In Silico Conformation of the Drug Colchicine into Tubulin Models and Acute Phytotoxic Activity on Cucumis sativus Radicles
Abstract
1. Introduction
2. Results and Discussion
2.1. Protein Sequence Alignment Comparison and Modeling of the αβ-Tubulin Heterodimers of C. sativus
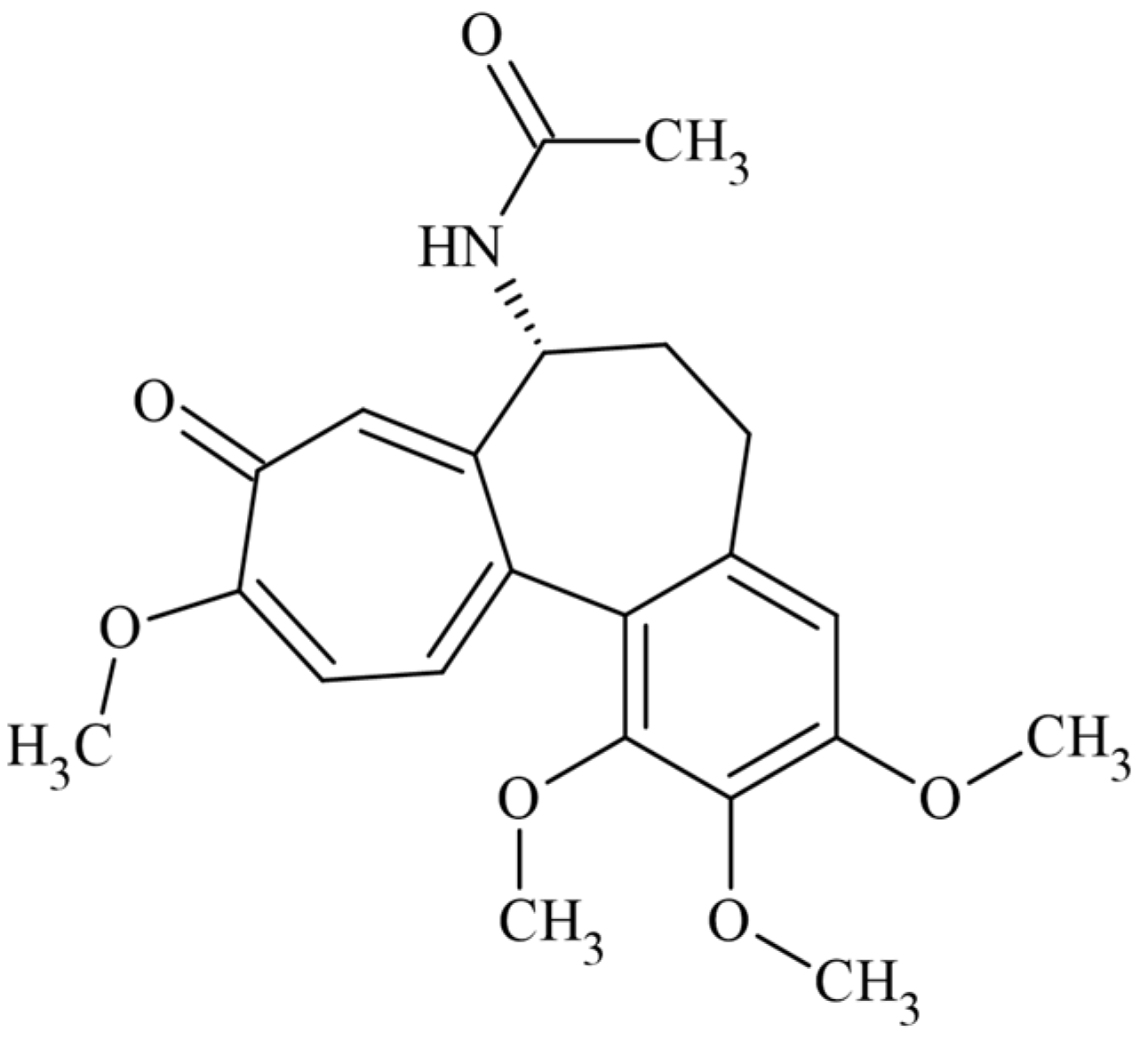
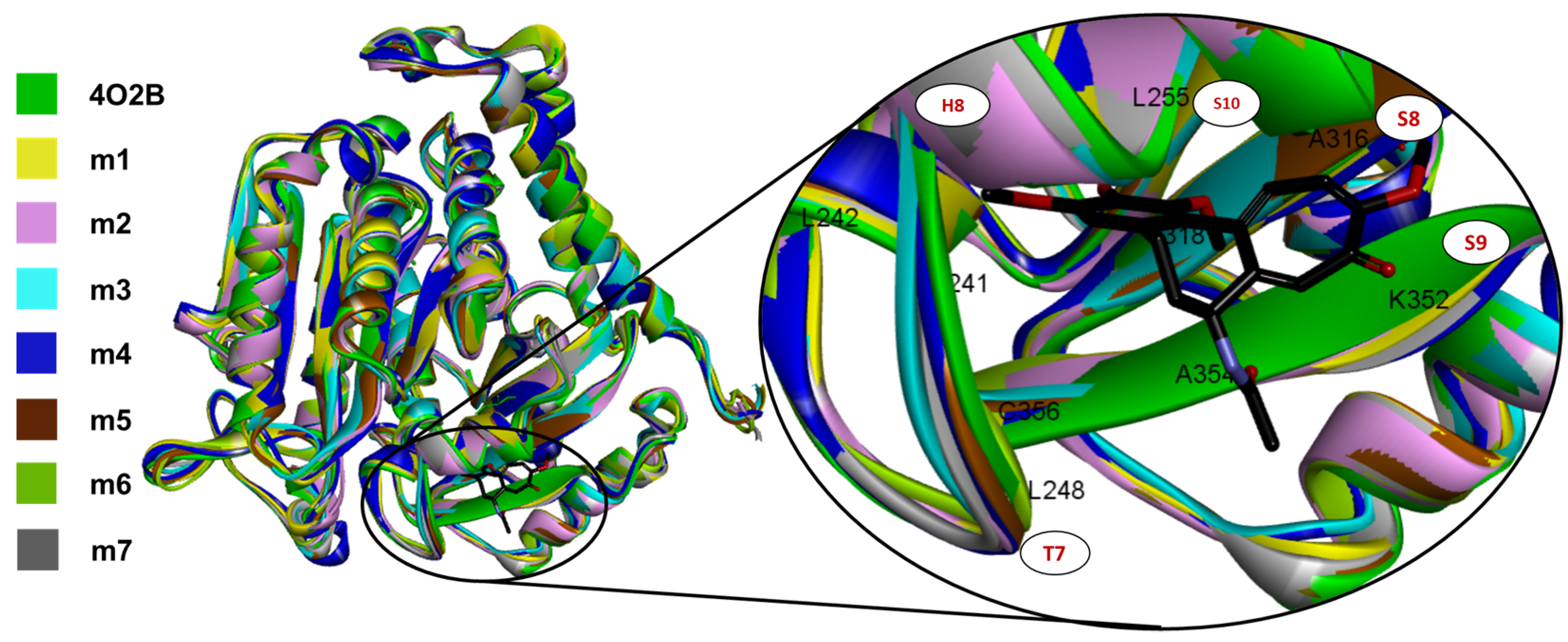
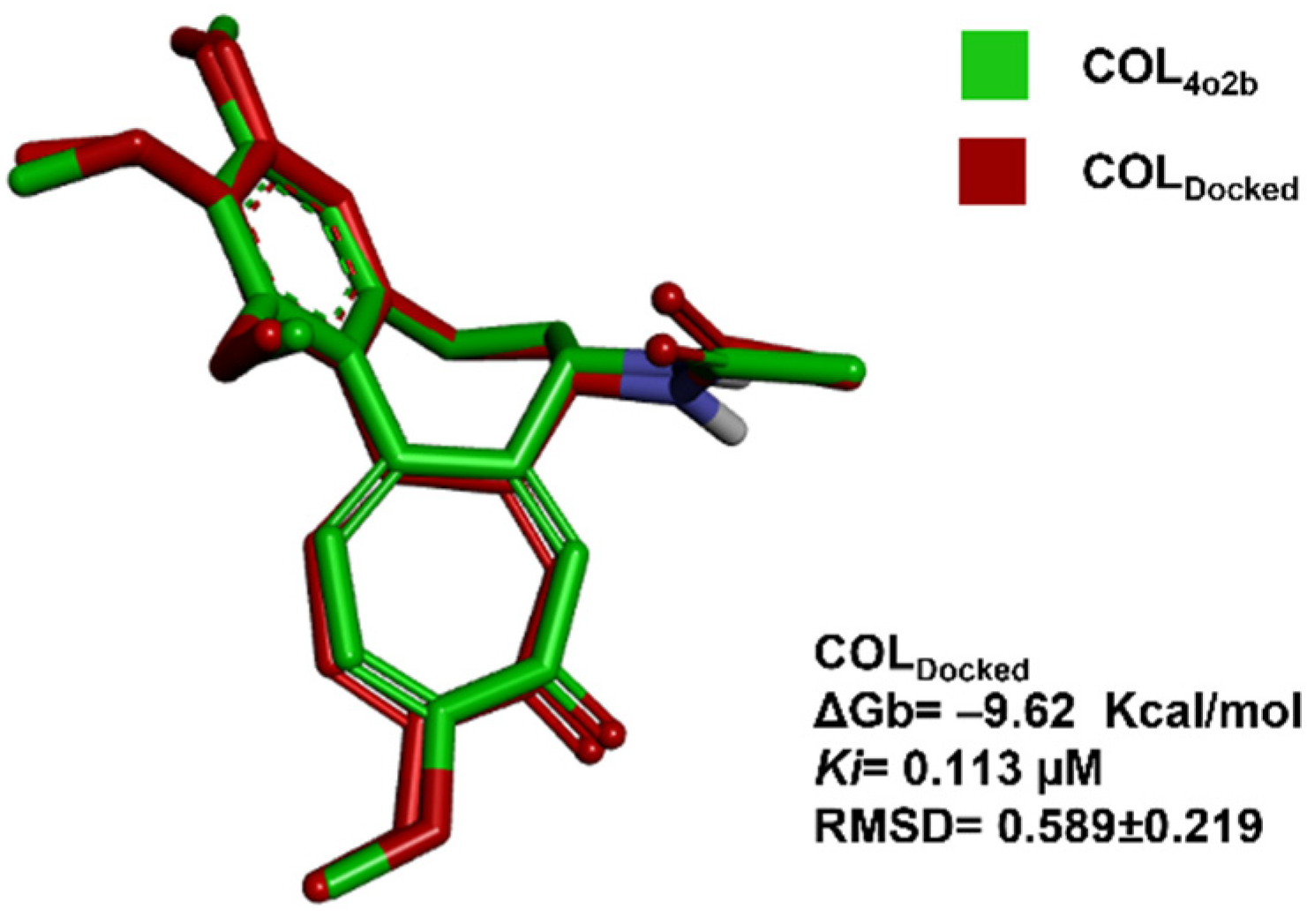
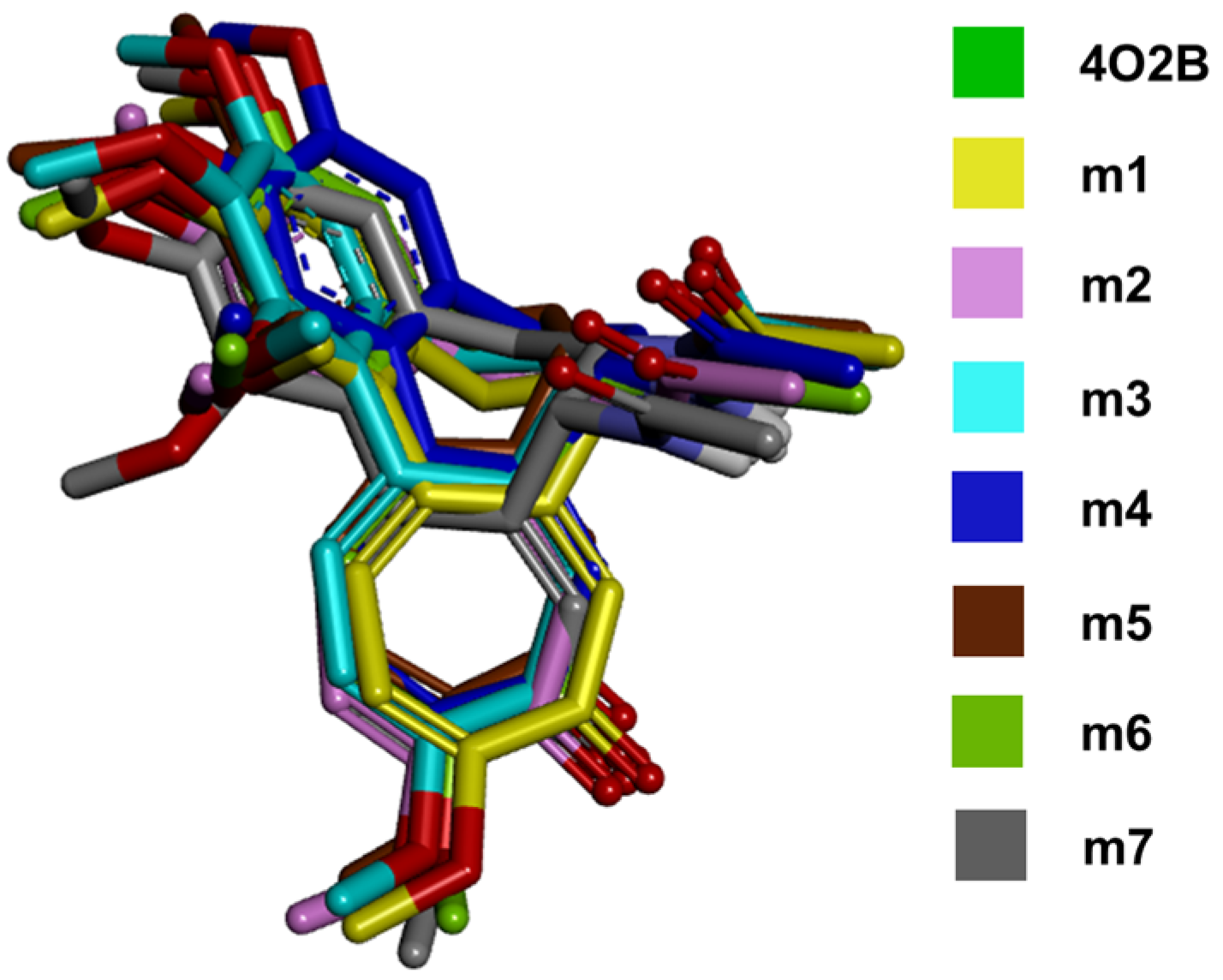
2.2. In Silico Docking Studies between Colchicine and the C. sativus αβ-Tubulin Models
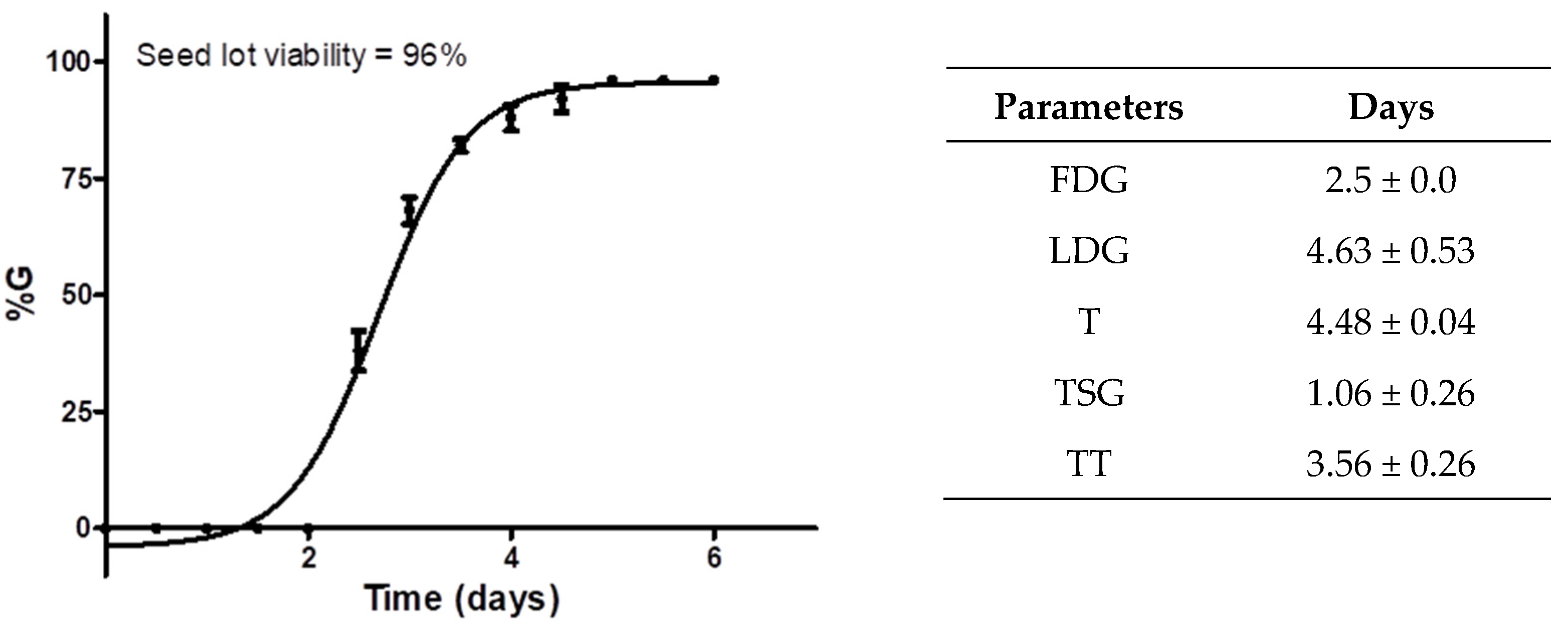
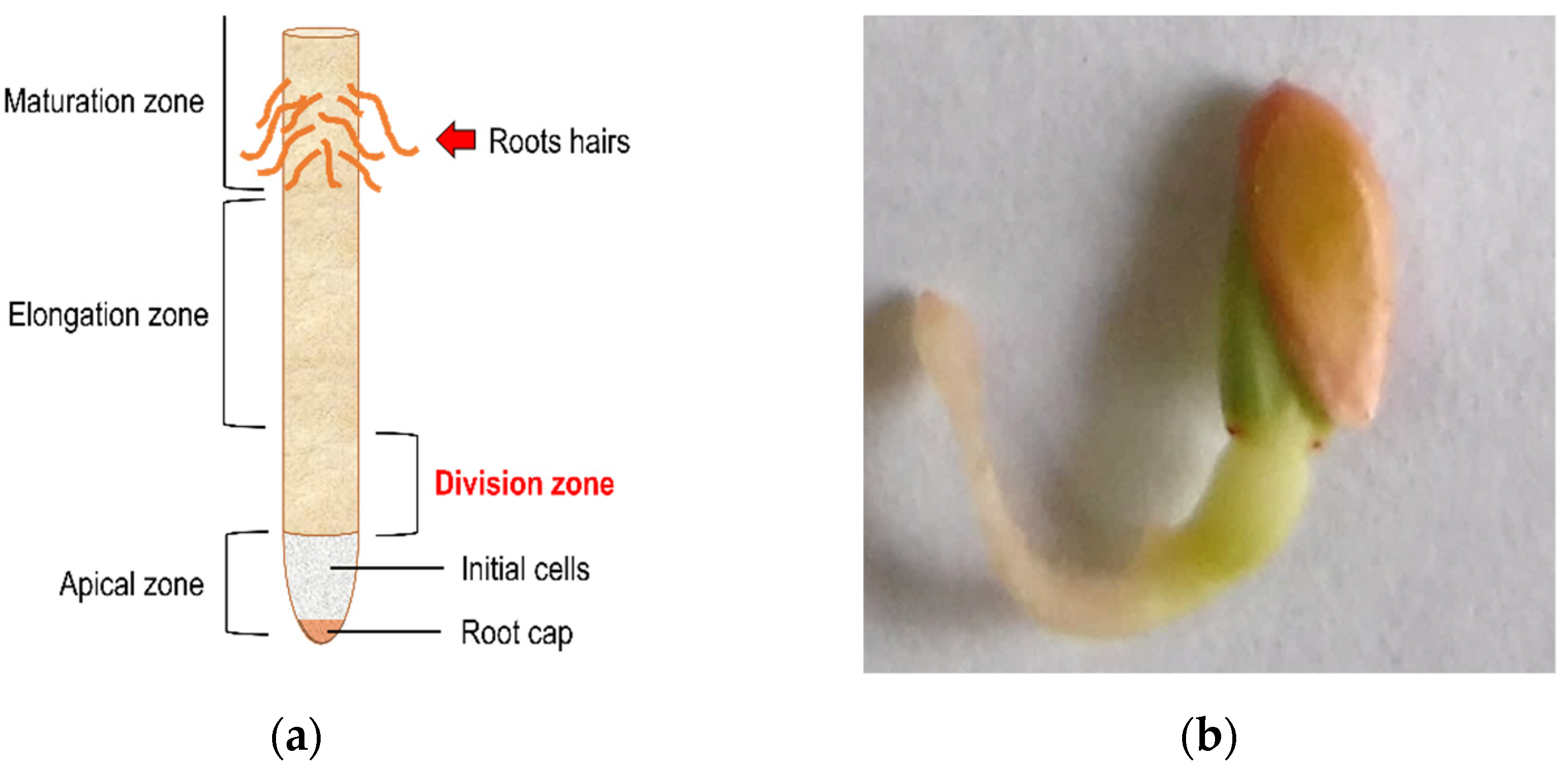
2.3. Technical Considerations before Starting the Phytotoxic Test
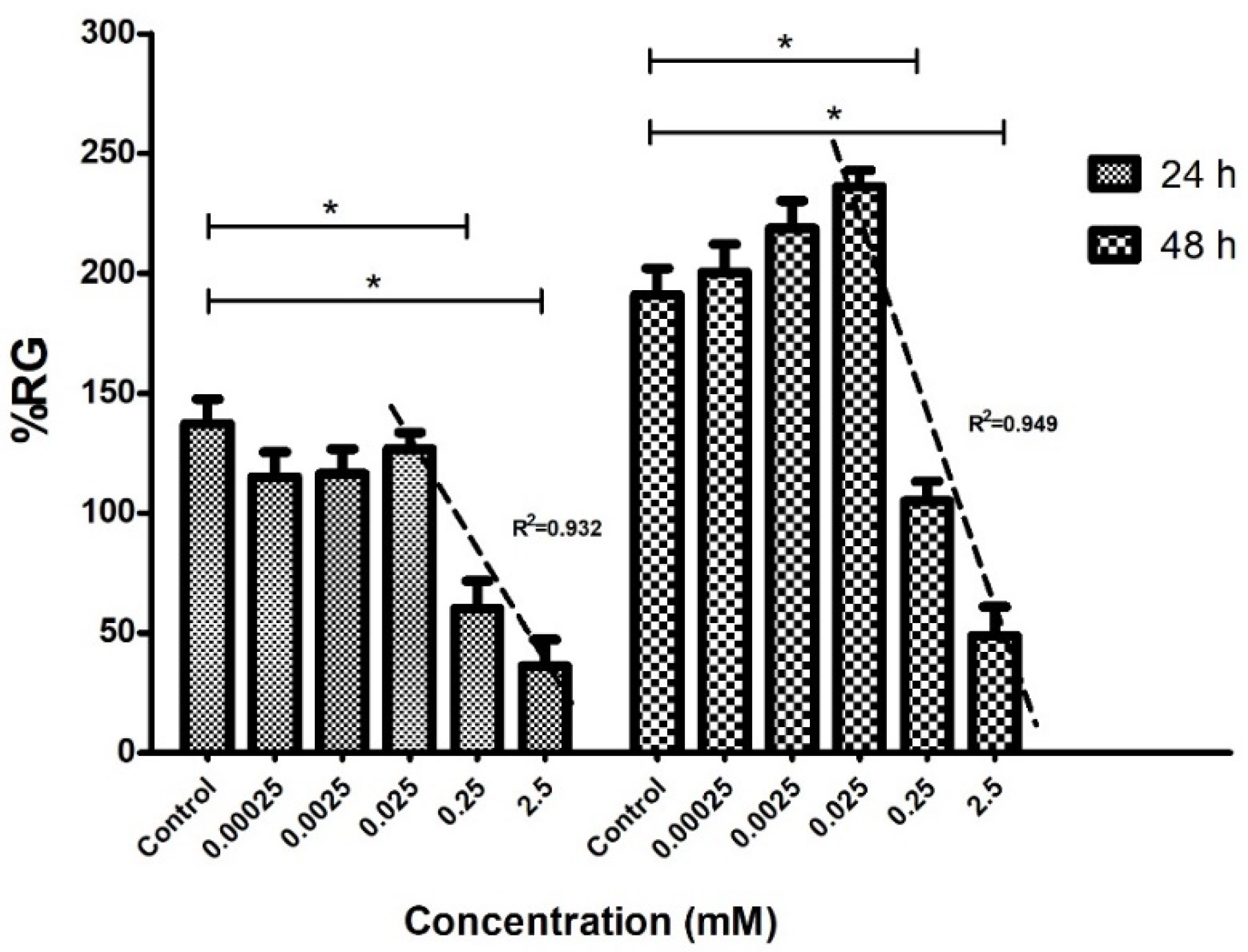
2.4. In Vitro Antimitotic Effect of Colchicine (Phytotoxicity)
2.5. Macroscopic Characteristics of Cucumber-Treated Seeds
3. Materials and Methods
3.1. Protein Sequence Alignment Comparison and Modeling of the αβ-Tubulin Heterodimer of C. sativus
3.2. In Silico Docking Studies between Colchicine and the C. sativus αβ-Tubulin Model
3.3. Technical Considerations before Starting the Phytotoxic Assay
3.3.1. Before Sowing
3.3.2. After Sowing
3.4. In Vitro Phytotoxic Effect of Colchicine
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Klatte, S.; Schaefer, H.-C.; Hempel, M. Pharmaceuticals in the environment—A short review on options to minimize the exposure of humans, animals and ecosystems. Sustain. Chem. Pharm. 2017, 5, 61–66. [Google Scholar] [CrossRef]
