Serial-Omics and Molecular Function Study Provide Novel Insight into Cucumber Variety Improvement
Abstract
:1. Introduction
2. Germplasm Resources and Molecular Markers
2.1. Origin and Varieties
2.2. Molecular Markers and Genetic Maps
3. Serial-Omics and Database
3.1. Transcriptome Research of Cucumber
3.2. Proteome Research of Cucumber
3.3. Metabolome Research of Cucumber
3.4. Information Resources for Cucumber Research
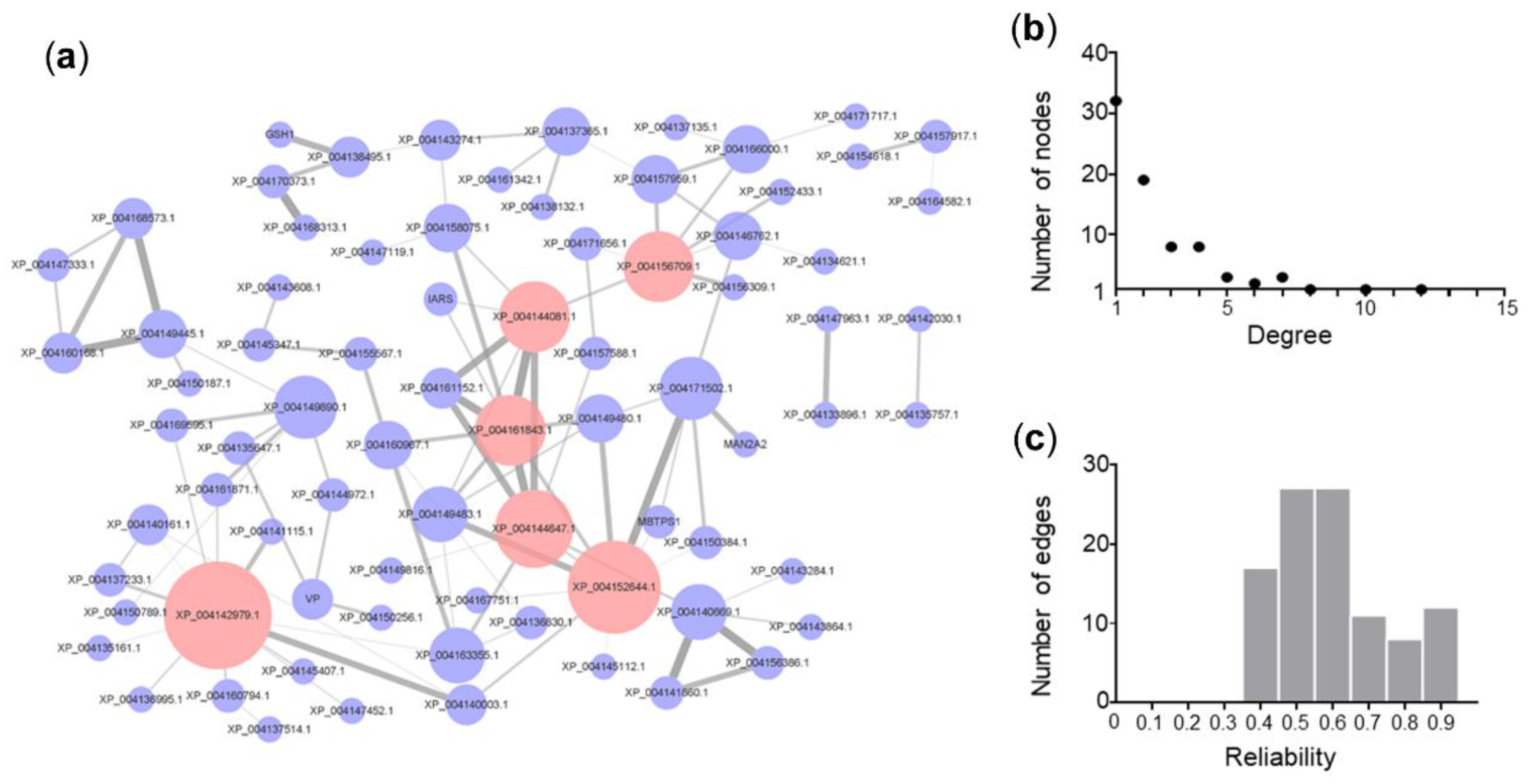
3.5. Salt Tolerance PPI Network of Cucumber
4. Gene Function Analysis and Trait Regulation
4.1. Functional Genes That Regulate Development and Quality
4.2. Functional Genes That Regulate Hormone Responses
4.3. Functional Genes That Resist Abiotic Stress
5. Application of Omics Techniques and Molecular Markers to Breeding
5.1. Genetic Diversity and Evaluation and Cucumber Germplasm Selection
5.2. Gene Mapping
5.3. Molecular Marker-Assisted Selection
6. Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bisht, I.S.; Bhat, K.V.; Tanwar, S.P.S.; Bhandari, D.C.; Joshi, K.; Sharma, A.K. Distribution and genetic diversity of Cucumis sativus var. hardwickii (Royle) Alef in India. J. Hortic. Sci. Biotechnol. 2004, 5, 783–791. [Google Scholar] [CrossRef]
- Sebastian, P.; Schaefer, H.; Telford, I.R.; Renner, S.S. Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc. Natl. Acad. Sci. USA 2010, 107, 14269–14273. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.Y.; Dong, S.Y.; Wei, S.; Wang, W.P.; Miao, H.; Bo, K.L.; GU, X.F.; Zhang, S.P. QTL mapping of heat tolerance in cucumber (Cucumis sativus L.) at adult stage. Plants 2021, 10, 324. [Google Scholar] [CrossRef] [PubMed]
- Fanourakis, N.E.; Simon, P.W. Analysis of genetic linkage in the cucumber. J. Hered. 1987, 78, 238–242. [Google Scholar] [CrossRef]
- Dijkhuizen, A.; Meglic, V.; Staub, J.E.; Havey, M.J. Linkages among RFLP, RAPD, isozyme, disease-resistance, and morphological markers in narrow and wide crosses of cucumber. Theor. Appl. Genet. 1994, 89, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Serquen, F.C.; Bacher, J.; Staub, J.E. Mapping and QTL analysis of horticultural traits in a narrow cross in cucumber (Cucumis sativus L.) using random-amplified polymorphic DNA markers. Mol. Breed. 1997, 3, 257–268. [Google Scholar] [CrossRef]
- Park, Y.H.; Sensoy, S.; Wye, C.; Antonise, R.; Peleman, J.; Havey, M.J. A genetic map of cucumber composed of RAPDs, RFLPs, AFLPs, and loci conditioning resistance to papaya ringspot and zucchini yellow mosaic viruses. Genome 2000, 43, 1003–1010. [Google Scholar] [CrossRef]
- Bradeen, J.M.; Staub, J.E.; Wye, C.; Antonise, R.; Peleman, J. Towards an expanded and integrated linkage map of cucumber (Cucumis sativus L.). Genome 2001, 44, 111–119. [Google Scholar] [CrossRef]
- Danin-Poleg, Y.; Reis, N.; Tzuri, G.; Katzir, N. Development and characterization of microsatellite markers in cucumis. Theor. Appl. Genet. 2008, 7, 19–31. [Google Scholar] [CrossRef]
- Kong, Q.S. Characterization, Development and Application of EST-SSR Markers in Cucumis Genus Based on Public Sequence Database; Huazhong Agricultural University: Wuhan, China, 2006. [Google Scholar] [CrossRef]
- Cheng, Z.C.; GU, X.F.; Zhang, S.P.; Miao, H.; Zhang, R.W.; Liu, M.M.; Yang, S.J. QTL analysis for fruit length of cucumber. China Veget. 2010, 12, 20–25. [Google Scholar]
- Miao, H.; Zhang, S.P.; Wang, X.W.; Zhang, Z.H.; Li, M.; Mu, S.Q.; Cheng, Z.C.; Zhang, R.W.; Huang, S.W.; Xie, B.Y.; et al. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 2011, 182, 167–176. [Google Scholar] [CrossRef]
