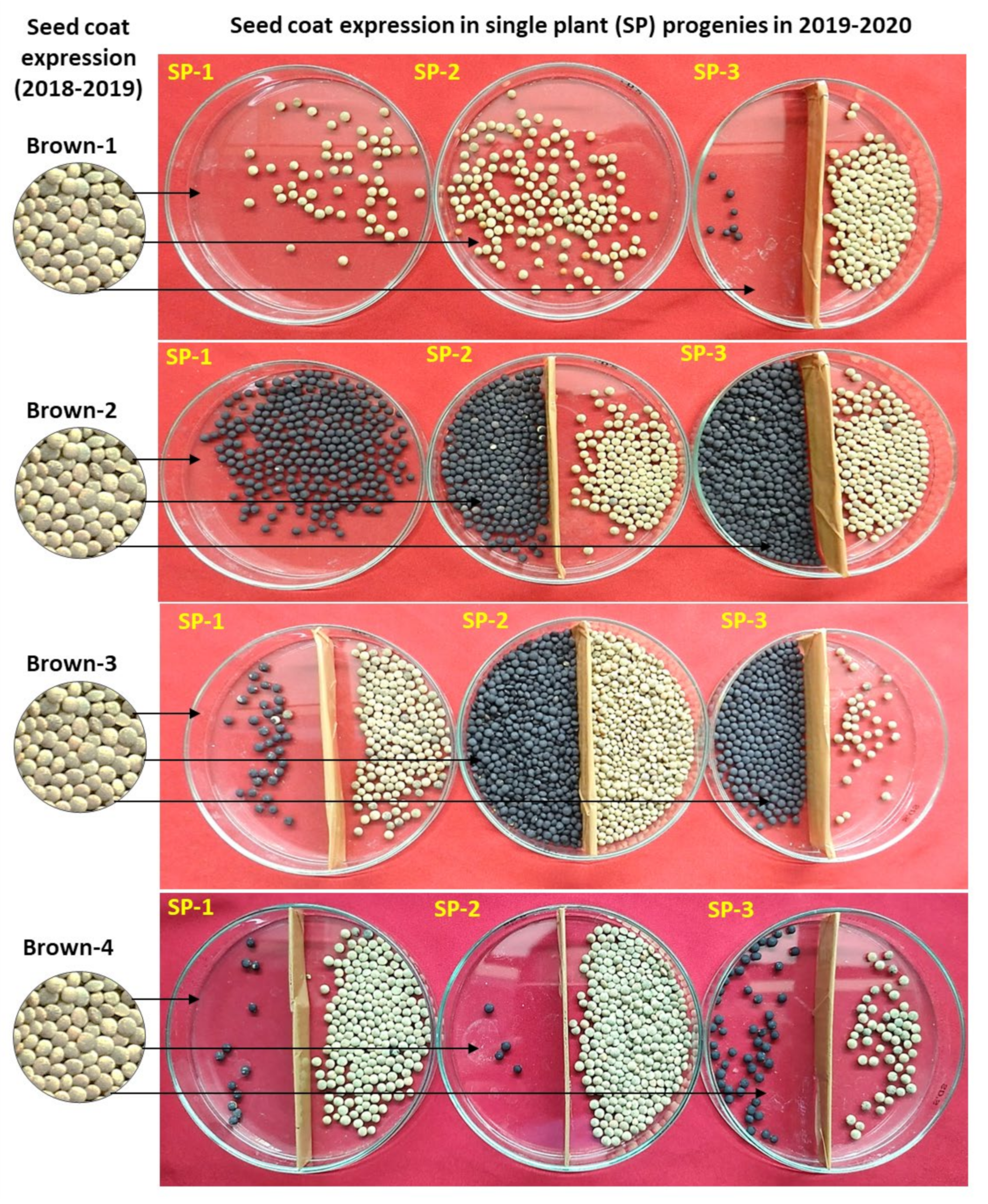
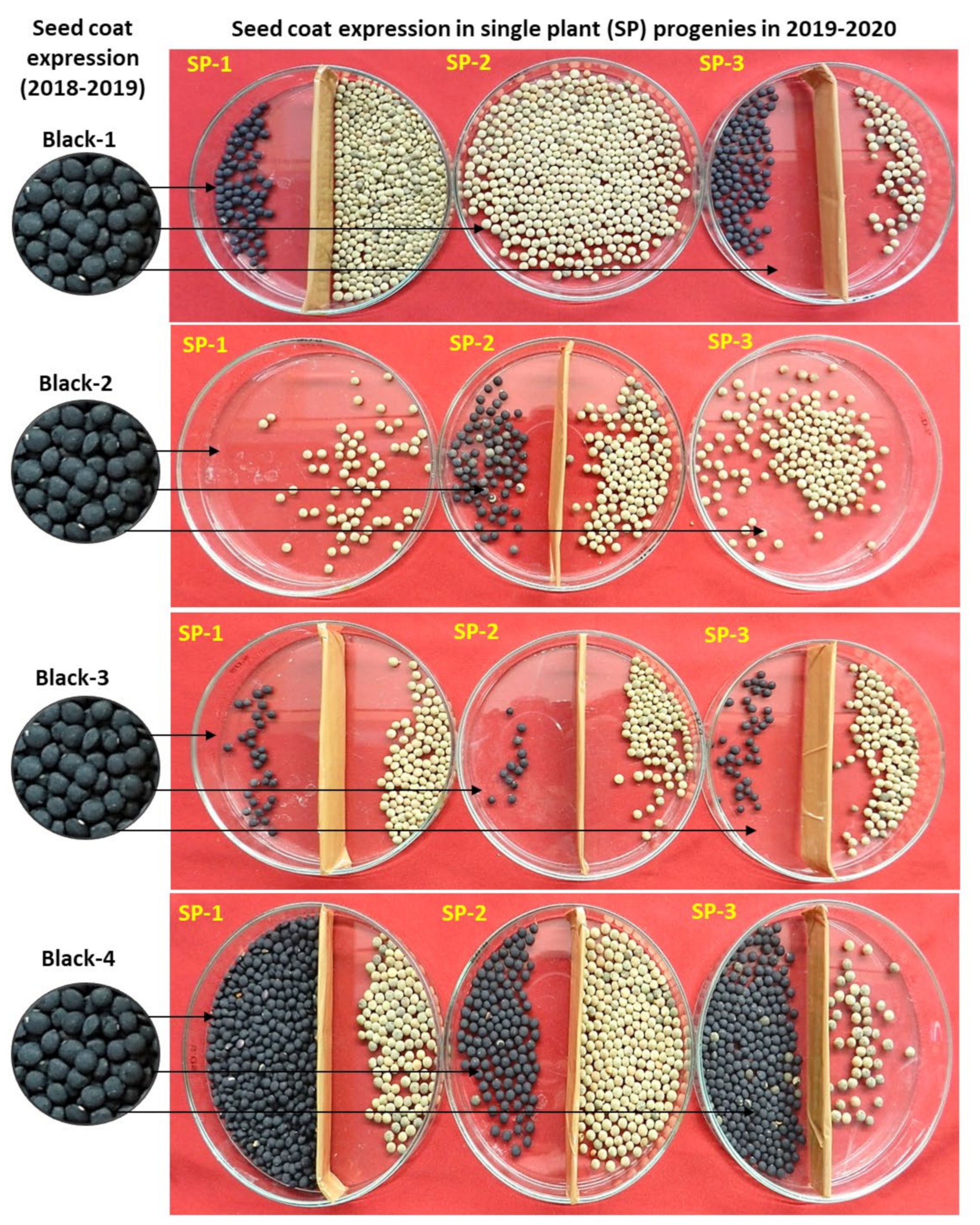




| Year | Seed-Coat Color-Based Plant Category | Total | ||
|---|---|---|---|---|
| Brown Seed-Coat Colored Plants | Black Seed-Coat Colored Plants | Mixed Seed-Coat Colored Plants | ||
| 2018–2019 | 36 | 01 | 75 | 112 |
| 2019–2020 | 75 | 03 | 164 | 242 |
| 2020–2021 | 65 | 02 | 102 | 169 |
| 2018–2019 (Brown Seed Coat) | 2019–2020 | 2018–2019 (Black Seed Coat) | 2019–2020 | 2018–2019 (Spotted Seed Coat) | 2019–2020 | |||
|---|---|---|---|---|---|---|---|---|
| Progenies | Seed-Coat Color | Progenies | Seed-Coat Color | Progenies | Seed-Coat Color | |||
| Plant-1 | SP-1 | Brown | Plant-1 | SP-1 | Mixed | Plant-1 | SP-1 | Brown |
| SP-2 | Brown | SP-2 | Brown | SP-2 | Mixed | |||
| SP-3 | Mixed | SP-3 | Mixed | SP-3 | Brown | |||
| Plant-2 | SP-1 | Black | Plant-2 | SP-1 | Brown | Plant-2 | SP-1 | Mixed |
| SP-2 | Mixed | SP-2 | Mixed | SP-2 | Mixed | |||
| SP-3 | Mixed | SP-3 | Brown | SP-3 | Mixed | |||
| Plant-3 | SP-1 | Mixed | Plant-3 | SP-1 | Mixed | Plant-3 | SP-1 | Mixed |
| SP-2 | Mixed | SP-2 | Mixed | SP-2 | Mixed | |||
| SP-3 | Mixed | SP-3 | Mixed | SP-3 | Mixed | |||
| Plant-4 | SP-1 | Mixed | Plant-4 | SP-1 | Mixed | Plant-4 | SP-1 | Brown |
| SP-2 | Mixed | SP-2 | Mixed | SP-2 | Mixed | |||
| SP-3 | Mixed | SP-3 | Mixed | SP-3 | Mixed | |||