- Walker, S.D.; McEldowney, S. Molecular docking: A potential tool to aid ecotoxicity testing in environmental risk assessment of pharmaceuticals. Chemosphere 2013, 93, 2568–2577. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M. Vicia faba bioassay for environmental toxicity monitoring: A review. Chemosphere 2016, 144, 785–802. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L. Bioassays as a tool for evaluating advanced oxidation processes in water and wastewater treatment. Water Res. 2011, 45, 4311–4340. [Google Scholar] [CrossRef]
- Rodríguez, R.A.J.; Robles, S.C.A.; Ruíz, P.R.A.; López, L.E.; Sedeño, D.J.E.; Rodríguez, D.A. Índices de germinación y elongación radical de Lactuca sativa en el biomonitoreo de la calidad del agua del río Chalma. Rev. Int. Contam. Ambie 2014, 30, 307–316. [Google Scholar]
- Irahola, P.; Giménez, A. Inhibition of germination of seeds and toxicity in Artemia salina, as indicators of activity antitumoral. Biofarbo 2000, 8, 81–85. [Google Scholar]
- Yan, Z.; Li, P.; Xiao, Y.; Cao, L.; Yao, L. Phytotoxic Effects of Allelochemical Acacetin on Seed Germination and Seedling Growth of Selected Vegetables and Its Potential Physiological Mechanism. Agronomy 2022, 12, 1038. [Google Scholar] [CrossRef]
- Cuéllar, C.L.; Morales, R.E.; Treviño, N.J.F. La germinación in vitro una alternativa para obtener explantes en cactáceas. Zonas Áridas 2016, 10, 129–133. [Google Scholar] [CrossRef]
- Martínez-Zamora, L.; Castillejo, N.; Artés-Hernández, F. UV-B Radiation as Abiotic Elicitor to Enhance Phytochemicals and Development of Red Cabbage Sprouts. Horticulturae 2021, 7, 567. [Google Scholar] [CrossRef]