- Hu, J.; Wang, L.; Li, J. Comparison of genomic SSR and EST-SSR markers for estimating genetic diversity in cucumber. Biol. Plant. 2011, 55, 577–580. [Google Scholar] [CrossRef]
- Miao, H.; Gu, X.F.; Zhang, S.P.; Zhang, Z.H.; Huang, S.W.; Wang, Y.; Cheng, Z.C.; Zhang, R.W.; Mu, S.Q.; Li, M.; et al. Mapping QTLs for fruit-associated traits in Cucumis sativus L. Sci. Agric. Sin. 2011, 44, 5031–5040. [Google Scholar] [CrossRef]
- Zhang, W.W.; Pan, J.S.; He, H.L.; Zhang, C.; Li, Z.; Zhao, J.L.; Yuan, X.J.; Zhu, L.H.; Huang, S.W.; Cai, R. Construction of a high density integrated genetic map for cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2012, 124, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.Y.; Miao, H.; Zhang, S.P.; Liu, M.M.; Wang, Y.; Gu, X.F. Genetic analysis and gene mapping of white fruit skin in cucumber (Cucumis sativus L.). Acta Bot. Bor-Occid. Sin. 2012, 32, 2177–2181. [Google Scholar] [CrossRef]
- Wei, Q.Z.; Wang, Y.Z.; Qin, X.D.; Zhang, Y.X.; Zhang, Z.T.; Wang, J.; Li, J.; Lou, Q.F.; Chen, J.F. An SNP-based saturated genetic map and QTL analysis of fruit-related traits in cucumber using specific-length amplified fragment (SLAF) sequencing. BMC Genom. 2014, 15, 1158. [Google Scholar] [CrossRef] [Green Version]
- Weng, Y.Q.; Colle, M.; Wang, Y.H.; Yang, L.M.; Rubinstein, M.; Sherman, A.; Ophir, R.; Grumet, R. QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 2015, 128, 1747–1763. [Google Scholar] [CrossRef]
- Yang, Y.T.; Liu, Y.; Qi, F.; Xu, L.L.; Li, X.Z.; Cong, L.J.; Guo, X.; Chen, S.X.; Fang, Y.L. Assessment of genetic diversity of cucumber cultivars in China based on simple sequence repeats and fruit traits. GMR Genet. Mol. Res. 2015, 14, 19028–19039. [Google Scholar] [CrossRef]
- Wei, Q.Z.; Fu, W.Y.; Wang, Y.Z.; Qin, X.D.; Wang, J.; Li, J.; Lou, Q.F.; Chen, J.F. Rapid identification of fruit length loci in cucumber (Cucumis sativus L.) using next-generation sequencing (NGS)-based QTL analysis. Sci. Rep. 2016, 6, 27496. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, T.; Li, L.; Xu, J.; Qin, X.D.; Zhang, T.L.; Cui, L.; Lou, Q.F.; Li, J.; Chen, J.F. Identification of a stable major-effect QTL (Parth 2.1) controlling parthenocarpy in cucumber and associated candidate gene analysis via whole genome re-sequencing. BMC Plant Biol. 2016, 16, 182. [Google Scholar] [CrossRef] [Green Version]
- Dar, A.A.; Mahajan, R.; Lay, P.; Sharma, S. Genetic diversity and population structure of Cucumis sativus L. by using SSR markers. 3 Biotech 2017, 7, 307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, J.J.; Zhang, L.; Luo, J.; Zhao, H.; Zhang, J.N.; Wen, C.L. A new SNP genotyping technology target SNP-seq and its application in genetic analysis of cucumber varieties. Sci. Rep. 2020, 10, 5623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adhikari, B.N.; Savory, E.A.; Vaillancourt, B.; Childs, K.L.; Hamilton, J.P.; Day, B.; Buell, C.R. Expression profiling of Cucumis sativus in response to infection by Pseudoperonospora cubensis. PLoS ONE 2012, 7, e34954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkhardt, A.; Day, B. Transcriptome and small RNAome dynamics during a resistant and susceptible interaction between cucumber and downy mildew. Plant Genome 2016, 9, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.Q.; Lu, X.H.; Sun, M.H.; Guo, R.J.; van Diepeningen, A.D.; Li, S.D. Transcriptome analysis of virulence-differentiated Fusarium oxysporum f. sp. cucumerinum isolates during cucumber colonisation reveals pathogenicity profiles. BMC Genom. 2019, 20, 570. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.P.; Wang, Y.A.; Xian, Q.Q.; Chen, X.H.; Xu, J. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant Biol. 2020, 20, 334. [Google Scholar] [CrossRef]