| S. No. | Samples | TPC (mg GAE/100 g DW) | TFC (mg/g DW) | DPPH scavenging (%) | FRAP (µmol Fe2+/g DW) | H2O2 (nmol/g DW) | TCC (mg/100 g DW) | Crude protein (g/100 g DW) | TAC (mg/g DW) | TAA (mg/100 g DW) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L4717 seeds | 1108.15 ± 32.12 h | 1.91 ± 0.07 d | 35.97 ± 4.67 d | 8.56 ± 0.66 e | 1.64 ± 0.05 c | 2.55 ± 0.09 e | 25.60 ± 0.05 ab | 0.016 ± 0.009 b | 27.94 ± 1.70 fg |
| 2 | Brown seed- coat seeds | 1206.24 ± 20.53 g | 1.97 ± 0.02 cd | 42.86 ± 3.63 cd | 9.23 ± 0.64 e | 1.76 ± 0.08 c | 2.57 ± 0.10 e | 25.04 ± 0.36 b | 0.018 ± 0.003 b | 29.41 ± 4.80 defg |
| 3 | Black seed- coat seeds | 1132.99 ± 44.25 h | 2.04 ± 0.17 cd | 36.48 ± 3.67 d | 7.35 ± 0.20 f | 1.78 ± 0.14 c | 2.61 ± 0.11 e | 24.88 ± 0.29 b | 0.042 ± 0.018 a | 28.68 ± 5.02 efg |
| 4 | Spotted seed- coat seeds | 1047.64 ± 32.29 i | 1.98 ± 0.29 cd | 44.90 ± 5.46 c | 8.82 ± 0.60 e | 1.79 ± 005 c | 2.57 ± 0.14 e | 26.43 ± 1.13 a | 0.033 ± 0.004 ab | 27.21 ± 2.82 g |
| 5 | L4717 sprouts | 1489.68 ± 11.86 f | 2.41 ± 0.12 bc | 59.45 ± 2.31 ab | 14.37 ± 0.09 d | 2.84 ± 0.14 a | 41.95 ± 0.30 bc | 21.75 ± 0.11 d | ND | 35.29 ± 0.00 cdef |
| 6 | Brown seed- coat sprouts | 1471.21 ± 12.72 f | 2.62 ± 0.13 b | 46.94 ± 3.23 c | 14.99 ± 0.10 c | 2.95 ± 0.23 a | 42.81 ± 0.21 b | 21.16 ± 0.45 d | ND | 38.24 ± 2.40 abc |