- Ibrahim, M.; Nawaz, S.; Iqbal, K.; Rehman, S.; Ullah, R.; Nawaz, G.; Almeer, R.; Sayed, A.A.; Peluso, I. Plant-Derived Smoke Solution Alleviates Cellular Oxidative Stress Caused by Arsenic and Mercury by Modulating the Cellular Antioxidative Defense System in Wheat. Plants 2022, 11, 1379. [Google Scholar] [CrossRef]
- An, Y.J.; Kim, Y.M.; Kwon, T.I.; Jeong, S.W. Combined effects of copper, cadmium, and lead upon Cucumis sativus growth and bioaccumulation. Sci. Total Environ. 2004, 326, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Munzuroglu, O.; Geckil, H. Effects of metals on seed germination, root elongation, and coleoptile and hypocotyls growth in Triticum aestivum and Cucumis sativus. Arch. Environ. Contam. Toxicol 2002, 43, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, C.; Gao, S.; Wang, L.; Shuokui, H. Validation of germination rate and root elongation as indicator to assess phytotoxicity with Cucumis sativus. Chemosphere 2001, 44, 1711–1721. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, S.; Jia, X.; Huang, Y.; Ji, R.; Zhao, L. Comparation of the phytotoxicity between chemically and green synthesized silver nanoparticles. Sci. Total Environ. 2021, 752, 142264. [Google Scholar] [CrossRef]
- Swiech, J.N.D.; Folquitto, D.G.; Bobek, V.B.; Urban, A.M.; Betim, F.C.M.; Oliveira, L.F.; Pereira, C.B.; Merino, F.J.Z.; Dias, J.D.F.G.; da Silva, R.Z.; et al. Phytotoxic and Enzymatic Study of Philodendron meridionale on seeds of Lactuca sativa L. Res. Soc. Dev. 2021, 10, e5610111336. [Google Scholar] [CrossRef]