- Słomnicka, R.; Olczak-Woltman, H.; Sobczak, M.; Bartoszewski, G. Transcriptome profiling of cucumber (Cucumis sativus L.) early response to Pseudomonas syringae pv. lachrymans. Int. J. Mol. Sci. 2021, 22, 4192. [Google Scholar] [CrossRef]
- Li, J.; Wu, Z.; Cui, L.; Zhang, T.L.; Guo, Q.W.; Xu, J.; Jia, L.; Lou, Q.F.; Huang, S.W.; Li, Z.G.; et al. Transcriptome comparison of global distinctive features between pollination and parthenocarpic fruit set reveals transcriptional phytohormone cross-talk in cucumber (Cucumis sativus L.). Plant Cell Physiol. 2014, 55, 1325–1342. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.N.; Cao, C.X.; Zheng, S.S.; Zhang, H.Y.; Liu, P.J.; Ge, Q.; Li, J.R.; Ren, Z.H. Transcriptomic analysis of short-fruit 1 (sf1) reveals new insights into the variation of fruit-related traits in Cucumis sativus. Sci. Rep. 2017, 7, 2950. [Google Scholar] [CrossRef]
- Jiang, L.; Yan, S.S.; Yang, W.C.; Li, Y.Q.; Xia, M.X.; Chen, Z.J.; Wang, Q.; Yan, L.Y.; Song, X.F.; Liu, R.Y.; et al. Transcriptomic analysis reveals the roles of microtubule-related genes and transcription factors in fruit length regulation in cucumber (Cucumis sativus L.). Sci. Rep. 2015, 5, 8031. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Di, Q.H.; Sun, T.S.; Li, Y.S.; Duan, Y.; Wang, J.; Yan, Y.; He, C.X.; Wang, C.L.; Yu, X.C. Integrated metabolome and transcriptome analysis provide insights into the effects of grafting on fruit flavor of cucumber with different rootstocks. Int. J. Mol. Sci. 2019, 20, 3592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Chen, L.; Liang, Z.J.; He, X.M.; Liu, W.R.; Jiang, B.; Yan, J.Q.; Sun, P.Y.; Cao, Z.Q.; Peng, Q.W.; et al. Metabolome and transcriptome analyses reveal chlorophyll and anthocyanin metabolism pathway associated with cucumber fruit skin color. BMC Plant Biol. 2020, 20, 386. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Jiang, B.; Peng, Q.W.; Liu, W.R.; He, X.M.; Liang, Z.J.; Lin, Y. Transcriptome analyses in different cucumber cultivars provide novel insights into drought stress responses. Int. J. Mol. Sci. 2018, 19, 2067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; He, X.M.; Peng, Q.; Liang, Z.J.; Peng, Q.W.; Liu, W.R.; Jiang, B.; Xie, D.S.; Chen, L.; Yan, J.Q.; et al. Understanding the heat resistance of cucumber through leaf transcriptomics. Funct. Plant Biol. 2020, 47, 704–715. [Google Scholar] [CrossRef]
- Xiao, X.M.; Lv, J.; Xie, J.M.; Feng, Z.; Ma, N.; Li, J.; Yu, J.H.; Calderón-Urrea, A. Transcriptome analysis reveals the different response to toxic stress in rootstock grafted and non-grafted cucumber seedlings. Int. J. Mol. Sci. 2020, 21, 774. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.L.; Ren, X.M.; Li, L.; Hou, R.P.; Sun, W.; Jiao, C.J.; Yang, N.; Dong, Y.X. H2S regulation of metabolism in cucumber in response to salt-stress through transcriptome and proteome analysis. Front. Plant Sci. 2020, 11, 1283. [Google Scholar] [CrossRef]
- Kęska, K.; Szcześniak, M.W.; Makałowska, I.; Czernicka, M. Long-term waterlogging as factor contributing to hypoxia stress tolerance enhancement in cucumber: Comparative transcriptome analysis of waterlogging sensitive and tolerant accessions. Genes 2021, 12, 189. [Google Scholar] [CrossRef]
- Xu, Q.; Guo, S.R.; Li, L.; An, Y.H.; Shu, S.; Sun, J. Proteomics analysis of compatibility and incompatibility in grafted cucumber seedlings. Plant Physiol. Biochem. 2016, 105, 21–28. [Google Scholar] [CrossRef]