| 7 | Black seed- coat sprouts | 1639.36 ± 17.94 e | 2.53 ± 0.27 b | 41.92 ± 2.36 cd | 14.75 ± 0.17 d | 2.93 ± 0.02 a | 45.35 ± 1.44 a | 20.80 ± 0.31 d | ND | 36.03 ± 1.47 bcde |
| 8 | Spotted seed- coat sprouts | 1628.54 ± 18.95 e | 2.39 ± 0.32 bc | 47.10 ± 3.12 c | 14.75 ± 0.40 d | 3.04 ± 0.16 a | 42.72 ± 0.29 b | 23.62 ± 0.54 c | ND | 36.76 ± 2.94 a bcd |
| 9 | L4717 seedlings | 2107.52 ± 21.72 d | 3.25 ± 0.26 a | 66.46 ± 1.00 a | 46.04 ± 0.34 a | 2.74 ± 0.17 a | 37.95 ± 0.30 bd | 9.82 ± 0.16 f | ND | 42.65 ± 3.80 abc |
| 10 | Brown seed- coat seedlings | 2332.36 ± 25.76 b | 3.09 ± 0.25 a | 56.55 ± 2.83 b | 44.70 ± 1.59 b | 2.81 ± 0.12 a | 38.81 ± 0.21 d | 9.52 ± 013 f | ND | 44.12 ± 3.40 a |
| 11 | Black seed- coat seedlings | 2434.27 ± 14.62 a | 3.15 ± 0.17 a | 61.89 ± 3.68 ab | 43.24 ± 0.68 c | 2.41 ± 0.07 b | 41.35 ± 1.44 c | 10.80 ± 0.15 e | ND | 43.97 ± 2.42 a |
| 12 | Spotted seed- coat seedlings | 2199.24 ± 16.25 c | 3.28 ± 0.07 a | 65.85 ± 1.22 a | 42.85 ± 0.36 c | 2.30 ± 0.07 b | 38.72 ± 0.29 d | 9.93 ± 0.57 ef | ND | 43.38 ± 4.52 ab |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, G.P.; Ankita; Aski, M.S.; Tontang, M.T.; Choudhary, P.; Tripathi, K.; Singh, A.; Kumar, R.R.; Thimmegowda, V.; Stobdan, T.; et al. Morphological, Molecular, and Biochemical Characterization of a Unique Lentil (Lens culinaris Medik.) Genotype Showing Seed-Coat Color Anomalies Due to Altered Anthocyanin Pathway. Plants 2022, 11, 1815. https://doi.org/10.3390/plants11141815