- Abd-ElGawad, A.M.; El Gendy, A.E.G.; Assaeed, A.M.; Al-Rowaily, S.L.; Alharthi, A.S.; Mohamed, T.A.; Nassar, M.I.; Dewir, Y.H.; Elshamy, A.I. Phytotoxic Effects of Plant Essential Oils: A Systematic Review and Structure-Activity Relationship Based on Chemometric Analyses. Plants 2021, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Krstin, L.; Katanić, Z.; Žuna, P.T.; Špoljarić, D.M.; Marinčić, D.; Martinović, A.; Štolfa, I.C. Phytotoxic effect of invasive species Amorpha ruticose L. on germination and the early growth of forage and agricultural crop plants. Ecol. Res. 2021, 36, 97–106. [Google Scholar] [CrossRef]
- Eze, M.O.; George, S.C.; Hose, G.C. Dose-response analysis of diesel fuel phytotoxicity on selected plant species. Chemosphere 2021, 263, 128382. [Google Scholar] [CrossRef]
- Khaldari, I.; Naghavi, M.R.; Motamedi, E. Synthesis of green and pure copper oxide nanoparticles using two plant resources via solid-state route and their phytotoxicity assessment. RSC Adv. 2021, 11, 3346–3353. [Google Scholar] [CrossRef]
- Infante, C.; Morales, G.F.A. Evaluación de la toxicidad en desechos y suelos petrolizados empleando semilla de Lactuca sativa L. Interciencia 2012, 37, 782–788. [Google Scholar]
- Lloyd, C.; Chan, J. Not so divided: The common basis of plant and animal cell division. Nat. Rev. Mol. Cell Biol. 2005, 7, 147–152. [Google Scholar] [CrossRef]
- Sablowski, R.; Gutierrez, C. Cycling in a crowd: Coordination of plant cell division, growth, and cell fate. Plant Cell. 2022, 34, 193–208. [Google Scholar] [CrossRef]