- Li, J.; Xu, J.; Guo, Q.W.; Wu, Z.; Zhang, T.; Zhang, K.J.; Cheng, C.Y.; Zhu, P.Y.; Lou, Q.F.; Chen, J.F. Proteomic insight into fruit set of cucumber (Cucumis sativus L.) suggests the cues of hormone-independent parthenocarpy. BMC Genom. 2017, 18, 896. [Google Scholar] [CrossRef] [Green Version]
- Yuan, M.; Huang, Y.Y.; Ge, W.N.; Jia, Z.H.; Song, S.S.; Zhang, L.; Huang, Y.L. Involvement of jasmonic acid, ethylene and salicylic acid signaling pathways behind the systemic resistance induced by Trichoderma longibrachiatum H9 in cucumber. BMC Genom. 2019, 20, 144. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.M.; Tang, S.Y.; Meng, X.H.; Zhu, H.; Zhu, Y.Y.; Liu, D.Y.; Shen, Q.R. Proteomic analysis demonstrates a molecular dialog between trichoderma guizhouense NJAU 4742 and cucumber (Cucumis sativus L.) roots: Role in promoting plant growth. Mol. Plant-Microbe Interact. 2021, 34, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhu, Y.Q.; Luo, X.J.; Zhou, S.J. Comparative proteomic analysis provides insights into the complex responses to Pseudoperonospora cubensis infection of cucumber (Cucumis sativus L.). Sci. Rep. 2019, 9, 9433. [Google Scholar] [CrossRef] [PubMed]
- Du, C.X.; Fan, H.F.; Guo, S.R.; Tezuka, T.; Li, J. Proteomic analysis of cucumber seedling roots subjected to salt stress. Phytochemistry 2010, 71, 1450–1459. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.F.; Xu, Y.L.; Du, C.X.; Wu, X. Phloem sap proteome studied by iTRAQ provides integrated insight into salinity response mechanisms in cucumber plants. J. Proteom. 2015, 125, 54–67. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.H.; Zhong, M.; Shu, S.; Du, N.S.; Sun, J.; Guo, S.R. Proteomic and physiological analyses reveal putrescine responses in roots of cucumber stressed by NaCl. Front. Plant Sci. 2016, 7, 1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sang, T.; Shan, X.; Li, B.; Shu, S.; Sun, J.; Guo, S.R. Comparative proteomic analysis reveals the positive effect of exogenous spermidine on photosynthesis and salinity tolerance in cucumber seedlings. Plant Cell Rep. 2016, 35, 1769–1782. [Google Scholar] [CrossRef] [PubMed]
- Donnini, S.; Prinsi, B.; Negri, A.S.; Vigani, G.; Espen, L.; Zocchi, G. Proteomic characterization of iron deficiency responses in Cucumis sativus L. roots. BMC Plant Biol. 2010, 10, 268. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sun, J.; Yang, Y.J.; Guo, S.R.; Glick, B.R. Identification of hypoxic-responsive proteins in cucumber roots using a proteomic approach. Plant Physiol. Biochem. 2012, 51, 74–80. [Google Scholar] [CrossRef]
- He, L.Z.; Lu, X.M.; Tian, J.; Yang, Y.J.; Li, B.; Li, J.; Guo, S.R. Proteomic analysis of the effects of exogenous calcium on hypoxic-responsive proteins in cucumber roots. Proteome Sci. 2012, 10, 42. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.W.; Ji, J.; Ma, X.T.; Xu, Q.; Qi, X.H.; Chen, X.H. Comparative proteomic analysis provides insight into the key proteins involved in cucumber (Cucumis sativus L.) adventitious root emergence under waterlogging stress. Front. Plant Sci. 2016, 7, 1515. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.Y.; Dai, H.X.; Wang, X.Y.; Wang, G.H. Physiological and proteomic analysis of selenium-mediated tolerance to Cd stress in cucumber (Cucumis sativus L.). Ecotoxicol. Environ. Saf. 2016, 133, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Bian, B.T.; Gong, T.Y.; Liao, W.B. Comparative proteomic analysis of key proteins during abscisic acid-hydrogen peroxide-induced adventitious rooting in cucumber (Cucumis sativus L.) under drought stress. J. Plant Physiol. 