Mishra GP, Ankita, Aski MS, Tontang MT, Choudhary P, Tripathi K, Singh A, Kumar RR, Thimmegowda V, Stobdan T, et al. Morphological, Molecular, and Biochemical Characterization of a Unique Lentil (Lens culinaris Medik.) Genotype Showing Seed-Coat Color Anomalies Due to Altered Anthocyanin Pathway. Plants. 2022; 11(14):1815. https://doi.org/10.3390/plants11141815
Chicago/Turabian StyleMishra, Gyan P., Ankita, Muraleedhar S. Aski, Mechiya Tomuilim Tontang, Priti Choudhary, Kuldeep Tripathi, Ajeet Singh, Ranjeet Ranjan Kumar, Vinutha Thimmegowda, Tsering Stobdan, and et al. 2022. "Morphological, Molecular, and Biochemical Characterization of a Unique Lentil (Lens culinaris Medik.) Genotype Showing Seed-Coat Color Anomalies Due to Altered Anthocyanin Pathway" Plants 11, no. 14: 1815. https://doi.org/10.3390/plants11141815
APA StyleMishra, G. P., Ankita, Aski, M. S., Tontang, M. T., Choudhary, P., Tripathi, K., Singh, A., Kumar, R. R., Thimmegowda, V., Stobdan, T., Kumar, A., Bhardwaj, R., Praveen, S., Yadava, D. K., Kumar, S., & Dikshit, H. K. (2022). Morphological, Molecular, and Biochemical Characterization of a Unique Lentil (Lens culinaris Medik.) Genotype Showing Seed-Coat Color Anomalies Due to Altered Anthocyanin Pathway. Plants, 11(14), 1815. https://doi.org/10.3390/plants11141815