- Atlas de Histología Vegetal y Animal. Available online: https://mmegias.webs.uvigo.es/2-organos-v/guiada_o_v_raiz.php (accessed on 24 May 2022).
- Alonso, P.J.R. Manual de Histología Vegetal; Ediciones Mundi-Prensa: Madrid, Spain, 2011; pp. 15–40. [Google Scholar]
- Janke, C. The tubulin code: Molecular components, readout mechanisms, and funtions. J. Cell Biol. 2014, 206, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Khabudaev, K.V.; Petrova, D.P.; Bedoshvili, Y.D.; Likhoshway, Y.V.; Grachev, M.A. Molecular Evolution of Tubulins in Diatoms. Int. J. Mol. Sci. 2022, 23, 618. [Google Scholar] [CrossRef] [PubMed]
- Logan, C.M.; Menko, A.S. Microtubules: Evolving roles and critical cellular interactions. Exp. Biol. Med. 2019, 244, 1240–1254. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, R.; Gigant, B.; Curmi, A.; Jourdain, I.; Lachkar, S.; Sobel, A.; Knossow, M. Insight into tubulin regulation from a complex with colchicines and a stathmin-like domain. Nature 2004, 428, 198–202. [Google Scholar] [CrossRef]
- Horio, T.; Murata, T. The role of dynamic instability in microtubule organization. Front. Plant Sci. 2014, 5, 511. [Google Scholar] [CrossRef]
- Cao, Y.-N.; Zheng, L.-L.; Wang, D.; Liang, X.-X.; Gao, F.; Zhou, X.-L. Recent advances in microtubule-stabilizing agents. Eur. J. Med. Chem 2018, 143, 806–828. [Google Scholar] [CrossRef]
- Rodríguez-López, V.; Millán-Pacheco, C.; González-Christen, J.; Anaya-Ruíz, M.; Peña-Morán, O.A. In Vitro Anti-Tubulin Activity on MCF10A Cell Line and In Silico Rigid/Semiflexible-Residues Docking, of Two Lignans from Bursera Fagaroides var. Fagaroides. Molecules 2021, 26, 6155. [Google Scholar] [CrossRef]