2018, 229, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.Q.; Li, Y.M.; He, X.R.; Li, S.H.; Zhong, X.; Liu, B.B.; Zhang, D.L.; Li, Q.M. Physiological and iTRAQ based proteomics analyses reveal the mechanism of elevated CO2 concentration alleviating drought stress in cucumber (Cucumis sativus L.) seedlings. Plant Physiol. Biochem. 2019, 143, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Mansfeld, B.N.; Colle, M.; Kang, Y.Y.; Jones, A.D.; Grumet, R. Transcriptomic and metabolomic analyses of cucumber fruit peels reveal a developmental increase in terpenoid glycosides associated with age-related resistance to Phytophthora capsici. Hortic. Res. 2017, 4, 17022. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.Y.; Zhao, H.Y.; Wang, W.; Xu, M.F.; Shi, J.X.; Nie, X.B.; Yang, G.L. Identification of conserved and diverse metabolic shift of the stylar, intermediate and peduncular segments of cucumber fruit during development. Int. J. Mol. Sci. 2018, 19, 135. [Google Scholar] [CrossRef] [Green Version]
- Hu, C.Y.; Zhao, H.Y.; Shi, J.X.; Li, J.; Nie, X.B.; Yang, G.L. Effects of 2,4-dichlorophenoxyacetic acid on cucumber fruit development and metabolism. Int. J. Mol. Sci. 2019, 20, 1126. [Google Scholar] [CrossRef] [Green Version]
- Bityutskii, N.P.; Yakkonen, K.L.; Petrova, A.I.; Shavarda, A.L. Interactions between aluminium, iron and silicon in Cucumber sativus L. grown under acidic conditions. J. Plant Physiol. 2017, 218, 100–108. [Google Scholar] [CrossRef]
- Zhao, L.J.; Huang, Y.X.; Paglia, K.; Vaniya, A.; Wancewicz, B.; Keller, A.A. Metabolomics reveals the molecular mechanisms of copper induced cucumber leaf (Cucumis sativus) senescence. Environ. Sci. Technol. 2018, 52, 7092–7100. [Google Scholar] [CrossRef]
- Li, M.; Li, Y.M.; Zhang, W.D.; Li, S.H.; Gao, Y.; Ai, X.Z.; Zhang, D.L.; Liu, B.B.; Li, Q.M. Metabolomics analysis reveals that elevated atmospheric CO2 alleviates drought stress in cucumber seedling leaves. Anal. Biochem. 2018, 559, 71–85. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, S.; Bai, Y.; Sun, H.H.; Jiao, C.; Guo, S.G.; Zhao, K.; Blanca, J.; Zhang, Z.H.; Huang, S.W.; et al. Cucurbit Genomics Database (CuGenDB): A central portal for comparative and functional genomics of cucurbit crops. Nucleic Acids Res. 2019, 47, D1128–D1136. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zhang, Q.B.; Liu, B.; Lin, K.; Zhang, Z.H.; Pang, E. CuAS: A database of annotated transcripts generated by alternative splicing in cucumbers. BMC Plant Biol. 2020, 20, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodstein, D.M.; Shu, S.Q.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Duvick, J.; Fu, A.; Muppirala, U.; Sabharwal, M.; Wilkerson, M.D.; Lawrence, C.J.; Lushbough, C.; Brendel, V. PlantGDB: A resource for comparative plant genomics. Nucleic Acids Res. 2008, 36, D959–D965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, C.Z.; Jaiswal, P.; Hebbard, C.; Avraham, S.; Buckler, E.S.; Casstevens, T.; Hurwitz, B.; McCouch, S.; Ni, J.J.; Pujar, A.; et al. Gramene: A growing plant comparative genomics resource. Nucleic Acids Res. 2008, 36, D947–D953. [Google Scholar] [CrossRef] [Green Version]
- Mosca, R.; Pons, T.; Céol, A.; Valencia, A.; Aloy, P. Towards a detailed atlas of protein-protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 929–940. [Google Scholar] [CrossRef] [PubMed]
- Hao, T.; Peng, W.; Wang, Q.; Wang, B.; Sun, J.S. Reconstruction and application of protein-protein interaction network. Int. J. Mol. Sci. 2016, 17, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vella, D.; Marini, S.; Vitali, F.; Di Silvestre, D.; Mauri, G.; Bellazzi, R. MTGO: PPI network analysis via topological and functional module identification. Sci. Rep. 2018, 8, 5499. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Huang, S.W.; Li, R.Q.; Zhang, Z.H.; Li, L.; Gu, X.F.; Fan, W.; Lucas, W.J.; Wang, X.W.; Xie, B.Y.; Ni, P.X.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.J.; Zhang, J.P.; Mu, Z.H.; Wang, Y.J.; Wen, C.L.; Wu, T.; Yu, C.; Li, Z.; Wang, H.S. Recent progress on the molecular breeding of Cucumis sativus L. in China. Theor. Appl. Genet. 2020, 133, 1777–1790. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.M.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.W.; Weng, Y.Q. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pramnoi, P.; Somta, P.; Chankaew, S.; Juwattanasomran, R.; Srinives, P. A single recessive gene controls fragrance in cucumber (Cucumis sativus L.). J. Genet. 2013, 92, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Wen, C.L.; Weng, Y.Q. Fine mapping of the pleiotropic locus B for black spine and orange mature fruit color in cucumber identifies a 50 kb region containing a R2R3-MYB transcription factor. Theor. Appl. Genet. 2013, 126, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.K.; Fang, X.X.; Li, X.H.; Chen, Y.; Wan, Z.J.; Xu, Y.J. Genetic study on immature fruit color of cucumber. Acta Hortic. Sin. 2013, 40, 479–486. [Google Scholar] [CrossRef]
- Zhou, Q.; Wang, S.H.; Hu, B.W.; Chen, H.M.; Zhang, Z.H.; Huang, S.W. An accumulation and replication of chloroplasts 5 gene mutation confers light green peel in cucumber. J. Integr. Plant Biol. 2015, 57, 936–942. [Google Scholar] [CrossRef] [Green Version]
- Lun, Y.Y.; Wang, X.; Zhang, C.Z.; Yang, L.; Gao, D.L.; Chen, H.M.; Huang, S.W. A CsYcf54 variant conferring light green coloration in cucumber. Euphytica 2016, 208, 509–517. [Google Scholar] [CrossRef]
- Liu, H.Q.; Jiao, J.Q.; Liang, X.J.; Liu, J.; Meng, H.W.; Chen, S.X.; Li, Y.H.; Cheng, Z.H. Map-based cloning, identification and characterization of the w gene controlling white immature fruit color in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1247–1256. [Google Scholar] [CrossRef]
- Hao, N.; Du, Y.L.; Li, H.Y.; Wang, C.; Wang, C.; Gong, S.Y.; Zhou, S.M.; Wu, T. CsMYB36 is involved in the formation of yellow green peel in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2018, 131, 1659–1669. [Google Scholar] [CrossRef]
- Liu, B.; Liu, X.W.; Yang, S.; Chen, C.H.; Xue, S.D.; Cai, Y.L.; Wang, D.D.; Yin, S.; Gai, X.S.; Ren, H.Z. Silencing of the gibberellin receptor homolog, CsGID1a, affects locule formation in cucumber (Cucumis sativus) fruit. New Phytol. 2016, 210, 551–563. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Sun, J.; Yang, F.; Weng, Y.Q.; Chen, P.; Du, S.L.; Wei, A.M.; Li, Y.H. CsKTN1 for a katanin p60 subunit is associated with the regulation of fruit elongation in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2021, 134, 2429–2441. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Wang, L.N.; Zheng, S.S.; Liu, Z.Z.; Wu, X.Q.; Gao, Z.H.; Cao, C.X.; Li, Q.; Ren, Z.H. A fragment substitution in the promoter of CsHDZIV11/CsGL3 is responsible for fruit spine density in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2016, 129, 1289–1301. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.W.; Wang, T.; Bartholomew, E.; Black, K.; Dong, M.M.; Zhang, Y.Q.; Yang, S.; Cai, Y.L.; Xue, S.D.; Weng, Y.Q.; et al. Comprehensive analysis of NAC transcription factors and their expression during fruit spine development in cucumber (Cucumis sativus L.). Hortic. Res. 2018, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Song, M.F.; Zhang, M.R.; Cheng, F.; Wei, Q.Z.; Wang, J.; Davoudi, M.; Chen, J.F.; Lou, Q.F. An irregularly striped rind mutant reveals new insight into the function of PG1β in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2020, 133, 371–382. [Google Scholar] [CrossRef] [PubMed]