- Vindya, N.G.; Sharma, N.; Yadav, M.; Ethiraj, K.R. Tubulins—The target for anticancer therapy. Curr. Top. Med. Chem. 2015, 15, 73–82. [Google Scholar] [CrossRef]
- McLoughlin, E.C.; O’Boyle, N.M. Colchicine-Binding Site Inhibitors from Chemistry to Clinic: A Review. Pharmaceuticals 2020, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.D.; Hwang, J.; Li, W.; Miller, D.D. Current advances of tubulin inhibitors in nanoparticle drug delivery and vascular disruption/angiogenesis. Molecules 2016, 21, 1468. [Google Scholar] [CrossRef]
- Leung, Y.Y.; Yao Hui, L.L.; Kraus, V.B. Colchicine—Update on mechanisms of action and therapeutic uses. Semin. Arthritis Rheum. 2015, 45, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Imazio, M.; Nidorf, M. Colchicine and the heart. Eur. Heart J. 2021, 42, 1–16. [Google Scholar] [CrossRef] [PubMed]
- DeLorenzo, M.E.; Scott, G.I.; Ross, P.E. Toxicity of pesticides to aquatic microorganisms: A review. Environ. Toxicol. Chem. 2001, 20, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Rauf, S.; Ortiz, R.; Malinowski, D.P.; Clarindo, W.R.; Kainat, W.; Shehzad, M.; Waheed, U.; Hassan, S.W. Induced Polyploidy: A Tool for Forage Species Improvement. Agriculture 2021, 11, 210. [Google Scholar] [CrossRef]
- CONABIO. Cucumis sativus. Sistema de Información de Organismos Vivos Modificados. Proyecto GEF-CIBIOGEM de Bioseguridad; CONABIO: Mexico city, Mexico, 2003.