- He, M.X. Fine QTL Mapping of Parthenocarpy in Cucumber (Cucumis sativus L.) and Validation of Candidate Gene Expression; Yangzhou University: Yangzhou, China, 2019. [Google Scholar] [CrossRef]
- Wu, J.; Liu, S.Y.; Guan, X.Y.; Chen, L.F.; He, Y.J.; Wang, J.; Lu, G. Genome-wide identification and transcriptional profiling analysis of auxin response-related gene families in cucumber. BMC Res. Notes 2014, 7, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.Q.; Zhang, W.W.; He, H.L.; Nie, J.T.; Bie, B.B.; Zhao, J.L.; Ren, G.L.; Li, Y.; Zhang, D.B.; Pan, J.S.; et al. Tuberculate fruit gene Tu encodes a C2H2 zinc finger protein that is required for the warty fruit phenotype in cucumber (Cucumis sativus L.). Plant J. 2014, 78, 1034–1046. [Google Scholar] [CrossRef]
- Hou, S.S.; Niu, H.H.; Tao, Q.Y.; Wang, S.H.; Gong, Z.H.; Li, S.; Weng, Y.Q.; Li, Z. A mutant in the CsDET2 gene leads to a systemic brassinosteriod deficiency and super compact phenotype in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2017, 130, 1693–1703. [Google Scholar] [CrossRef]
- Wen, H.F.; Chen, Y.; Du, H.; Zhang, L.Y.; Zhang, K.Y.; He, H.L.; Pan, J.S.; Cai, R.; Wang, G. Genome-wide identification and characterization of the TCP gene family in cucumber (Cucumis sativus L.) and their transcriptional responses to different treatments. Genes 2020, 11, 1379. [Google Scholar] [CrossRef]
- Wang, W.J.; Liu, X.W.; Gai, X.S.; Ren, J.J.; Liu, X.F.; Cai, Y.L.; Wang, Q.; Ren, H.Z. Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol. 2015, 56, 1339–1354. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.S.; Che, G.; Ding, L.; Chen, Z.J.; Liu, X.F.; Wang, H.Y.; Zhao, W.S.; Ning, K.; Zhao, J.Y.; Tesfamichael, K.; et al. Different cucumber CsYUC genes regulate response to abiotic stresses and flower development. Sci. Rep. 2016, 6, 20760. [Google Scholar] [CrossRef]
- Lü, J.G.; Sui, X.L.; Ma, S.; Li, X.; Liu, H.; Zhang, Z.X. Suppression of cucumber stachyose synthase gene (CsSTS) inhibits phloem loading and reduces low temperature stress tolerance. Plant Mol. Biol. 2017, 95, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wang, G.; Shen, F.; Zhu, S.J. A glycine-rich RNA-binding protein, CsGR-RBP3, is involved in defense responses against cold stress in harvested cucumber (Cucumis sativus L.) Fruit. Front. Plant Sci. 2018, 9, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.Z.; Miao, L.; Huang, B.; Gao, L.H.; He, C.X.; Yan, Y.; Wang, J.; Yu, X.C.; Li, Y.S. Genome-wide identification and characterization of cucumber BPC transcription factors and their responses to abiotic stresses and exogenous phytohormones. Int. J. Mol. Sci. 2019, 20, 5048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.H.; Chen, X.Q.; Han, J.; Lu, W.L.; Ren, Z.H. Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 2020, 20, 443. [Google Scholar] [CrossRef] [PubMed]
- Dan, Y.Y.; Niu, Y.; Wang, C.L.; Yan, M.; Liao, W.B. Genome-wide identification and expression analysis of the trehalose-6-phosphate synthase (TPS) gene family in cucumber (Cucumis sativus L.). PeerJ 2021, 9, e11398. [Google Scholar] [CrossRef] [PubMed]
- Li, S.T.; Wang, Z.R.; Wang, F.; Lv, H.M.; Cao, M.; Zhang, N.; Li, F.J.; Wang, H.; Li, X.S.; Yuan, X.W.; et al. A tubby-like protein CsTLP8 acts in the ABA signaling pathway and negatively regulates osmotic stresses tolerance during seed germination. BMC Plant Biol. 2021, 21, 340. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.; Zhu, C.X.; Hu, Z.Y.; Liu, S.Q.; Wu, H.; Zhou, Y. Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) Family Genes in cucumber. Biochem. Genet. 2021, 59, 884–901. [Google Scholar] [CrossRef]