- Aydin, S.S.; Gökçe, E.; Büyük, I.; Aras, S. Characterization of stress induced by copper and zinc on cucumber (Cucumis sativus L.) seedlings by means of molecular and population parameters. Mutat. Res. 2012, 746, 49–55. [Google Scholar] [CrossRef]
- Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000137285-TUBB2B/tissue (accessed on 18 May 2022).
- Lu, Y.; Liu, C.; Xu, Y.F.; Cheng, H.; Shi, S.; Wu, C.T.; Yu, X.J. Stathmin destabilizing microtubule dynamics promotes malignant potential in cancer cells by epithelial-mesenchymal transition. Hepatobiliary Pancreat Dis. Int. 2014, 13, 386–394. [Google Scholar] [CrossRef]
- Prota, A.E.; Danel, F.; Bachmann, F.; Bargsten, K.; Buey, R.M.; Pohlmann, J.; Reinelt, S.; Lane, H.; Steinmetz, M.O. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J. Mol. Biol. 2014, 426, 1848–1860. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [PubMed]
- Prota, A.E.; Bargsten, K.; Zurwerra, D.; Field, J.J.; Díaz, J.F.; Altmann, K.H.; Steinmetz, M.O. Molecular mechanism of action of microtubule-stabilizing anticancer agents. Science 2013, 339, 587–590. [Google Scholar] [CrossRef] [PubMed]
- Erofeeva, E.A. Plant hormesis and Shelford’s tolerance law curve. J. For. Res. 2021, 32, 1789–1802. [Google Scholar] [CrossRef]
- Thompson, D.; Anderson, N.; Staden, J. Colchicine-induced Somatic Polyploids from In Vitro-germinated Seeds of South African Watsonia Species. HortScience 2010, 45, 1398–1402. [Google Scholar] [CrossRef]
- Li, W.; Sun, H.; Xu, S.; Zhu, Z.; Xu, J. Tubulin inhibitors targeting the colchicine binding site: A perspective of privileged structures. Future Med. Chem. 2017, 9, 1765–1794. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Xiao, M.; Li, W.; Miller, D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res. 2012, 29, 2943–2971. [Google Scholar] [CrossRef]
- Péret, B.; De Rybel, B.; Casimiro, I.; Benková, E.; Swarup, R.; Laplaze, L.; Beeckman, T.; Bennett, M.J. Arabidopsis lateral root development: An emerging story. Trends. Plant. Sci. 2009, 14, 399–408. [Google Scholar] [CrossRef]
- Eng, W.H.; Ho, W.S. Polyploidization using colchicine in horticultural plants: A review. Sci. Horticult. 2019, 246, 604–617. [Google Scholar] [CrossRef]
- Koonin, E.V. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010, 11, 209. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Dagan, T.; Talmor, Y.; Graur, D. Ratios of Radical to Conservative Amino Acid Replacement are Affected by Mutational and Compositional Factors and May Not Be Indicative of Positive Darwinian Selection. Mol. Biol. Evol. 2002, 19, 1022–1025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ti, S.C.; Pamula, M.C.; Howes, S.C.; Duellberg, C.; Cade, N.I.; Kleiner, R.E.; Forth, S.; Surrey, T.; Nogales, E.; Kapoor, T.M. Mutations in Human Tubulin Proximal to the Kinesin-Binding Site Alter Dynamic Instability at Microtubule Plus- and Minus-Ends. Dev. Cell. 2016, 37, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef]
- BIOVIA Discovery Studio. Discovery Studio Modeling Environment; Dassault Systemes: San Diego, CA, USA, 2015. [Google Scholar] [CrossRef]
- Schrödinger Release 2020-4: Maestro, Schrödinger; LLC: New York, NY, USA, 2020.