- Wang, J.; Pan, C.T.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.F.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef] [Green Version]
- Han, X.Y.; Li, P.X.; Zou, L.J.; Tan, W.R.; Zheng, T.; Zhang, D.W.; Lin, H.H. Golden2-like transcription factors coordinate the tolerance to cucumber mosaic virus in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 477, 626–632. [Google Scholar] [CrossRef]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.F.; Zhang, L.; Cao, Y.Y.; Qi, C.D.; Li, S.T.; Liu, L.; Wang, G.L.; Mao, A.J.; Ren, S.X.; Guo, Y.D. CsATAF1 positively regulates drought stress tolerance by an ABA-dependent pathway and by promoting ROS scavenging in cucumber. Plant Cell Physiol. 2018, 59, 930–945. [Google Scholar] [CrossRef]
- Sharif, R.; Xie, C.; Wang, J.; Cao, Z.; Zhang, H.Q.; Chen, P.; Li, Y.H. Genome wide identification, characterization and expression analysis of HD-ZIP gene family in Cucumis sativus L. under biotic and various abiotic stresses. Int. J. Biol. Macromol. 2020, 158, 502–520. [Google Scholar] [CrossRef] [PubMed]
- Park, G.; Choi, Y.; Jung, J.K.; Shim, E.J.; Kang, M.Y.; Sim, S.C.; Chung, S.M.; Lee, G.P.; Park, Y. Genetic diversity assessment and cultivar identification of cucumber (Cucumis sativus L.) using the fluidigm single nucleotide polymorphism assay. Plants 2021, 10, 395. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.Y.; Dong, X.; Wang, J.K.; Xia, J.H.; Xie, F.; Zhang, Y.; Yao, X.; Xu, Y.J.; Wang, Z.J. Fine mapping and candidate gene prediction for white immature fruit skin in cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2018, 19, 1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.K.; Zhao, F.Y.; Gao, S.; Wang, X.Y.; Wei, A.M.; Chen, Z.W.; Liu, N.; Tong, X.Q.; Fu, X.M.; Wen, C.L.; et al. Fine mapping of a male sterility gene ms-3 in a novel cucumber (Cucumis sativus L.) mutant. Theor. Appl. Genet. 2018, 131, 449–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahveci, E.; Devran, Z.; Özkaynak, E.; Hong, Y.G.; Studholme, D.J.; Tör, M. Genomic-assisted marker development suitable for Cscvy-1 selection in cucumber breeding. Front. Plant Sci. 2021, 12, 691576. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; Li, H.; Gao, Z.H.; Wang, L.N.; Ren, Z.H. Localization of quantitative trait loci for cucumber fruit shape by a population of chromosome segment substitution lines. Sci. Rep. 2020, 10, 11030. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, D.; Ma, X.; Zhang, L.; Zhang, S.; Sun, Q.; Li, P.; Shu, J.; Zhao, Y. Serial-Omics and Molecular Function Study Provide Novel Insight into Cucumber Variety Improvement. Plants 2022, 11, 1609. https://doi.org/10.3390/plants11121609
Han D, Ma X, Zhang L, Zhang S, Sun Q, Li P, Shu J, Zhao Y. Serial-Omics and Molecular Function Study Provide Novel Insight into Cucumber Variety Improvement. Plants. 2022; 11(12):1609. https://doi.org/10.3390/plants11121609
Chicago/Turabian StyleHan, Danni, Xiaojun Ma, Lei Zhang, Shizhong Zhang, Qinghua Sun, Pan Li, Jing Shu, and Yanting Zhao. 2022. "Serial-Omics and Molecular Function Study Provide Novel Insight into Cucumber Variety Improvement" Plants 11, no. 12: 1609. https://doi.org/10.3390/plants11121609
APA StyleHan, D., Ma, X., Zhang, L., Zhang, S., Sun, Q., Li, P., Shu, J., & Zhao, Y. (2022). Serial-Omics and Molecular Function Study Provide Novel Insight into Cucumber Variety Improvement. Plants, 11(12), 1609. https://doi.org/10.3390/plants11121609