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Hanwell, D.M.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 16, 2785–2791. [Google Scholar] [CrossRef]
- Jiménez-Pérez, J. Estandarización del Ensayo de Crecimiento Radicular Para Evaluar Toxicidad Aguda, en Semillas de Cucumis sativus, Medicago sativa y Lens culinaris. Bachelor Thesis, Universidad Juárez Autónoma de Tabasco, Cunduacán, Tabasco, Mexico, 2021. [Google Scholar]
- Al-Mudaris, M. Notes on various parameters recording the speed of seed germination. J. Agr. Rural Dev. Trop. 1998, 99, 147–154. [Google Scholar]
- González, Z.L.; Orozco, S.A. Métodos de análisis de datos en la germinación de semillas, un ejemplo: Manfreda brachystachya. Bol. Soc. Bot. México. 1996, 58, 15–30. [Google Scholar]
- Luna, C.V. Establecimiento de un método eficiente de estandarización de la germinación in vitro de Moringa oleifera (Moringaceae). Acta Bot. Mex. 2019, 126, e1496. [Google Scholar] [CrossRef]
- GIMP. GNU Image Manipulation Program; GNOME Foundation: Orinda, CA, USA, 2018. [Google Scholar]

| Entry | Length | Identity (%) | Similarity (%) |
|---|---|---|---|
| α-tubulin | |||
| A0A0A0K6A8 | 450 | 84.7 | 97.6 |
| A0A0A0LFM5 | 449 | 81.8 | 96.7 |
| A0A0A0KWR8 | 449 | 81.4 | 96.7 |
| A0A0A0KWB5 | 450 | 84.0 | 98.0 |
| A0A0A0KIM4 | 415 | 77.8 | 97.6 |
| Mean | 442.6 ± 15.4 | 82.0 ± 2.7 | 97.3 ± 0.6 |
| β-Tubulin | |||
| A0A0A0L2I9 | 446 | 84.5 | 95.7 |
| A0A0A0LTS3 | 445 | 83.6 | 96.4 |
| A0A0A0LCY8 | 446 | 83.0 | 95.5 |
| A0A0A0LPG6 | 449 | 83.3 | 96.4 |
| A0A0A0LXT7 | 447 | 83.4 | 94.6 |
| A0A0A0LVT8 | 443 | 86.1 | 96.6 |
| A0A0A0KQW7 | 441 | 82.9 | 97.1 |
| Mean | 445.3 ± 2.6 | 83.8 ± 1.1 | 96.1 ± 0.8 |
| Uniprot ID | β-Tubulin Residues in the CBS | Model | Z-Score | RP 1 (%) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 238 | 241 | 242 | 248 | 250 | 251 | 254 | 255 | 258 | 259 | 314 | 315 | 316 | 318 | 350 | 352 | 378 | ||||
| Q9BVA1 | V | C | L | L | A | D | K | L | N | M | T | V | A | I | N | K | I | - | - | - |
| A0A0A0L2I9 | V | C | L | L | S | D | K | L | N | L | T | A | S | M | N | K | I | m1 | −9.36 | 96.2 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| A0A0A0LTS3 | V | C | L | L | S | D | K | L | N | L | T | A | S | M | N | K | I | m2 | −10.01 | 96.4 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| A0A0A0LCY8 | V | C | L | L | S | D | K | L | N | L | T | A | S | M | N | K | I | m3 | −9.65 | 96.4 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| A0A0A0LPG6 | V | C | L | L | S | D | K | L | N | L | T | A | S | M | N | K | I | m4 | −9.95 | 96.9 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| A0A0A0LXT7 | V | C | L | L | S | D | K | L | N | L | T | A | S | M | N | K | I | m5 | −9.39 | 96.6 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| A0A0A0LVT8 | V | C | L | L | S | D | K | L | N | L | T | A | S | M | N | K | I | m6 | −9.48 | 96.6 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| A0A0A0KQW7 | V | C | L | L | S | D | K | L | N | L | T | A | S | L | N | K | I | m7 | −9.81 | 97.5 |
| * | * | * | * | : | * | * | * | * | : | * | . | : | : | * | * | * | ||||
| Model | Poses/Clusters 1 | ΔGb (Kcal/mol) | Ki (µM) | RMSD |
|---|---|---|---|---|
| m1 | 28/8 | −8.99 ± 0.59 | 0.412 | 1.12 ± 0.39 |
| m2 | 26/10 | −8.79 ± 0.45 | 0.606 | 1.11 ± 0.31 |
| m3 | 30/9 | −9.56 ± 0.51 | 0.145 | 1.01 ± 0.35 |
| m4 | 41/8 | −9.65 ± 0.21 | 0.091 | 1.18 ± 0.79 |
| m5 | 2/10 | −8.44 ± 1.01 | 1.195 | 1.02 ± 0.57 |
| m6 | 2/14 | −8.72 ± 1.02 | 0.744 | 1.01 ± 0.51 |
| m7 | 4/10 | −8.03 ± 0.67 | 1.988 | 1.58 ± 0.26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peña-Morán, O.A.; Jiménez-Pérez, J.; Cerón-Romero, L.; Rodríguez-Aguilar, M. In Silico Conformation of the Drug Colchicine into Tubulin Models and Acute Phytotoxic Activity on Cucumis sativus Radicles. Plants 2022, 11, 1805. https://doi.org/10.3390/plants11141805
Peña-Morán OA, Jiménez-Pérez J, Cerón-Romero L, Rodríguez-Aguilar M. In Silico Conformation of the Drug Colchicine into Tubulin Models and Acute Phytotoxic Activity on Cucumis sativus Radicles. Plants. 2022; 11(14):1805. https://doi.org/10.3390/plants11141805
Chicago/Turabian StylePeña-Morán, Omar Aristeo, Jesús Jiménez-Pérez, Litzia Cerón-Romero, and Maribel Rodríguez-Aguilar. 2022. "In Silico Conformation of the Drug Colchicine into Tubulin Models and Acute Phytotoxic Activity on Cucumis sativus Radicles" Plants 11, no. 14: 1805. https://doi.org/10.3390/plants11141805
APA StylePeña-Morán, O. A., Jiménez-Pérez, J., Cerón-Romero, L., & Rodríguez-Aguilar, M. (2022). In Silico Conformation of the Drug Colchicine into Tubulin Models and Acute Phytotoxic Activity on Cucumis sativus Radicles. Plants, 11(14), 1805. https://doi.org/10.3390/plants11141805






